Abstract
There is compelling evidence that proliferating cell nuclear antigen (PCNA), a DNA sliding clamp, co-ordinates the processing and joining of Okazaki fragments during eukaryotic DNA replication. However, a detailed mechanistic understanding of functional PCNA:ligase I interactions has been incomplete. Here we present the co-crystal structure of yeast PCNA with a peptide encompassing the conserved PCNA interaction motif of Cdc9, yeast DNA ligase I. The Cdc9 peptide contacts both the inter-domain connector loop (IDCL) and residues near the C-terminus of PCNA. Complementary mutational and biochemical results demonstrate that these two interaction interfaces are required for complex formation both in the absence of DNA and when PCNA is topologically linked to DNA. Similar to the functionally homologous human proteins, yeast RFC interacts with and inhibits Cdc9 DNA ligase whereas the addition of PCNA alleviates inhibition by RFC. Here we show that the ability of PCNA to overcome RFC-mediated inhibition of Cdc9 is dependent upon both the IDCL and the C-terminal interaction interfaces of PCNA. Together these results demonstrate the functional significance of the β-zipper structure formed between the C-terminal domain of PCNA and Cdc9 and reveal differences in the interactions of FEN-1 and Cdc9 with the two PCNA interfaces that may contribute to the co-ordinated, sequential action of these enzymes.
INTRODUCTION
DNA ligases seal nicks in double-stranded DNA. The budding yeast Saccharomyces cerevisiae contains two different DNA ligases encoded by the CDC9 and DNL4 genes that are homologs of human DNA ligases I and IV, respectively (1–6). Dnl4 appears to function only in the repair of DNA double strand breaks by non-homologous end joining (3,4,6). In contrast, genetic and biochemical studies have shown that Cdc9 DNA ligase is multifunctional. It is an essential DNA replication protein that joins Okazaki fragments generated during lagging strand DNA synthesis (2,7). When grown at the permissive temperature, temperature-sensitive cdc9 strains exhibit sensitivity to killing by various DNA damaging agents, implicating Cdc9 DNA ligase in DNA repair (2,8). In accord with this phenotype, extracts from the cdc9 mutant strains are defective in the completion of both base and nucleotide excision repair (9). Finally, two polypeptides are generated from the CDC9 gene by alternative translation initiation (10). The shorter polypeptide participates in the nuclear transactions described above whereas the longer polypeptide is targeted to the mitochondria and plays a critical role in mitochondrial DNA metabolism (10).
The participation of human DNA ligase I in DNA replication and DNA repair is mediated by an interaction with proliferating cell nuclear antigen (PCNA) (11,12). PCNA belongs to the family of DNA sliding clamps, which also includes the β clamp of E. coli and bacteriophage T4 gp45. These ring proteins encircle the DNA duplex and serve as processivity factors for DNA polymerases (13–16). Despite only sharing 35% amino acid sequence identity, the overall structures of the human and yeast PCNA (Pol30) homotrimers are superimposable and closely resemble the dimeric β clamp of E. coli (17–19).
A rapidly growing number of PCNA interacting proteins have been identified, implicating PCNA in functions as varied as DNA replication, DNA repair, cell cycle checkpoint control, chromatin assembly and apoptosis (as reviewed in (16,20–23)). Many of these proteins contain a conserved motif called the PCNA interacting protein (PIP) box that interacts with the inter-domain connector loop (IDCL) of PCNA. In human DNA ligase I, the PIP box is located at the N-terminus (residues 2–9) (12) whereas in Cdc9 the PIP box is located at residues 38–45 (24,25).
Since PCNA is a homotrimer, it is possible that complexes containing up to three PCNA-interacting proteins can be assembled on a single PCNA ring. Recently, it has been shown that each of the subunits in a human PCNA trimer can simultaneously bind to a different flap endonuclease 1 (FEN-1) molecule, a DNA structure-specific endonuclease involved in Okazaki fragment processing (26). By contrast, the complex formed between human DNA ligase I and PCNA topologically linked to DNA contained only one DNA ligase I molecule per PCNA trimer (27). This result, together with the size and shape of the ring formed by the catalytic domain of DNA ligase I when it encircles nicked DNA (28), suggests that the binding of one molecule of DNA ligase I to a PCNA trimer occludes the other IDCL-binding sites. In accord with this model, the binding of yeast Cdc9 and FEN-1 (Rad27) to a yeast PCNA (yPCNA) trimer appear to be mutually exclusive (25). Moreover, genetic instability induced by Cdc9 overexpression is dependent upon a functional PIP box, presumably because Cdc9 sequesters yPCNA molecules away from other PCNA interacting proteins, thereby disrupting the PCNA-dependent co-ordination of Okazaki fragment processing (25).
Although the IDCL of PCNA mediates the binding to FEN-1 in the absence of DNA (29,30), amino acid residues in the C-terminal domain of yPCNA are required to stimulate the flap cleavage activity of FEN-1 (Rad27) (31). In a recent X-ray structure of a peptide containing the PIP box of archaeal FEN-1 complexed with an archaeal PCNA, amino acid residues immediately adjacent to the PIP box form a β-zipper structure with residues in the C-terminal domain of PCNA, suggesting that these contacts hold FEN-1 on the PCNA ring in a suitable position to cleave the DNA single strand flap (32).
Unlike FEN-1 (31–34), human DNA ligase I catalytic activity is not stimulated by PCNA (27,33). However, human DNA ligase I interacts with and is inhibited by replication factor C (RFC), a heteropentameric complex that loads PCNA onto DNA (35). Interestingly, the presence of PCNA alleviates inhibition by RFC but only if the PIP box of DNA ligase I is functional (35). This suggests that pairwise interactions among RFC, PCNA and DNA ligase I may co-ordinate the ligation step that joins Okazaki fragments and completes DNA excision repair events. However, these pairwise interactions have not been completely defined at high resolution.
Here, we present the co-crystal structure of yPCNA with a peptide encompassing the PIP box of Cdc9. As FEN-1 (Rad27) generates the ligatable nick that is sealed by Cdc9 DNA ligase, the striking similarities of the complexes formed by PCNA with the PIP box of Cdc9 and with the FEN-1 PIP box region indicate that these interactions have significant functional implications (32). Structural, mutational and biochemical results indicate that the Cdc9 PIP box region makes physical and functional contacts with both the IDCL and C-terminal domain of yPCNA. In addition, we show that interplay among the replicative DNA ligase, sliding clamp and clamp loader is conserved in eukaryotes (35) and that residues within the C-terminal domain of PCNA are critical for these functional interactions.
MATERIALS AND METHODS
Overexpression and purification of yPCNA for crystallization
yPCNA was purified following overexpression in E. coli BL21 (DE3) cells (Invitrogen) containing the plasmid pT7-PCNA essentially as described (36). Briefly, E. coli cells harboring pT7-PCNA were grown at 37°C to an OD600 of 0.35 in Luria Broth (LB) media supplemented with ampicillin (100 µg/ml), and yPCNA expression was induced by the addition of 1 mM isopropylthiogalactoside (IPTG) to the media followed by incubation for 3 h at 37°C. Cells were lysed on ice for 1 h with lysozyme in 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.01% NP40 and 1 mM dithiothreitol (DTT) plus protease inhibitors (5 mM each of pepstatin, leupeptin and chymostatin and 0.5 mM p-methylphenylsulfonyl fluoride), followed by sonication. yPCNA was purified from the cleared lysate by sequential steps of Q-sepharose, SP-sepharose, hydroxylapatite and phenyl-sepharose column chromatography. Next, yPCNA-containing fractions were pooled and dialyzed overnight against 25 mM Tris-HCl (pH 7.4), 1 mM EDTA, 200 mM NaCl, 0.01% NP-40, 10% glycerol, 5 mM DTT plus protease inhibitors. After loading onto a Q-sepharose column, yPCNA was eluted with a 200–700 mM NaCl gradient. Fractions containing yPCNA were dialyzed overnight against 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 5 mM 2-mercaptoethanol and then concentrated using a Centricon-10 column (Amicon). Approximately 41 mg of >95% homogenous yPCNA were purified from 3 l of culture. The final concentration of yPCNA was determined by absorbance at 280 nm to be 68 mg/ml.
Crystallization, data collection and refinement of the yPCNA-Cdc9 peptide complex
A peptide corresponding to the PCNA-binding region of Cdc9 (AGKKPKQATLARFFTSMKNKPT, residues 32–53) was synthesized by the Protein Core Facility at The Scripps Research Institute. Lyophilized peptide was dissolved in 20 mM Tris-HCl (pH 8.0), 50 mM NaCl to a final concentration of 1.2 mM. The yPCNA-peptide complex was formed by mixing 1 mM yPCNA with 1.2 mM peptide in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 5 mM 2-mercaptoethanol. After the addition of an equal volume of precipitant (1.6 M (NH4)2SO4, sodium citrate pH 5.6), crystals were grown by hanging-drop vapor diffusion at 21°C over a period of 1 week. The crystals belonged to the space group I23 with unit cell dimensions a = b = c = 139.
Single crystals of the yPCNA-peptide complex were harvested from the drop, and soaked in mother liquor supplemented with 20% ethylene glycol for 10–20 s. X-ray diffraction data were obtained from a single crystal cooled to 100K at beamline 9–1 of the Stanford Synchrotron Radiation Laboratory (SSRL). The structure of the yPCNA–peptide complex was determined by molecular replacement with the program AMoRe (37), using the structure of yPCNA (PDB code 1PLQ, (18)) as a starting model. To reduce model bias, the IDCL (residues 121–132) and the C-terminal 8 residues (residues 251–258) of yPCNA were deleted from the starting model prior to phasing. Molecular replacement with AMoRe yielded a single solution with a correlation coefficient of 48.6 and an R-factor of 40.8 using data from 4–8 Å. The next best solution had a correlation coefficient of 35.5 and an R-factor of 45.5. Rigid-body, restrained B-factor refinement, followed by simulated annealing and grouped B-factor refinement was carried out in CNS (38). The Cdc9 peptide and the IDCL and C-terminus of yPCNA were clearly visible in σA-weighted Fo-Fc maps contoured at 2.5 or 3σ. These maps were sufficient to model 11 of the 22 residues of the Cdc9 peptide, the entire IDCL region (residues 121–132) and all but the 3 C-terminal residues of PCNA (residues 256–258), which are not included in the model. After initial model-building in Xfit (39), the structure was refined by cycles of maximum-likelihood restrained refinement with TLS against all data to 2.8 Å resolution with REFMAC5 (40) followed by manual building in Xfit. For TLS refinement, the peptide residues were defined as a separate group from yPCNA. The electron density for the refined structure clearly shows the position of the peptide backbone, and critical side-chains such as Gln38, Leu41 and Phe44. However, no electron density was observed for the side chains of residues Lys35 and Lys37. The side-chain of the final conserved Phe of the Cdc9 peptide (Phe45) is weak, but is present at 2.5σ contour level. The current model of the complex is refined to a crystallographic R-value of 0.247 (R-free = 0.285), with 90.5% of the residues in the most preferred phi/psi regions and no residues in the generously allowed or disallowed regions. Coordinates and structure factors for the Cdc9 peptide:PCNA complex have been deposited in the RCSB protein data bank with accession code 2OD8.
Proteins and antibodies
Untagged wild-type yPCNA and the mutant versions, pcna-79 and pcna-90 were expressed in and purified from E. coli cells by a modified version of a published purification scheme (41). Briefly, an overnight culture (10 ml) of M15 cells (Qiagen) harboring the PCNA expression plasmid pQE16-PCNA was diluted in 1 l of LB containing ampicillin and kanamycin and grown at 37°C. At an OD600 of 0.8, 0.5 mM IPTG was added and incubation continued for 3 h at 37°C. Cells were harvested and then resuspended in HEG buffer (30 mM HEPES-NaOH (pH 7.5), 0.5 mM EDTA, 10% glycerol, 0.01% NP-40, 1 mM DTT, 5 mM each of proteinase inhibitors pepstatin, leupeptin, chymostatin and 0.5 mM p-methylphenylsulfonyl fluoride) containing 200 mM KCl and lysed by sonication. The mutant versions of yPCNA, pcna-79 and pcna-90 (30) were expressed in the strain BL21(DE3). Wild-type and mutant versions of PCNA were purified from cleared lysates by Q sepharose (GE Healthcare), hydroxyapatite (Bio-Rad), Mono Q and Superdex 200 (GE Healthcare) column chromatography. Approximately 10 mg of >95% homogeneous PCNA was obtained from a 1-l culture.
Glutathione-S-transferase (GST) fusion proteins of wild-type Cdc9 and a PIP box mutant version PIP box (FF44,45AA) were expressed and purified as described previously (35). The expression of his-tagged versions of wild-type Cdc9 and the PIP box mutant in E. coli M15 cells was induced by the addition of 0.5 mM IPTG at an OD600 of 0.8 followed by growth for 4 h at 25°C. Harvested cells were resuspended in HEG buffer containing 100 mM KCl and lysed by sonication. The his-tagged versions of Cdc9 were purified from the cleared lysates to near homogeneity by SP Sepharose, Source S, Mono Q and Mono S (GE Healthcare) column chromatography.
Yeast RFC (yRFC) was expressed in and purified from E. coli cells as described previously (42,43) except that a Superdex 200 26/60 (GE Healthcare) gel filtration column was included as the final step.
Polyclonal antibodies against yPCNA were kindly provided by Drs Satya Prakash and Stefan Jentsch. Cdc9 polyclonal antibodies have been described previously (44).
Surface plasmon resonance
yPCNA, pcna-79 or pcna–90 were immobilized on a Biacore CM5 chip at comparable densities by carbodiimide coupling according to the manufacturer's instructions. The running buffer contained 30 mM HEPES-NaOH (pH 7.5), 0.5 mM EDTA, 10% glycerol, 0.1 mM EDTA, 0.01% NP-40 and 150 mM NaCl. GST-Cdc9 wild-type, GST-Cdc9 PIP box mutant (FF44,45AA) and FEN-1 (Rad27) were injected (100 nM, 60 μl total at 20 μl/min) and monitored for association and dissociation (response units, RU) as a function of time. The surface plasmon resonance studies were carried out on a Biacore 300 Biosensor in the Biacore Core Facility of the University of Maryland School of Medicine.
GST pull-down assays
To prepare beads with GST, GST-Cdc9p or GST-Cdc9 PIP box mutant (FF44,45AA) as the ligand, 10 µg of each purified protein was incubated with a 50 µl slurry of Glutathione Sepharose beads (GE Healthcare) equilibrated in binding buffer (50 mM KH2PO4 (pH7.5), 10% glycerol, 150 mM NaCl, 0.2% NP-40, 1 mM 2-mercaptoethanol, and 1 mg/ml bovine serum albumin (BSA)) for 30 min at 4°C with constant agitation. After washing with binding buffer, the beads were resuspended in 100 µl binding buffer. GST, GST-Cdc9p or GST-Cdc9 PIP box mutant beads (5 pmol of each protein) were incubated with 5 pmol of wild-type yPCNA, pcna-79 or pcna-90 at room temperature for 30 min with constant agitation. Beads were collected by centrifugation and then washed extensively in binding buffer prior to being resuspended in 10 µl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer. After separation by SDS-PAGE, proteins were detected by immunoblotting. Similar pull-down assays with the GST fusion proteins described above were performed with 5 pmol of purified yRFC. After separation by SDS-PAGE, proteins were detected by staining with coomassie blue.
Preparation of biotin-labeled linear DNA substrate
This substrate was prepared as described previously (35). Briefly a 5′ biotinylated 90 mer was annealed with two complementary 15 mer to generate a partial duplex of 30 bp with a single nick in the middle flanked by single-strand regions of 30 nt. For ligation assays, one of the 15-mer oligonucleotides was 5′ end-labeled with [γ32P]ATP using T4 polynucleotide kinase to generate a ligatable central nick.
DNA joining reaction with biotin-labeled linear DNA substrate
DNA joining assays were carried out as described previously (35). Streptavidin-agarose beads (10 μl, Pierce) were incubated with 1.6 pmol of the biotinylated linear DNA substrate in phosphate-buffered saline for 30 min at room temperature. After washing three times with ligation buffer (50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 0.25 mg/ml BSA and 1 mM ATP), the beads were incubated with 2 pmol of E. coli single strand binding (SSB) protein (Sigma) per pmol of DNA in the same buffer for 15 min at room temperature. The beads liganded by SSB-coated DNA were washed three times and finally resuspended in ligation buffer containing 150 mM NaCl. The binding of SSB to the single strand ends of the partial duplex prevents PCNA molecules loaded at the central nick from sliding off the DNA. The SSB-blocked substrate (1 pmol) was incubated with 1 pmol yRFC in the presence and absence of wild-type yPCNA, pcna-79 or pcna-90 (1 pmol trimer) at 30°C for 2 min prior to the addition of Cdc9 (1 pmol of either wild-type or Cdc9 PIP box mutant). After incubation at room temperature for 5 min, the beads were spun down and the reaction terminated by addition of 10 µl of stop mix (10% glycerol, 0.2% SDS, 4 mM EDTA and 0.01% bromophenol blue) and heating at 100°C for 3 min to denature DNA. Aliquots (2 μl) were mixed with 2 µl of denaturing PAGE dye (95% formamide, 0.05% bromophenol and 0.05% xylene cyanol) prior to electrophoresis through a 12% denaturing polyacrylamide gel. After drying, labeled oligonucleotides in the gel were detected and quantitated by PhosphorImager analysis (Molecular Dynamics).
Analysis of DNA–protein complexes by streptavidin pull-down assays
The DNA substrate used in assays to detect DNA protein complexes formed by yRFC, yPCNA and Cdc9 was prepared as described above except for the absence of a 5′ phosphate group at the central nick. The SSB-blocked substrate (1 pmol) was incubated in ligation buffer with 150 NaCl, with various combinations of yRFC, Cdc9 and wild-type and mutant versions of yPCNA for 15 min at 25°C. The streptavidin beads were collected by centrifugation and then washed twice with ligation buffer containing 150 mM salt. Bound proteins were released from the beads by the addition of 10 µl of SDS sample buffer. After heating at 100°C for 3 min, proteins (5 µl) were separated by SDS-PAGE and detected by immunoblotting.
RESULTS
Structure of a Cdc9 peptide bound to yPCNA
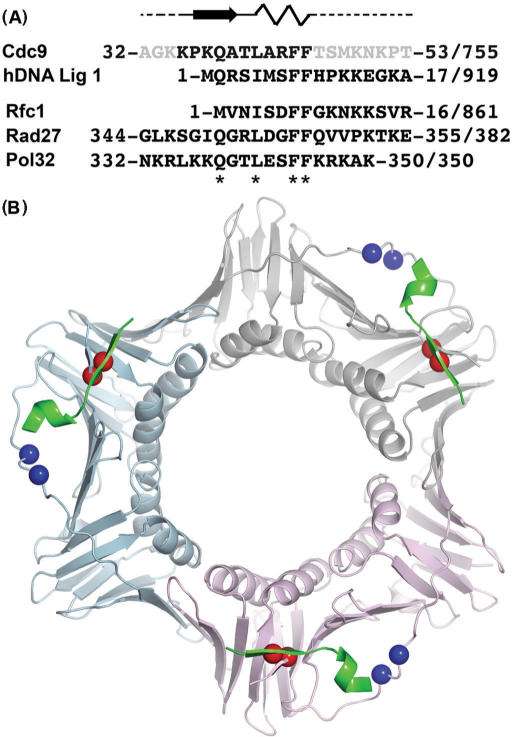
To characterize the binding interface between yPCNA and Cdc9 DNA ligase, we determined the structure of a complex between a peptide (32-AGKKPKQATLARFFTSMKNKPT-53, Figure 1A) encompassing the PIP box (bold) of Cdc9 DNA ligase bound to yPCNA (Pol30) to 2.8 Å resolution by X-ray crystallography (Table 1). The structure contains one peptide bound to one subunit of yPCNA in the asymmetric unit, which form trimers through crystallographic symmetry (Figure 1B). yPCNA exhibits a two-domain, α/β fold that is characteristic of sliding clamp homologs (45) and closely resembles the existing yPCNA structure (rmsd = 1.1 Å for 252 out of the 255 comparable Calpha atoms) (18). Electron density for the peptide was sufficient to model 11 residues (35-KPKQATLARFF-45) that encompass the PIP box (bold) and interact with both the IDCL and C-terminus of yPCNA (Figure 2A). The side chains of peptide residues Gln38, Leu41 and Phe44 were well defined by the omit electron density (Figure 2A), and used to anchor the overall position of the peptide in the binding site. Peptide residues with weaker side chain density (Phe45) were modeled based on a combination of the electron density, previous PCNA:peptide structures and biochemical data (see Figure 5,7–8). The interactions between the Cdc9 peptide and the two regions of PCNA account for most of the differences between the yPCNA:peptide complex and the free yPCNA structure.
Figure 1.
Crystal structure of yPCNA complexed with a Cdc9 peptide encompassing the PIP box. (A) Cdc9 peptide sequence used for crystallization showing the secondary structure of residues observed in the co-crystal structure (bold black letters) in relation to disordered residues (gray letters) that had no observed electron density. The Cdc9 peptide is aligned with the PIP box and surrounding residues from human DNA ligase I (hDNA Lig I), yeast Rfc-1 (Rfc1), yeast FEN-1 (Rad27) and yeast Pol δ (Pol32). The starting and ending residue numbers, along with the total sequence length is shown for each protein. Key residues of the PIP box are denoted by asterisks (*) below the sequence. (B) Structure of the Cdc9 peptide:yPCNA complex shown as a trimer based on crystallographic symmetry. The yPCNA trimer is shown as a ribbon (subunits colored beige, blue and pink) with colored spheres marking the position of mutant residues in the pcna-79 (blue spheres) and pcna-90 (red spheres) mutants. The Cdc9 peptide (green) is bound to each of the subunits.
Table 1.
X-ray diffraction data and refinement statistics
| X-ray diffraction data | PCNA:Cdc9 peptide |
|---|---|
| Wavelength (Å) | 0.9790 |
| Space group | I23 |
| Resolution (Å) | 40-2.8 (2.9-2.8) |
| Observations | 35,241 |
| Unique reflections | 11,287 (1,104) |
| Completeness (%) | 99.2 (99.2) |
| Rsyma (%) | 3.9 (41.8) |
| Wilson B-factor (Å2) | 94 |
| Model refinement | |
| Resolution (Å) | 30-2.8 |
| Number of reflections | 11,219 |
| Rcrystb | 0.247 |
| Rfreec | 0.285 |
| r.m.s.d bonds | 0.007 |
| r.m.s.d angles | 1.081 |
| Number of PCNA atomsd | 2003 (38) |
| Number of peptide atomsd | 92 (40) |
| Number of solvent atomsd | 17 (70) |
Values in parentheses indicate the high-resolution shells, unless otherwise noted.
aRsym = Σh Σi | I(h,i) − <I(h)> |/Σh Σi I(h,i), where I(h,i) is the intensity of the i th measurement of reflection h and <I(h)> is the average value over multiple measurements.
bRcryst = Σ‖Fo| − |Fc‖/Σ|Fo| for all reflections (no σ cutoff), where Fo and Fc are the observed and calculated structure factors, respectively.
cRfree was calculated as Rcryst against 5% of the reflections removed at random.
dValues in parentheses indicate the average B-factor (Å2).
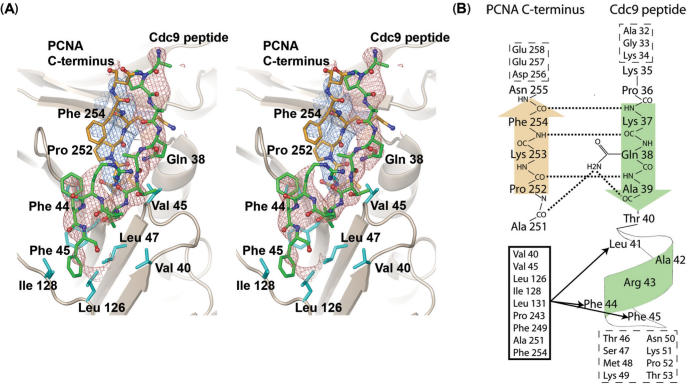
Figure 2.
Structure of the PCNA:Cdc9 interface. (A). Stereo view showing omit Fo-Fc electron density for the Cdc9 peptide (green carbons, red density contoured at 2.5σ calculated by omitting the peptide residues) and the C-terminal residues of PCNA (251–255; orange carbons, blue density contoured at 2.5σ calculated by omitting residues 251–255 from the model). The omit density shows the general position of the peptide backbone and most side chains for both the peptide and C-terminus of PCNA. The side chains of peptide residues Gln38, Leu41 and Phe44 were well defined by the omit electron density, and used to anchor the overall position of the peptide in the binding site. However, no density was observed for the side chains of peptide residues Lys 35 and Lys 37, and only weak density was observed for the Phe 45 side chain. These residues were modeled based on a combination of the electron density, previous PCNA:peptide structures, and biochemical data. In addition to interactions with residues near the C-terminus of PCNA, Cdc9 binds to a hydrophobic pocket formed by several PCNA residues (cyan) surrounding the IDCL (Leu126, Ile128). (B). Schematic showing hydrogen bonds and hydrophobic interactions between residues near the C-terminus of PCNA (orange) and residues from the Cdc9 peptide (green). Several hydrogen bonds (dotted lines) form between the peptide backbones of Pro252, Lys253 and Phe254 with peptide residues Lys37, Gln38 and Ala39. The side chain of Gln38 also forms hydrogen bonds with Ala251 from PCNA and Ala39 from Cdc9. Residues from the hydrophobic pocket on PCNA (black box) form hydrophobic interactions (black lines with arrows) with Leu41, Phe44 and Phe45, located on the 310 in the Cdc9 peptide. Residues with no observed electron density are listed inside boxes with dash-line borders.
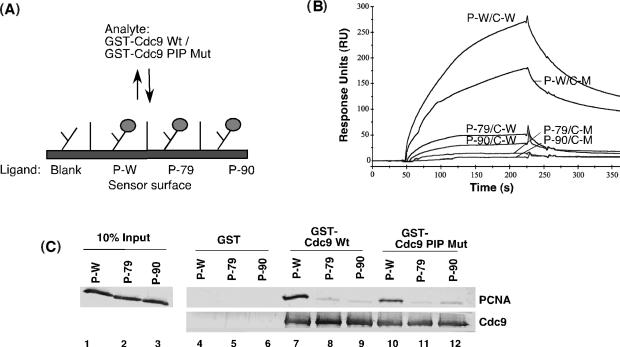
Figure 5.
Interaction of Cdc9 with wild-type and mutant versions of PCNA in the absence of DNA. (A). Schematic of sensor chip used in surface plasmon resonance analysis of association of Cdc9 with PCNA. PCNA wild-type PCNA (P-W), pcna-79 (P-79) and pcna-90 (P-90) were immobilized to a CM5 chip and 100 nM of GST fused to either wild-type Cdc9 (Wt) or Cdc9 FF44,45AA mutant (PIP box Mut) were passed over the chip. (B). Sensorgram showing the association and disassociation curves of GST-Cdc9 wild-type (C-W) with PCNA wild-type (P-W/C-W), pcna79 (P-79/C-W) and pcna-90 (P-90/C-W) and of GST Cdc9 FF44,45AA mutant (PIP box Mutant; C-M) with PCNA wild-type (P-W/C-M), pcna79 (P-79/C-M) and pcna-90 (P-90/C-M). (C) Pull-down assays. Lanes 1–3 represent 10% of input PCNA used in the pull-down assay. Purified PCNA (P-W), pcna-79 (P-79) and pcna-90 (P-90) (5 pmol of PCNA trimer) were incubated with glutathione sepharose beads liganded by 5 pmol of GST, GST–Cdc9 wild-type (GSTCdc9 Wt) or GST-Cdc9-PIP box mutant (GST Cdc9 PIP Mut). Proteins bound to GST beads (lanes 4–6), GST–Cdc9 Wt beads (lanes 7–9) and GST–Cdc9p PIP Mut beads (lanes 10–12) were separated by SDS-PAGE. PCNA (top panel) and GST-Cdc9p (bottom panel) were detected by immunoblotting with Cdc9 and PCNA antibodies, respectively.
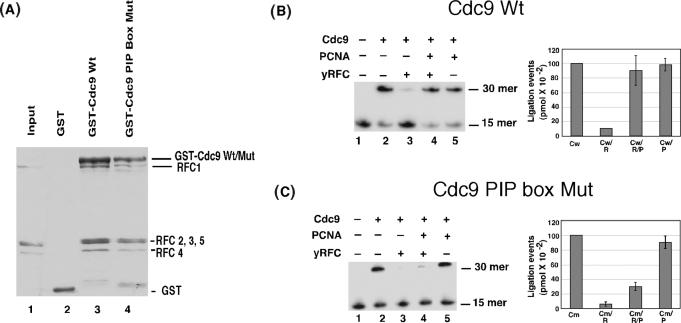
Figure 7.
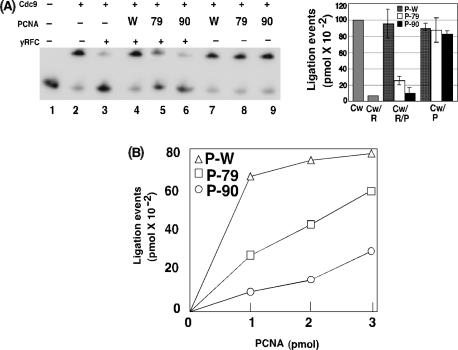
Interaction between Cdc9 and yRFC. Effect of yRFC and yPCNA on the DNA joining activity of Cdc9. (A). Purified yRFC (0.5 pmol) complex was incubated with glutathione sepharose beads liganded by; lane 2, GST; lane 3, GST fused to wild-type Cdc9 (GST-Cdc9-Wt); and lane 4, GST fused to the PIP box mutant version of Cdc9 (GST-Cdc9 PIP box mutant). Proteins bound to the beads were separated by SDS-PAGE gel and then stained with coomassie blue. Lane 1 contains 5% of the RFC input. The positions of bands corresponding to GST, the GST-Cdc9 fusion proteins, the large subunit of yRFC (RFC1) and the four small yRFC subunits (RFC2-5) are indicated on the right. (B). The linear end-blocked DNA substrate (1 pmol) was incubated as described in materials and methods section with equimolar ratios (1 pmol) of yRFC (R), yPCNA (P) and wild-type Cdc9 (Cw). (C). The linear end-blocked DNA substrate (1 pmol) was incubated as described in experimental procedures section with equimolar ratios (1 pmol) of yRFC (R), yPCNA (P) and a mutant version of Cdc9 with a defective PIP (Cm) as indicated. Labeled oligonucleotides were separated by denaturing PAGE and quantitated by PhosphorImager analysis (molecular dynamics). The positions of labeled 15-mer substrate and 30-mer product are indicated on the right of a representative gel (left panel). The results of three independent experiments are shown graphically (right panel).
Figure 8.
Amino acid residues within both the IDCL and C-terminus of PCNA are required to overcome RFC-mediated inhibition of Cdc9. (A). DNA-joining reactions were carried out as described in the legend of Figure 5 except that pcna–79 (79) and pcna-90 (90) were included in addition to wild-type PCNA (W) as indicated. Left panel, a representative gel and, right panel, a bar graph show the results of three independent experiments. (B). DNA-joining assays with RFC (1 pmol) and Cdc9 (1 pmol) and increasing amounts of wild-type PCNA (P-W) and the mutant versions of PCNA, pcna–79 (P-79) and pcna-90 (P-90), were carried out as described in the legend to Figure 5 and are shown graphically.
Similar to other PCNA-peptide structures, conserved residues in the PIP-box of the Cdc9 peptide (LARFF) adopt a helical conformation that positions the hydrophobic residues on the same face of the helix. This conserved structural motif (Figure 1A) acts as a hydrophobic anchor that facilitates binding to the surface of PCNA. The hydrophobic-binding site on yPCNA is formed primarily by conserved residues (Leu126, Ile128) in the IDCL together with Val40, Val45, and Leu47 from the N-terminal domain (Figure 2). These residues are located at the inter-domain interface of yPCNA (Figure 2A, 3) and provide a conserved binding site for several proteins that interact with PCNA (46,47).
Figure 3.
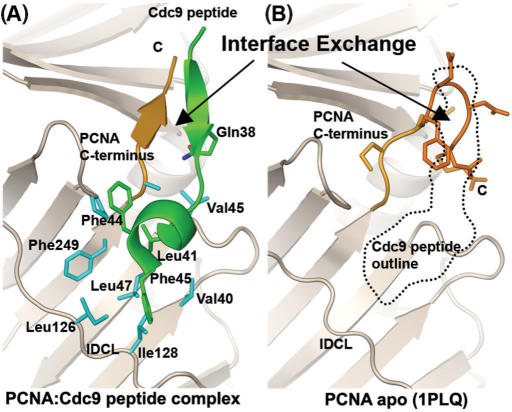
Interface exchange at the C-terminus of PCNA. (A). Cdc9 peptide fold (green) bound to PCNA (beige and orange) shown as ribbons indicating that the N-terminus of the Cdc9 peptide forms a β-sheet with the PCNA C-terminus (orange). Conserved hydrophobic residues (green, labeled) interact with a hydrophobic pocket on the surface of PCNA (blue side chains). (B). The location of the Cdc9 peptide (dotted black outline) relative to the structure of PCNA determined without a peptide (PCNA apo; PDB code 1PLQ). Without bound peptide, the C-terminus of PCNA (orange) loops back to interact with itself by forming a tight turn. Binding of the Cdc9 peptide (dotted outline) exchanges the PCNA intra-molecular beta-turn structure with an inter-molecular β-sheet interface (A and B) formed by residues at the N-terminus of the Cdc9 peptide hydrogen bonding to residues near the C-terminus of PCNA.
As noted above, there are significant differences between the structures of the yPCNA:peptide and free yPCNA (rmsd of 2.2 Å for the Calpha atoms of yPCNA residues 121–132; Figure 4). In the yPCNA apo structure, the IDCL region contacts the IDCL of neighboring PCNA molecules in the crystal (18) whereas the Cdc9 peptide:yPCNA complex crystallizes in a different space group, in which the IDCL only contacts the Cdc9 peptide. Thus, the yPCNA IDCL region exhibits structural plasticity and becomes ordered due to interactions with PIP box containing peptides (Figure 4A).
Figure 4.
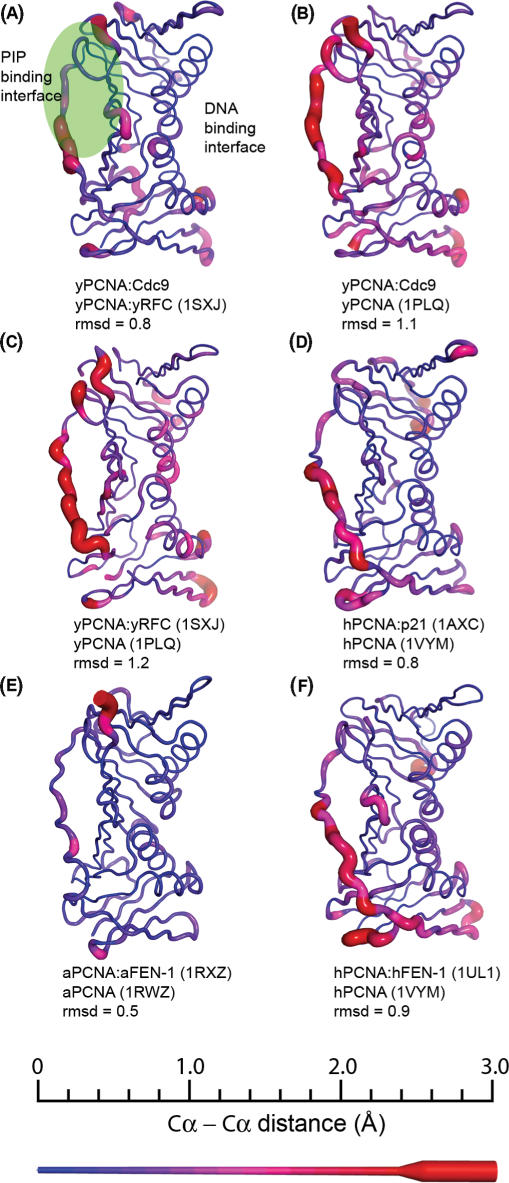
Structural changes caused by binding to PCNA. PCNA subunits from structure of PCNA:peptide or PCNA:enzyme complexes were aligned with structures of the apo form of PCNA from the same organism. The distance between Cα residues for each pair of structures is shown mapped onto the structure of PCNA as both a color gradient and ribbon thickness. The distances were sorted as a histogram with 0.2 Å bins ranging from 0 Å (blue, thin ribbon) to ≥3.0 Å (red, thick ribbon) as indicated by the scale bar in the bottom section. The PDB codes for the structures used in each comparison are shown in parentheses, along with the rmsd values for each comparison. The majority of the differences are localized to flexible loop regions, with minor differences in the helices that contact DNA. Both the inter-domain connecting loop (IDCL), as well as the C-terminal tail make up a large proportion of the structural changes. These regions form the structural interface responsible for binding the conserved PIP-motif (green region) found in several enzymes that interact with PCNA. (A) Yeast PCNA (yPCNA):Cdc9 peptide complex compared to the yRFC:yPCNA complex (1SXJ). (B) yPCNA:Cdc9 peptide complex compared to apo yPCNA (1PLQ). (C) yPCNA:yRFC complex (1SXJ) compared to apo yPCNA (1PLQ). (D) human PCNA (hPCNA):p21 peptide complex (1AXC) compared to apo hPCNA (1VYM). (E) Archaeoglobus fulgidus PCNA (aPCNA): A. fulgidus FEN-1 (aFEN-1) peptide complex (1RXZ) compared to apo aPCNA (1RWZ). (F) hPCNA:human FEN-1 complex (1UL1) compared to apo hPCNA (1VYM).
A structural comparison of human, yeast and archaeal PCNA with PCNA complexes from the same organisms shows that localized structural changes in PCNA are significant and well conserved (Figure 4). For example, the structural differences between yPCNA:Cdc9 and yPCNA are similar to those between yPCNA:yRFC and yPCNA (Figure 4B and C). Comparison of human PCNA and human PCNA complexes shows a similar trend (Figure 4D and F). In general, most of the structural differences greater than 2 Å (between identical Calpha atoms in each structure) are localized to the IDCL and the C-terminal tail of PCNA. Structural changes in solvent-exposed loops, and in residues at the trimer interface are also observed, consistent with recent results highlighting the flexible nature of interactions among PCNA subunits (48). Notably, yPCNA from our yPCNA:Cdc9 peptide complex is more similar to yPCNA from the yRFC:yPCNA complex than to yPCNA from either complex compared to the unbound structure of yPCNA (Figure 4A–C), suggesting that conserved structural changes occur in PCNA when enzymes associate with PCNA via the conserved PIP motif.
In addition to the conserved hydrophobic-anchor motif (Figure 1A), residues near the N-terminus of the Cdc9 peptide (KQA) form a β-sheet with residues in the C-terminal domain of yPCNA (Figures 2 and 3A). The β-sheet backbone interactions are supplemented by two additional hydrogen bonds formed between the side chain of Gln38 and the peptide backbone of both Cdc9 and PCNA (Figure 2B). The side-chain of Gln38 (Nε) within the Cdc9 peptide forms a hydrogen bond with the carbonyl oxygens of Ala251 from yPCNA and Ala39 from the Cdc9 peptide (Figure 2B). These interactions appear to stabilize the β-sheet and compensate for the presence of the conserved Pro252, which would otherwise disrupt continuous hydrogen bond interactions between the peptide backbones of Cdc9 and yPCNA.
The β-sheet interactions observed in the yPCNA:Cdc9 peptide complex are almost identical to those observed in the structure of human FEN-1 bound to human PCNA (26) and similar to those observed in structures of FEN-1 peptides bound to archaeal PCNA (32). It should be noted that the β-sheet formed between the FEN-1 peptide and the C-terminus of archael PCNA was slightly longer than the similar structures in the complexes formed by eukaryotic PCNA. This may reflect differences in the size of archaeal and eukaryotic PCNAs, in particular the C-terminal domain of archaeal PCNA is shorter than that of eukaryotic PCNA (26,32,49) (Supplementary Figure 1). For example, the shorter archaeal PCNA lacks two of the three residues at the C-terminal end of yPCNA (residues 256–258) that fold back to form a hairpin in the apo structure of yPCNA and interact with the same PCNA residues (residues 252–255) contacted by the Cdc9 peptide (Figure 3B). In the presence of the Cdc9 peptide, the three C-terminal residues of yPCNA (residues 256–258) are disordered (no observed electron density), and the peptide occupies the β-sheet binding interface (Figure 3).
Although many studies on PCNA interacting proteins have focused on interactions with the IDCL of PCNA (46,47), there is emerging evidence that regions adjacent to the PIP box are involved in functionally important contacts with the C-terminal domain of PCNA (31,32,50,51). While the functional role of the β-zipper interaction is not completely understood, the formation of the same structure with peptides from both FEN-1 and Cdc9 DNA ligase, suggests that it plays a key functional role in interactions between PCNA and its partner proteins.
In the case of yeast FEN-1 (Rad27), the interaction with the IDCL of yPCNA is required for complex formation in the absence of DNA whereas the C-terminal residues of PCNA involved in formation of the β-zipper are critical for the stimulation of the DNA structure-specific endonuclease activity of FEN-1 (Rad27) (29,31,34). This prompted us to examine the roles of the IDCL and C-terminal interfaces of PCNA on physical and functional interactions with Cdc9 in the absence or presence of DNA.
Interaction between Cdc9 and PCNA in the absence of DNA
To determine the contribution of the IDCL region of yPCNA to Cdc9 binding, we used pcna-79, a mutant in which Leu126 and Ile128 are substituted with alanine. These residues are located in the IDCL and form the hydrophobic pocket on yPCNA that binds the hydrophobic anchor motif formed by PIP box residues of Cdc9 (Figures 2 and 3). The IDCL mutations severely disrupt Cdc9 binding to pcna-79 as measured both by surface plasmon resonance (Figure 5A and B) and GST pull-down assays (Figure 5C, compare lanes 7 and 8). In addition, we also replaced the conserved phenylalanine residues (F44F45) in the PIP box of Cdc9 with alanine residues. Based on the Cdc9 peptide:yPCNA structure, these residues interact directly with the conserved hydrophobic-binding site that is disrupted in the pcna-79 mutant. Consistent with the complex structure (Figures 1–4) and previous biochemical studies (25), substitution of these PIP box residues in Cdc9 also reduced complex formation with yPCNA (Figure 5B and C, compare lanes 7 and 10). Notably, changes in the PIP box of Cdc9 had a much lesser effect on the interaction between Cdc9 and yPCNA compared with changes in the IDCL of yPCNA. In contrast, replacement of the adjacent phenylalanine residues in the FEN-1 (Rad27) PIP box had a slightly more severe impact on the interaction between PCNA and FEN-1 compared with the amino changes in the IDCL of yPCNA (31).
It should be noted that, in the FEN-1 (Rad27) study described above (33), the conserved adjacent Phe residues in the PIP box were replaced with Gly and Ala whereas, in the Cdc9 PIP box, they were replaced with two Ala residues. The energetic difference between burying an Ala or a Gly at this interface is not significant compared with the loss of the Phe side chain (52). However, Gly is known to interfere more with helical stability compared to Ala. Thus, the differences in the interactions of residues surrounding the PIP boxes of FEN-1 and Cdc9 with yPCNA might underlie the differences observed following the replacement of the conserved Phe residues in the PIP boxes of Cdc9 and FEN-1. For example, the interface between human FEN-1 and PCNA extends beyond both sides of the PIP box to include β-sheet interactions with both the C-terminus and the IDCL of human PCNA (26). In contrast, the structure of the Cdc9 peptide:yPCNA complex does not show additional interactions with the IDCL beyond the PIP box. Disruption of the helical structure that positions the Phe residues would also disrupt the positioning of FEN-1 residues following the mutation amplifying the affect of the Phe substitutions in FEN-1 (Rad27).
An alternative explanation for this difference in mutational results is the presence of additional PCNA interactions between Cdc9 and PCNA that do not involve the N-terminal PIP Box of Cdc9. A recent structure of a DNA Ligase I homolog from Sulfolobus solfataricus identified a PIP box located in the DNA-binding domain (53). Whilst the DNA binding domains of hLig1 and Cdc9 do not contain a canonical PIP-box, further binding experiments revealed an additional weak interaction between PCNA and DNA binding domain of human DNA Ligase I (hLig1) (53). If the IDCL and C-terminal tail of yPCNA also interact weakly with the DNA-binding domain of Cdc9, then mutations in yPCNA would affect both interfaces. Thus, mutations in yPCNA would have a greater effect on the interaction between Cdc9 and yPCNA than mutations in the PIP box of Cdc9.
To determine the contribution of the C-terminal region of yPCNA to Cdc9 binding, we used pcna-90, a mutant in which Pro252 and Lys253 at the base of the C-terminal tail of yPCNA are substituted with alanine. Surprisingly, these changes severely disrupted binding of Cdc9 to pcna-90 as measured by surface plasmon resonance (Figure 5A and B) and GST pull-down assays (Figure 5C, compare lanes 7 and 9). Thus, amino acid changes in the IDCL and C-terminal domain of yPCNA reduce complex formation with Cdc9 to a similar extent (Figure 5), demonstrating that Cdc9, unlike FEN-1 (Rad27), requires interactions with both the IDCL and C-terminal domain interfaces of yPCNA to form a complex in the absence of DNA.
Interaction between Cdc9 and PCNA in the presence of DNA
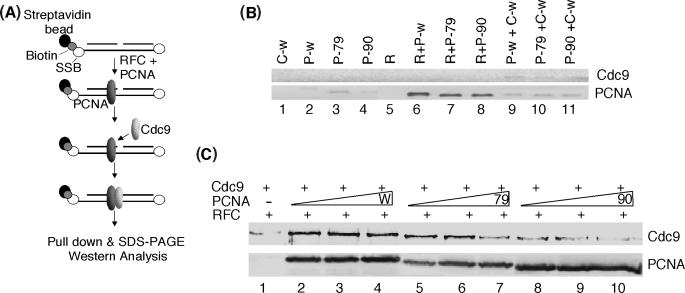
To determine the contributions of the IDCL and C-terminal regions of PCNA to Cdc9 binding in the presence of DNA, we used yRFC to load wild-type and mutant versions of PCNA onto biotinylated DNA substrates with blocked ends (Figure 6A). In initial experiments using a ligatable substrate, we were unable to detect Cdc9 binding to yPCNA on the DNA-streptavidin beads (data not shown), presumably because Cdc9 rapidly dissociates from the DNA after ligation. Using a non-ligatable DNA substrate (Figure 6B), we found that, as expected, yPCNA was only retained by the DNA beads after loading by yRFC (Figure 6B, compare lanes 2–4 with lanes 6–8) and that Cdc9 binding to the beads was barely detectable in the absence of loaded yPCNA (Figure 6B, lanes 1 and 9–11). In accord with the results of studies with the functionally homologous human proteins (27), Cdc9 was retained by the beads when wild-type yPCNA was topologically linked to the DNA molecule (Figure 6C, lanes 2–4). A similar amount of Cdc9 was retained by the DNA beads when yRFC was removed from the reaction after PCNA loading and prior to incubation with Cdc9 (data not shown), indicating that yRFC does not influence complex formation between yPCNA and Cdc9 on DNA under these conditions. The amount of bound Cdc9 was markedly reduced in assays with pcna-90 (Figure 6C, compare lanes 2–4 with 8–10), but not with pcna-79 (Figure 6C, compare lanes 2–4 with 5–7). Increasing the concentration of PCNA did not increase the amount of Cdc9 bound to the beads and, in fact reduced the amount retained in assays with the mutant versions of yPCNA. Together, these results demonstrate that the interaction between Cdc9 and the C-terminal domain of yPCNA plays a major role in complex formation both on and off DNA.
Figure 6.
Interaction of Cdc9 with PCNA topologically linked to duplex DNA. (A). Schematic illustration of the assay. Streptavidin beads bound by biotinylated, non-ligatable nicked DNA molecules with blocked ends were prepared as described in materials and methods section. PCNA was loaded onto the DNA by incubation with RFC and then Cdc9 was incubated with the PCNA/DNA complexes. (B). The DNA substrate (1 pmol) was incubated with 1 pmol of each of the following proteins; RFC (R), wild-type PCNA (P-w), pcna-79 (P-79) pcna-90 (P-90) and wild-type Cdc9 (C-w) either alone or in the combinations indicated. (C). The DNA substrate (1 pmol) and RFC (1 pmol) were incubated with; wild-type PCNA (W), lane 2, 1 pmol; lane 3, 2 pmol; lane 4, 3 pmol; pcna-79 (79), lane 5, 1 pmol; lane 6, 2 pmol; lane 7, 3 pmol; and pcna-90 (90), lane 8, 1 pmol; lane 9, 2 pmol; lane 10, 3 pmol; for 1 min followed by the addition of wild-type Cdc9 (1 pmol) and further incubation for 15 min at room temperature. After centrifugation to collect the DNA beads, the specific retention of PCNA and Cdc9 by the beads was detected by immunoblotting.
A conserved physical and functional interaction between the replicative clamp loader and DNA ligase of eukaryotes
Previously we showed that human DNA ligase I binds directly to human RFC (35). To determine whether this interaction also occurs between the functionally homologous yeast proteins, we immobilized GST-fusions of wild-type and mutant versions of Cdc9 on glutathione beads and assayed for retention of purified, recombinant yRFC. In these pull-down assays, yRFC bound specifically to beads liganded by wild-type GST-Cdc9 (Figure 7A, lane 3). Moreover, replacement of the adjacent phenylalanine residues in the Cdc9 PIP box with alanine residues that disrupted binding with yPCNA (Figure 5) had no significant effect on the interaction with yRFC (Figure 7A, lane 4), suggesting that Cdc9 binds to yRFC and yPCNA via different interfaces.
Next we asked whether yRFC modulates the activity of Cdc9 in the presence or absence of PCNA. yRFC inhibited the joining of a central nick within a linear DNA duplex (35) by Cdc9 DNA ligase (Figure 7B, compare lanes 2 and 3) whereas yPCNA had no effect (Figure 7B, compare lanes 2 and 5). Notably, co-incubation of yPCNA with yRFC abolished the inhibitory effect of yRFC (Figure 7B, lanes 2–4) but only when Cdc9 had a functional PIP box (Figure 7C). In assays with the ATP analog, ATPγS, which cannot be hydrolyzed by RFC but can be used as a co-factor by Cdc9 DNA ligase, RFC still inhibits Cdc9, but the inclusion of PCNA does not alleviate this inhibition (data not shown). This suggests that PCNA loading by RFC and/or the dissociation of the RFC/PCNA complex following loading is also necessary to recover DNA joining activity. The results shown in Figure 7 recapitulate those obtained with the functionally homologous human proteins (35), indicating that the physical and functional interactions among the replicative DNA sliding clamp, clamp loader and DNA ligase are conserved among eukaryotes.
Both the IDCL region and C-terminal domain of yPCNA are important for the functional interplay among Cdc9, yRFC and yPCNA
The results of studies with the functionally homologous human (35) and yeast proteins (Figure 7) have shown that the PCNA-dependent recovery of DNA joining activity requires adjacent phenylalanine residues within the PIP box of the DNA ligase. If the physical interaction between the Cdc9 PIP box and the IDCL of yPCNA is critical for the functional interplay among yRFC, yPCNA and Cdc9 DNA ligase, then amino acid changes within the IDCL of yPCNA that disrupt the binding with the Cdc9 PIP box should have a similar effect to inactivation of the Cdc9 PIP box. In accord with this prediction, the mutant version of yPCNA, pcna-79 that contains amino acid substitutions of key residues within the IDCL was unable to completely restore DNA joining in the presence of yRFC (Figure 8A). At equimolar ratios of yRFC and the pcna-79 mutant, ligation efficiency was reduced to 25% of that in comparable assays with wild-type Cdc9 (Figure 8A, compare lanes 4 and 5).
Our analysis of complex formation between Cdc9 and yPCNA revealed that an interaction between the C-terminal domain of PCNA and amino acid residues adjacent to the Cdc9 PIP box is required for complex formation both in the absence (Figure 5) and presence of DNA (Figure 6). Therefore, we examined the activity of pcna-90 that contains amino acid substitutions in C-terminal residues required for complex formation with Cdc9 in DNA-joining assays containing yRFC. At equimolar ratios of yRFC and the pcna-90 mutant, ligation efficiency was reduced to ∼10% of that in comparable assays with wild-type Cdc9 (Figure 8, lanes 4–6). Taken together these results show that interactions with both the IDCL and C-terminal regions of yPCNA are required for Cdc9 to overcome inhibition by yRFC.
Given the large number of PIP-box-containing proteins, it appears likely that the PIP box/IDCL interface may serve as the primary docking site that in turn facilitates interactions between other regions of PCNA and its partner proteins. If Cdc9 binding to the IDCL promotes functional contacts with the C-terminus of PCNA, then increasing the amount of the PCNA with a defective IDCL should overcome the requirement for the initial binding step. In accord with this model, when the molar ratio of pcna-79 to Cdc9 was increased from 1:1 to 3:1, the amount of DNA joining was almost restored to the levels observed with a wild-type yPCNA: Cdc9 ratio of 1:1 (Figure 8B). The pcna-90 mutant version is more defective in complex formation with Cdc9 when topologically linked to DNA and in alleviating inhibition by RFC compared with pcna-79, suggesting that formation of the β-zipper between Cdc9 and yPCNA plays a critical role in positioning Cdc9 on the yPCNA ring during the functional interplay with yRFC. This scenario predicts that increasing the amount of pcna-90 will be less effective in overcoming inhibition by yRFC compared with pcna-79. In accord with this model, DNA joining was still markedly inhibited, at a 3:1 ratio of pcna-90: Cdc9 (Figure 8B). Thus, our results have shown that the Cdc9 PIP box region makes critical contacts with both the IDCL and C-terminus of yPCNA and strongly suggest that these two protein–protein interfaces have distinct functional roles.
DISCUSSION
PCNA plays a central role in DNA metabolism via interactions with a large number of proteins involved in different DNA transactions and cell cycle regulation (reviewed in (16,23)). For example, the action of enzymes involved in the DNA synthesis, end processing and ligation steps during the maturation and joining of Okazaki fragments and completion of certain DNA excision repair pathways are coordinated by protein–protein interactions with PCNA. Many PCNA interacting proteins contain a conserved PIP box motif that interacts with the IDCL of PCNA (47). Crystal structures of PCNA complexed with PIP box peptides from several PCNA interacting proteins have shown that the highly conserved residues of the various PIP boxes make remarkably similar interactions with PCNA in the form of a 310 helix that interacts with the hydrophobic residues of the IDCL of PCNA (19,32,49,54). Since PIP-box-containing proteins bind to the same site on PCNA, the key question is how the interactions with different PCNA-binding proteins are regulated and co-ordinated. There is emerging evidence from the structural studies that the amino acid sequences adjacent to the PIP box contribute to the affinity and specificity of the protein–protein interaction by contacting different regions of PCNA (19,32,49). For example, the residues C-terminal to the PIP box of the cyclin-dependent protein kinase inhibitor p21 forms a β-sheet with the IDCL of PCNA and contribute to the high affinity interaction between p21 and PCNA (19,49). However, the functional and biological significance of protein–protein interactions involving amino acid sequences adjacent to the PIP box in PCNA-binding proteins have not been demonstrated unequivocally. In this study, our structural, biochemical and mutational results demonstrate that the PIP box region of Cdc9 contacts two distinct regions of yPCNA and that both of these protein–protein interfaces are critical for physical and functional interactions between Cdc9 and PCNA.
The structure of yPCNA complexed with a peptide encompassing the Cdc9 PIP box revealed that, in addition to the contacts made by conserved residues within the Cdc9 PIP box, amino acid residues N-terminal of the PIP box interact with residues within the C-terminus of PCNA to form a β-zipper structure. A similar structure was observed in complexes of intact human FEN-1 with human PCNA (26) and in the complex formed by an archaeal FEN-1 peptide and archaeal PCNA (32). Deletion analysis of the C-terminus of human PCNA has shown that whilst the 5 terminal residues (equivalent to residues 256–258 in yPCNA) are not required for physical and functional interactions with FEN-1, removal of the 8 terminal residues (equivalent to residues 253–258 in yPCNA) caused substantially reduced stimulation of FEN-1 activity by PCNA but had no affect on FEN-1 binding affinity for PCNA (26) (Supplementary Figure 1). Moreover, substitution of residues in the C-terminus of yPCNA that are involved in β-zipper formation result in hypersensitivity to hydroxyurea, MMS and UV light, and a defect in DNA-dependent physical and functional interactions between FEN-1 and PCNA (31,50). Thus, our results demonstrating that the same C-terminal residues of yPCNA are critical for physical and functional interactions with Cdc9 provide further evidence that the β-zipper structure is biologically significant.
Since the action of the DNA-processing enzyme FEN-1 generates the ligatable nick for Cdc9 DNA ligase during Okazaki fragment processing, a comparison of how these enzymes interact with PCNA should provide insights into the mechanisms by which PCNA co-ordinates the sequential action of these enzymes. Although the structures of peptides containing the PIP box of Cdc9 (Figures 1–4) and FEN-1 (34) complexed with PCNA reveal similar protein–protein interfaces involving both the IDCL and the C-terminal domain of PCNA (Figure 3), there are differences in the contributions of the two PCNA interaction interfaces to protein–protein interactions with FEN-1 and Cdc9 in the presence and absence of DNA. The IDCL of yPCNA is critical for complex formation with FEN-1 in the absence of DNA whereas FEN-1 binding to and stimulation by yPCNA topologically linked to DNA are dependent upon residues within the C-terminal domain of yPCNA (31). In contrast, the IDCL and the C-terminal domain interaction interfaces of yPCNA are each required for Cdc9 binding to yPCNA, both unlinked and when topologically linked to DNA, and for functional interactions among yRFC, yPCNA and Cdc9.
Because of the trimeric structure of the PCNA ring, it is possible that up to three proteins can bind simultaneously to the PCNA ring via the IDCL of the three PCNA monomers. Notably, the complex of human FEN-1 and PCNA that formed crystals contained 3 molecules of FEN-1 and 3 molecules of PCNA monomers with each FEN-1 molecule adopting a different conformation relative to the PCNA monomer (26). This suggests that FEN-1 is flexibly tethered to the PCNA ring and that, when a single intact FEN-1 molecule binds to a PCNA trimer, the IDCL-binding sites in the other two subunits of the PCNA trimer are potentially available for binding to other partners (26,32). In this scenario, the PCNA sliding clamp acts as a toolbelt and coordinates the actions of different proteins by holding them together on the same ring, thereby allowing them to engage the appropriate DNA substrate. Recent computational results suggest that PCNA associates with DNA asymmetrically, such that one PCNA subunit interacts specifically with the DNA minor groove, consistent with coordinated interactions among PCNA-binding partners that engage DNA (55). This type of mechanism appears to underlie the ability of translesion DNA polymerases to copy past a lesion that has stalled the replicative polymerase/clamp complex and then permit the replicative polymerase to re-engage on the undamaged template (56). A similar mechanism may also explain the efficient co-ordination of Pol δ and FEN-1 (Rad27) activities during Okazaki fragment processing (57).
In contrast, the complexes formed between human DNA ligase I and a DNA-linked PCNA trimer contained 1 molecule of DNA ligase I per PCNA trimer (27). Moreover, experiments with purified yFEN-1, yPCNA and Cdc9 have failed to detect PCNA complexes containing both FEN-1 and Cdc9 (25). These results strongly suggest that the binding of the replicative DNA ligase to a PCNA monomer within the PCNA trimer occludes the two unoccupied IDCL-binding sites within the PCNA ring. The notion that the binding of Cdc9 to PCNA prevents interactions with other PCNA-binding proteins is further supported by the observation that the induction of genetic instability by overexpression of Cdc9 does not require Cdc9 catalytic activity but instead is dependent upon a functional PIP box (25).
Biophysical and biochemical studies of Cdc9 DNA ligase and human DNA ligase I have shown that these proteins have a very asymmetric shape and that their non-catalytic N-terminal regions containing the PIP box are very sensitive to proteolysis (28,44). This suggests the N-terminal PIP box of the replicative DNA ligase resides within a flexible, unstructured region. When complexed with nicked DNA, the catalytic domain of human DNA ligase I forms a ring structure that is similar in size and shape to the PCNA ring and also encircles the DNA duplex (28). Thus, it is logical to hypothesize that the initial docking of the DNA ligase with a PCNA trimer via its PIP box region, which retains DNA ligase in its elongated form (53), is then promoted by PCNA association with DNA to form a ring structure that may involve more extensive contacts with the surface of the PCNA ring. Thus, multiple modular interactions between PCNA and DNA ligase may be important for function. Surprisingly, we and others have observed that PCNA does not stimulate the catalytic activity of the replicative DNA ligase (11,27,33,35). Thus, in addition to the differences in the stoichiometry of the complexes formed by FEN-1 and the replicative DNA ligase with PCNA, these interactions do not have the same effect on catalytic activity. We suggest that the DNA ligase interaction with PCNA blocks interactions with other PCNA-binding proteins because it is the last step in Okazaki fragment joining and DNA repair whereas interactions with proteins acting earlier in these multi-step pathways will involve a handover to the next protein.
Interestingly, there is a conserved interaction between the replicative DNA ligase and RFC, the loader of the PCNA sliding clamp (35). Although RFC inhibits DNA joining, the presence of PCNA alleviates this inhibition. This interplay among RFC, PCNA and the replicative DNA ligase is conserved and is dependent upon the physical interaction between the PIP box region of the DNA ligase and both the IDCL and C-terminal domain of PCNA. If RFC and Cdc9 recognize parts of a common interface on PCNA, then partner handoffs might be mediated by exchange of a common structural motif, such as the β-zipper, which resembles the Rad51 interface exchange between BRCA2 binding and filament assembly (59). Based on the horseshoe shape of RFC and the multiple contacts between the DNA ligase and the RFC subunits (35,58), we suggest that RFC may hold the DNA ligase in a C-shaped structure and then deliver the DNA ligase to PCNA rings at DNA nicks.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Peter Burgers, Stefan Jentsch, Mike O’Donnell and Satya Prakash for reagents. This work was supported by National Institutes of Health Grants GM57479 (to A.E.T.), CA081967 (to J.A.T.) and the Structural Cell Biology of DNA Repair Program Grant (P01 CA92584 to R.D.K., J.A.T. and A.E.T.). Funding to pay the Open Access publication charge was provided by the Structural Cell Biology of DNA Repair Grant (P01 CA92584).
Conflict of interest statement. None declared.
REFERENCES
- 1.Barnes DE, Johnston LH, Kodama K, Tomkinson AE, Lasko DD, Lindahl T. Human DNA ligase I cDNA: cloning and functional expression in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston LH, Nasmyth KA. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 3.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei YF, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, Shell BK, Nash RA, et al. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 7.Johnston LH. The Cdc9 ligase joins completed replicons in baker's yeast. Mol. Gen. Genet. 1983;190:315–317. doi: 10.1007/BF00330657. [DOI] [PubMed] [Google Scholar]
- 8.Montelone BA, Prakash S, Prakash L. Spontaneous mitotic recombination in mms8-1, an allele of the CDC9 gene of Saccharomyces cerevisiae. J. Bacteriol. 1981;147:517–525. doi: 10.1128/jb.147.2.517-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Braithwaite E, Wang Z. DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry. 1999;38:2628–2635. doi: 10.1021/bi982592s. [DOI] [PubMed] [Google Scholar]
- 10.Willer M, Rainey M, Pullen T, Stirling CJ. The yeast CDC9 gene encodes both a nuclear and a mitochondrial form of DNA ligase I. Curr. Biol. 1999;9:1085–1094. doi: 10.1016/s0960-9822(99)80477-1. [DOI] [PubMed] [Google Scholar]
- 11.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 12.Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, et al. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 14.Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J. Biol. Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 15.Prelich G, Tan C-K, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase δ auxillary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 16.Frouin I, Montecucco A, Spadari S, Maga G. DNA replication: a complex matter. EMBO Rep. 2003;4:666–670. doi: 10.1038/sj.embor.embor886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 18.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 19.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 20.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Paunesku T, Mittal S, Protic M, Oryhon J, Korolev SV, Joachimiak A, Woloschak GE. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int. J. Radiat. Biol. 2001;77:1007–1021. doi: 10.1080/09553000110069335. [DOI] [PubMed] [Google Scholar]
- 22.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic. Acid. Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 23.Kelman Z, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biochem. Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 24.Refsland EW, Livingston DM. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics. 2005;171:923–934. doi: 10.1534/genetics.105.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian J, Vijayakumar S, Tomkinson AE, Arnheim N. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics. 2005;171:427–441. doi: 10.1534/genetics.105.042861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin DS, Bai W, Yao N, O'Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 29.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 30.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 33.Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 35.Levin DS, Vijayakumar S, Liu X, Bermudez VP, Hurwitz J, Tomkinson AE. A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J. Biol. Chem. 2004;279:55196–55201. doi: 10.1074/jbc.M409250200. [DOI] [PubMed] [Google Scholar]
- 36.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 38.Brunger K, Henry A. Processing load and the extended optional infinitive stage. Int. J. Lang. Commun. Disord. 1998;33(Suppl. 422):427. doi: 10.3109/13682829809179462. [DOI] [PubMed] [Google Scholar]
- 39.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 40.Winn M, Isupov M, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 41.Bauer GA, Burgers PM. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7506–7510. doi: 10.1073/pnas.85.20.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, Hingorani MM, O'Donnell M. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]
- 43.Finkelstein J, Antony E, Hingorani MM, O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 44.Tomkinson AE, Tappe NJ, Friedberg EC. DNA ligase from Saccharomyces cerevisiae: physical and biochemical characterization of the CDC9 gene product. Biochemistry. 1992;31:11762–11771. doi: 10.1021/bi00162a013. [DOI] [PubMed] [Google Scholar]
- 45.Hingorani MM, O'Donnell M. A tale of toroids in DNA metabolism. Nat. Rev. Mol. Cell Biol. 2000;1:22–30. doi: 10.1038/35036044. [DOI] [PubMed] [Google Scholar]
- 46.Jonsson ZO, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 47.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 48.Tsutakawa SE, Hura GL, Frankel KA, Cooper PK, Tainer JA. Structural analysis of flexible proteins in solution by small angle X-ray scattering combined with crystallography. J. Struct. Biol. 2006 doi: 10.1016/j.jsb.2006.09.008. in press, doi:10.1016/j/jsb.2006.1009.1008. [DOI] [PubMed] [Google Scholar]
- 49.Bruning JB, Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol. Cell. Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson RE, Kovvali GK, Guzder SN, Amin NS, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 52.Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 53.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Matsumiya S, Ishino S, Ishino Y, Morikawa K. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells. 2002;7:911–922. doi: 10.1046/j.1365-2443.2002.00572.x. [DOI] [PubMed] [Google Scholar]
- 55.Ivanov I, Chapados BR, McCammon JA, Tainer JA. Proliferating cell nuclear antigen loaded onto double-stranded DNA: dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006;20:6023–6033. doi: 10.1093/nar/gkl744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN-1 and Dna2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 58.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 59.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, Volkmann N, Tsuruta H, Blundell TL, Venkitaraman AR, Tainer JA. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.