Abstract
The role of the transcription factor Yin Yang 1 (YY1) in development is largely unknown. Here we show that specific ablation of YY1 in mouse B cells caused a defect in somatic rearrangement in the immunoglobulin heavy-chain (IgH) locus and a block in the progenitor-B-to-precursor-B-cell transition, which was partially rescued by a prerearranged IgH transgene. Three-dimensional DNA fluorescence in situ hybridization analysis revealed an important function for YY1 in IgH locus contraction, a process indispensable for distal VH to DHJH recombination. We provide evidence that YY1 binds the intronic Eiμ enhancer within the IgH locus, consistent with a direct role for YY1 in VHDHJH recombination. These findings identified YY1 as a critical regulator of early B-cell development.
Keywords: YY1, immunoglobulin gene, B-cell development, V(D)J recombination
B lymphocyte development proceeds through the progenitor (pro-B), the precursor (pre-B), immature, and mature B-cell stages (Hardy 1989). Each stage is defined by the recombination status and expression pattern of the immunoglobulin heavy- and light-chain (IgH and IgL) genes and cell surface markers (Hardy et al. 2000; Hardy and Hayakawa 2001; Cancro 2004; Jung and Alt 2004). Ig genes (also known as B-cell receptor or antibody genes) are assembled by nonhomologous rearrangement from Variable (V), Diversity (D) (H only), and Joining (J) gene segments, mediated by the recombination-activating gene (RAG) recombinase (Jung and Alt 2004). Rearrangement of Ig loci is an ordered process (Alt et al. 1984; Chowdhury and Sen 2004). In pro-B cells, DH to JH rearrangement occurs before VH to DHJH recombination. Upon the generation of a productively rearranged VHDHJH fragment and the expression of a μ chain, pro-B cells proceed to the pre-B-cell stage of development. The expressed μ chain, in combination with the surrogate light chain, λ5 and VpreB, and the Ig-α/β heterodimers, forms the pre-B-cell receptor (pre-BCR), which signals pre-B-cell expansion and subsequent light-chain gene rearrangement (Fleming and Paige 2001). Upon completion of light-chain rearrangement, two identical heavy chains and two identical light chains, together with the Ig-α/β heterodimers, form the B-cell receptor and pre-B cells transit to immature B cells, which exit the bone marrow (BM) to become mature peripheral B cells.
The lineage and developmental stage specificity of V(D)J recombination relies on the local accessibility of the gene loci to the common RAG recombinase, which functions in both B- and T-cell receptor gene rearrangements (Chowdhury and Sen 2004; Jung and Alt 2004). The IgH locus becomes accessible to RAG recombinase in pro-B cells, accompanied by a series of changes including a periphery-to-center nuclear repositioning, locus contraction mediated by DNA looping, germline transcript expression, and covalent modifications of histones at specific sites (Yancopoulos and Alt 1985; Chowdhury and Sen 2001; Kosak et al. 2002; Morshead et al. 2003; Su et al. 2003; Bolland et al. 2004; Fuxa et al. 2004; Johnson et al. 2004; Roldan et al. 2005; Sayegh et al. 2005). The relationship, if any, among the multiple changes occurring at the IgH locus, and their exact roles in VHDHJH recombination, remain to be determined. Previous studies have identified several cis-acting elements within the IgH locus that are important for VHDHJH recombination, including the variable gene promoters, the intronic enhancer (Eiμ), and the 3′ enhancer (Sleckman et al. 1996). Studies using knockout mice confirmed an important role of the core intronic Eiμ in mediating VHDHJH recombination (Sakai et al. 1999; Perlot et al. 2005). However, it is unclear whether the Eiμ enhancer regulates VHDHJH recombination by controling IgH locus nuclear repositioning, contraction, and/or chromosomal changes. The Eiμ enhancer contains binding sites for multiple transcription factors including YY1 (Ernst and Smale 1995; Sleckman et al. 1996). The role of these proteins in VHDHJH recombination is largely unknown.
YY1 is a zinc finger protein that functions as a transcriptional activator, repressor, or transcription-initiator element-binding protein, depending on the promoter context (Liu and Shi 2005). Recently, YY1 has also been shown to regulate p53 stability independent of its transcriptional activity (Gronroos et al. 2004; Sui et al. 2004). YY1 is evolutionarily conserved from Drosophila to human and has been suggested to function as a Polycomb Group (PcG) protein during development (Brown et al. 1998, Brown et al. 2003; Atchison et al. 2003; Srinivasan and Atchison 2004). Animal studies indicate a role for YY1 in embryogenesis and in neuronal development (Donohoe et al. 1999; Satijn et al. 2001; Kwon and Chung 2003; Morgan et al. 2004). In vitro biochemical and cell-based analyses suggest that YY1 may play important roles in a number of biological and pathological processes, including B-cell development and function (Thomas and Seto 1999; Gordon et al. 2003; Patrone et al. 2004; Su et al. 2004; Liu and Shi 2005) However, the early embryonic lethality of YY1 knockout mice precluded the investigation of YY1 in specific developmental pathways in vivo.
To address the role of YY1 during later stage development, we generated mice carrying conditional yy1 alleles (yy1f). To investigate the role of YY1 in B-cell development, we took advantage of the novel mb1-Cre transgenic mouse (Hobeika et al. 2006), which recombines LoxP-flanked sequences in early B-cell progenitors. Phenotypic analyses of the B-cell-specific yy1 knockout mice (mb1-Cre yy1f/f) demonstrated that YY1 plays a critical role in controling the pro-B-to-pre-B-cell transition. Analysis of recombination events in the IgH locus of YY1-deficient pro-B cells revealed normal DH to JH, but impaired VH to DHJH recombination. A prerearranged IgH transgene inserted into the IgH locus partially rescued the pro-B to pre-B block caused by loss of YY1. This indicates that YY1-dependent VH to DHJH recombination is important for pro-B-cell differentiation and that YY1 also plays additional roles in the pro-B-to-pre-B-cell transition. Three-dimensional DNA fluorescence in situ hybridization (3D FISH) showed a significantly increased pro-B population unable to undergo IgH locus contraction upon loss of YY1. Chromatin immonoprecipiation (ChIP) showed YY1 binding to the Eiμ enhancer within the IgH locus. Taken together, our study identifies a novel function for YY1 in early B-cell development by controling IgH locus contraction and VHDHJH recombination, possibly through direct interaction with the IgH Eiμ enhancer.
Results
B-cell-specific deletion of yy1 with the mb1-Cre transgenic mice
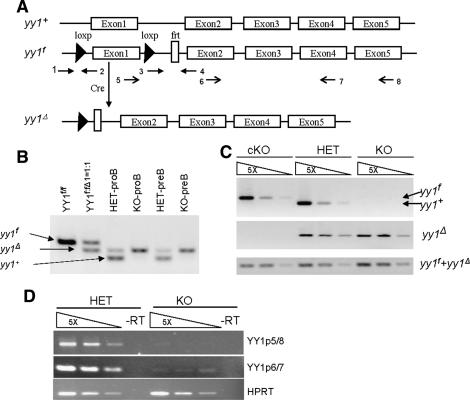
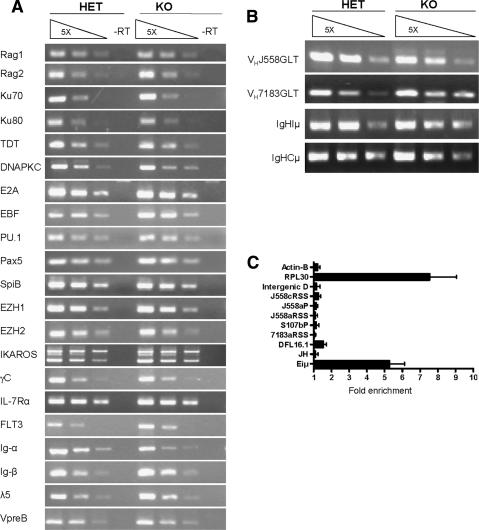
In order to study the role of YY1 in lineage development, we generated a conditional yy1 knockout allele (yy1f) by flanking the yy1 promoter region and exon1 with loxP-sites (Fig. 1A; Affar et al. 2006). The yy1f allele expresses normal levels of YY1 protein and Cre recombinase-mediated recombination yields a yy1-null allele (yy1Δ) similar to the constitutive null allele described before (Fig. 1A; Donohoe et al. 1999; Affar et al. 2006; data not shown). To achieve B-cell-specific ablation of YY1, we intercrossed yy1f mice with mice carrying the mb1-Cre transgene, which facilitates deletion of loxP-flanked sequences at the earliest stages of B-cell development with high efficiency (Hobeika et al. 2006). PCR analysis failed to detect loxP-flanked yy1f alleles in purified BM pro-B (CD19+CD43+sIgM−) and pre-B (CD19+CD43−sIgM−) cells of mb1-Cre yy1f/f (knockout/KO) and mb1-Cre yy1f/+ (heterozygous/HET) mice (Fig. 1B,C). In addition, YY1 mRNA was essentially undetectable by RT–PCR in pro-B cells purified from the KO mice (Fig. 1D), indicating almost complete ablation of YY1 expression in early B cells.
Figure 1.
B-cell-specific deletion of yy1 with the mb1-Cre transgenic mice. (A) Schematic diagram of the yy1 locus. The wild-type yy1 allele (yy1+) contains five exons. The conditional yy1 allele (yy1f) was constructed by inserting a pair of LoxP sites flanking the exon1 and the promoter region, which will be excised in the presence of Cre recombinase, thereby generating a null allele of yy1 (yy1Δ). The arrows indicate the location of the primers used for PCR detection of deletion efficiency, PCR genotyping, and RT–PCR to detect YY1mRNA. Primers 1 and 2 detect both yy1+ (223 bp) and yy1f (369 bp). Primers 1 and 4 detect yy1Δ (292 bp). Primers 3 and 4 detect both yy1f and yy1Δ (138 bp). Primers 5 and 8 detect a 480-bp YY1 mRNA, and primers 6 and 7 detect a 205-bp YY1 mRNA. (B) PCR detection of deletion efficiency in sorted pro-B (CD19+CD43+sIgM−) and pre-B (CD19+CD43−sIgM−) from mb1-Cre yy1f/+ (HET) and mb1-Cre yy1f/f (KO) mice. A sample of YY1f:Δ=1:1 was used to show the similar amplifying efficiency of the yy1f and yy1Δ alleles in the mixed primers of 1, 2, and 4. (C) PCR detection of deletion efficiency in sorted pro-B (CD19+CD43+sIgM−) cells from yy1f/f (cKO), HET, and KO mice. Primers 1 and 2 were used to detect the yy1f allele in the top panel. Primers 1 and 4 were used to detect the yy1Δ allele in the middle panel. The bottom panel showed the total yy1f and yy1Δ allele to serve as control for equal loading. (D) RT–PCR detection of YY1 mRNA. Two different primer combinations, primers 5 and 8 (p5/8) and 6 and 7 (p6/7), were used to identify YY1 mRNA in both control and KO mice. Input cDNA was normalized using the HPRT mRNA.
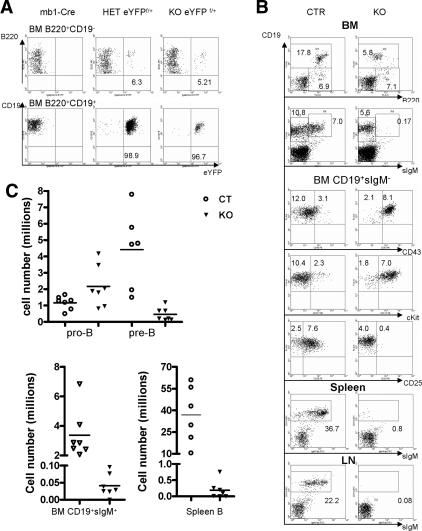
To identify cells in which Cre-mediated recombination occurred by flow cytometric analysis (FACS), we used the Rosa26-eYFP (R26-eYFP) allele (Srinivas et al. 2001). Cells carrying this allele fluoresce green light upon Cre-mediated excision of a loxP-flanked stop cassette. Therefore, deletion efficiency of the R26eYFP allele as reflected by the percentage of green fluorescent cells serves as an indirect measurement of recombination efficiency of other loxP-flanked alleles in the same cell population. The B220+CD19− population in the BM, comprising the earliest B-cell progenitors, contained a relatively low percentage of green fluorescent cells (5%–6%) (Fig. 2A). In contrast, >95% BM CD19+ B cells were green in mb1-Cre/R26eYFP, HET/R26eYFP, and KO/R26eYFP mice, which is consistent with the deletion efficiency detected by PCR and RT–PCR. These results confirmed successful generation of a B-cell-specific yy1 knockout mouse.
Figure 2.
Loss of YY1 caused a pro-B to pre-B differentiation block. (A) Detection of Cre-mediated recombination by FACS analysis of green-fluorescing cells at different stages of BM B-cell development with the R26eYFP Cre reporter. The mb1-Cre mice were used as a negative control for the eYFP signal. A total of three mice from each group were analyzed. (B) FACs analysis of B-cell development in mb1-Cre yy1f/f knockout mice (KO). Pro-B cells were characterized as B220+CD19+CD43+cKit+CD25−sIgM−, and pre-B cells were characterized as B220+CD19+CD43−cKit− CD25+sIgM−. Immature and mature B cells were CD19+sIgM+. In general, KO mice had a two- to threefold increase in pro-B percentage and a significant decrease of pre-B percentage. The numbers shown on the dot plot are the average percentage of each subpopulation in the total cells from BM, spleen, or LNs. A total of three to six experiments were performed. (C) Total cell number of each B-cell subpopulation in KO and control (CTR) mice. Pro-B cells were identified as CD19+CD43+sIgM− and pre-B cells were identified as CD19+CD43−sIgM−. BM immature (IMB) and mature B (MB) were CD19+sIgM+. Cells from two femurs and two tibias were counted. Spleen B cells were defined as CD19+. Bars indicate the average number of six to seven experiments.
Loss of YY1 blocks pro-B-cell differentiation
Mice genotyped as yy1f/+, yy1f/f, mb1-Cre, and mb1-Cre yy1f/+ (HET) were indistinguishable from wild-type mice and were subsequently grouped as controls (CTR), suggesting that a single yy1 allele is sufficient to support B-cell development. Consistent with the very low percentage of eYFP+ cells at the earliest B220+CD19− stage, no significant difference was detected between control and KO mice at this stage of B-cell development. In contrast, compared with control mice, KO mice contained twice as many pro-B cells (B220loCD19+CD43+cKit+ CD25−sIgM−), but a markedly reduced number of pre-B cells (B220medCD19+cKit−CD25+CD43−sIgM−) in the BM, and hardly any immature and mature B (CD19+sIgM+) cells in BM, spleen, and lymph nodes (LNs) (Fig. 2B,C), suggesting a critical role for YY1 during differentiation of pro-B cells to pre-B cells.
YY1-deficient pro-B cells exhibit impaired VH to DHJH recombination
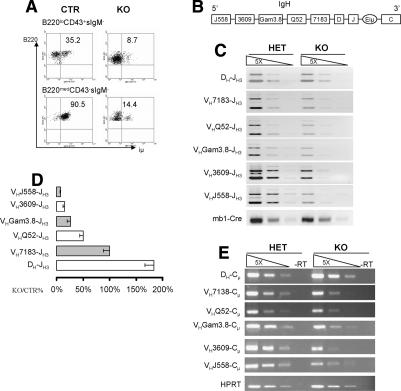
Successful rearrangement of the IgH gene and subsequent expression of Igμ is essential for pro-B-cell differentiation (Jung and Alt 2004). As shown in Figure 3A, while ∼35% of pro-B (B220+CD43+sIgM−) and 90% of pre-B cells expressed intracellular μ (iμ) chain in the BM of control mice, the percentage of iμ+ cells was 8% and 14% in the BM of yy1−/− pro-B and pre-B cells, respectively. To determine whether reduced iμ expression was due to a defect in VHDHJH recombination, we compared the recombination frequency of DH to JH and VH to DHJH in pro-B cells purified by cell sorting from control and yy1−/− mice using degenerative PCR primers as described previously (Fuxa et al. 2004). We found that pro-B cells without YY1 underwent DH to JH recombination normally (Fig. 3B–D). In contrast, YY1-null pro-B cells had a gradually decreased frequency of VH to DHJH rearrangement, which was inversely proportional to the distance separating VH families from the DHJH region (Fig. 3B–D). The recombined VH gene fragments from the most proximal VH families VH7183 and VHDQ52 in the KO pro-B cells were 50%–100% of those in the control pro-B cells. The recombination frequency of the distal VH3609 and VHJ558 segment in the KO pro-B cells was 6%–20% of that in the control pro-B cells. Consistently, RT–PCR showed that the expression of IgH μ mRNA corresponded to the genomic DNA recombination frequencies (Fig. 3D), indicating that loss of YY1 did not prevent transcription of the recombined IgH alleles. These findings suggest that YY1 plays a critical role in VH to DHJH recombination, thus identifying a novel role for YY1 in pro-B-cell differentiation.
Figure 3.
Loss of YY1 caused a defect in VH to DHJH, but not DH to JH recombination. (A) Detection of iμ-chain expression in pro-B and pre-B cells from CTR and KO mice. Results are representative of three experiments. (B) Schematic drawing of the IgH locus to show the relative location of the different VH gene clusters. Only the VH gene families analyzed are shown. The most distal VH family is J558 and the most proximal VH family is 7183. (C) PCR detection of DH-JH and VH-DHJH recombination in sorted HET and KO pro-B cells. Input DNA was normalized by PCR amplification of the Cre DNA from the Ig-α locus. Fivefold serial dilution was used for PCR. Degenerative forward primers are located in the respective V regions and the reverse primer was in the JH3 segment. (D) Quantitation of DH-JH and VH-DHJH recombination in sorted pro-B cells. The average recombination frequency with its standard errors is shown as the relative percentage of YY1KO compared with control pro-B cells. Six independent experiments were conducted. (E) RT–PCR to detect the mRNA levels of the μ transcripts. Same forward primers used as in C, and the reverse primer was located in the first exon of the μ-constant region. Equal loading of input cDNA was shown by the amplification of the HPRT mRNA.
YY1 regulates VHDHJH recombination by controling IgH locus contraction
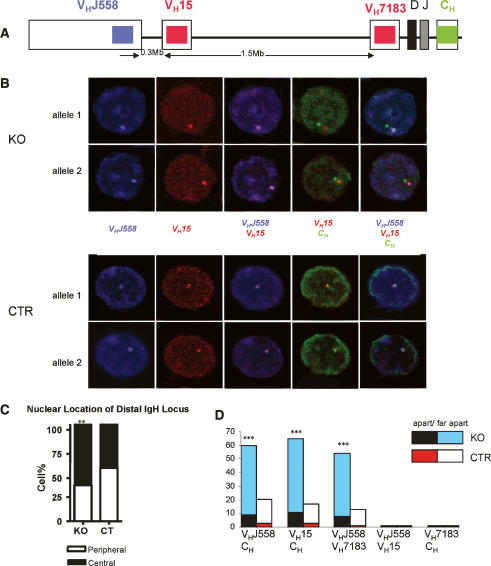
The murine IgH locus spans ∼3 Mb and the V gene region occupies the 5′ 2.5 Mb (Johnston et al. 2006). The IgH locus relocates from the periphery to the center of the nucleus in pro-B cells, a process believed to facilitate VHDHJH recombination (Kosak et al. 2002). Locus contraction brings the distal and mid VH gene segments into close proximity with the DHJH region. This enables RAG recombinase-mediated VH to DHJH recombination (Kosak et al. 2002; Roldan et al. 2005; Sayegh et al. 2005). Locus contraction, mediated by DNA looping, has been observed by three-color 3D DNA FISH experiments (Roldan et al. 2005; Sayegh et al. 2005) To investigate mechanisms by which YY1 regulates VH to DHJH recombination, and to specifically address the question of whether YY1 plays a role in IgH locus relocation and/or contraction, we performed 3D DNA FISH with ex vivo purified pro-B cells (CD19+cKit+) from control and KO mice. The nuclear location of different IgH gene segments in three-dimensionally preserved pro-B cells was detected with three differentially labeled locus-specific probes. Gene segments were scored as colocalized if the two fluorescence signals were overlapping or separated by a distance of <0.3 μm. In contrast, if the two signals were separated by 0.3–0.5 or 0.5–1.5 μm, they were scored as apart or far apart, respectively. Figure 4A shows the position and color of the three probes in the IgH locus and the approximate distance (in base pairs) separating the probes. As shown in Figure 4C, the percentage of pro-B cells with a centrally localized VHJ558 signal in the YY1 KO mice was increased compared with that of the control (YY1KO 63% and CTR 45%, p < 0.01), suggesting that loss of YY1 did not prevent relocation of the distal IgH locus from the periphery to the center of the nucleus.
Figure 4.
Loss of YY1 prevents IgH locus contraction. 3D DNA FISH was performed to show the subnuclear localization and contraction of the IgH locus in CTR and KO pro-B cells. (A) Schematic diagram indicating the probe positions in the IgH locus (not drawn to scale). (B) Confocal images of different IgH segments in pro-B nuclei. Pro-B cells were prepared from CTR and KO mice and sorted as CD19+cKit+ cells. The VHJ558 probe was directly labeled with Cy5-dUTP (blue) and synthesized from BAC526A21. The VH15 probe (BAC 243G9) and VH7183 (BAC167C1) probe were directly labeled with Cy3-dUTP (red). The CH probe (BAC C34H6) was directly labeled with FITC-dUTP (green). (C) Statistical analysis of the nuclear position of the VHJ558 gene segments in CTR and KO pro-B cells. (D) Statistical analysis of the distance separating different IgH gene segments in CT and KO pro-B cells. A distance of 0.3–0.5 μm was defined as “apart,” and a distance of 0.5–1.5 μm was referred to as “far apart.” (**) P < 0.01; (***) P < 0.001.
We next analyzed the relative distance separating the distal VH probes (VHJ558 and VH15) from the proximal VH probe (VH7183) or the constant region probe (CH). Consistent with the linear distance separating the gene segments along the chromosome, we found that the signals of the two distal VH gene families, VHJ558 and VH15, were colocalized, as were the signals of the proximal VH gene family VH7183 and the CH signal (Fig. 4B,D). While the colocalization of the distal VH probes and the CH probes can be a consequence of either IgH locus contraction or VH-DHJH recombination due to the deletion of DNA segments between the recombined VH and DHJH genes, only IgH locus contraction will result in the convergence of the distal VHJ558 and the proximal VH7183 probes. If recombination occurs 5′ of VH7183 genes, no VH7183 signal will be detected in pro-B cells due to the deletion of VH7183 gene segments. If recombination occurs within the VH7183 to DHJH region, it will not change the linear distance between distal VHJ558 and proximal VH7183 segments. In the majority of the control pro-B cells, the distal VH gene segments were colocalized with the proximal VH or CH region (Colocalization/Apart/Far apart percentage: VHJ558/CH 79.6/2.9/17.5, total 121 alleles; VH15/CH 83/3/14, total 61 alleles; VHJ558/VH7183 87/1/12, total 60 alleles) (Fig. 4D; Supplementary Table 1). In contrast, loss of YY1 resulted in a significant increase in alleles with separated distal and proximal IgH signals (Colocalization/Apart/Far apart percentage: VHJ558/CH 40/9/51, total 175 alleles; VH15/CH 35/11/54, total 98 alleles; VHJ558/VH7183 46/8/46, total 78 alleles, p < 0.001) (Fig. 4D; Supplementary Table 1). This indicated that loss of YY1 adversely affected IgH locus contraction, providing a mechanistic explanation for the observed defects in VH-DHJH recombination in the YY1 KO pro-B cells.
Loss of YY1 does not change RNA transcript levels of many molecules required for pro-B-cell differentiation and VHDHJH recombination
What is the mechanism by which YY1 regulates IgH locus contraction? To address this issue, we first asked whether loss of YY1 affected transcription of genes whose products are known to be important for IgH locus contraction. Pax5 and EZH2 are known to play a role in IgH locus contraction (Fuxa et al. 2004; A. Tarakhovsky, pers. comm.). As shown in Figure 5A, loss of YY1 did not change the mRNA level of Pax5 and EZH2. We also examined the mRNA levels of other molecules known to be important for V(D)J recombination and/or pro-B-cell differentiation. RT–PCR confirmed that loss of YY1 did not alter the mRNA levels of the main components of the V(D)J recombination machinery, including RAG1 and RAG2, intranuclear terminal deoxynucleotidyl transferase (TdT), the DNA-dependent protein kinase c (PKC), and Ku70/Ku80 (Fig. 5A; Jung and Alt 2004). We also examined the mRNA levels of several transcription factors previously shown to be critical for early B-cell development, including E2A, EBF, PU.1, spiB, EZH1/2, IKAROS, and surface receptors including Interleukin-7 (IL-7) receptor α, the common γ chain (γc), FLT3, and components of the pre-BCR, including Ig-α, Ig-β, λ5, and VpreB (Fleming and Paige 2002; Busslinger 2004; Corcoran et al. 2005). With the exception of the Ig-α (reduced by 50%) and FLT3 (two- to threefold increase), none of them appeared to be affected by the loss of YY1 (Fig. 5A). Finally, KO pro-B cells expressed normal to higher levels of Iμ, Cμ, VH7183, and VHJ558 germline transcripts (Fig. 5B), which are markers for an accessible IgH locus (Yancopoulos and Alt 1985), indicating that loss of YY1 did not interfere with the initial chromatin opening of the IgH locus. Taken together, loss of YY1 did not appear to affect the expression of genes known to be important for IgH locus contraction, suggesting that YY1 may play a direct role in this process.
Figure 5.
YY1 binds to IgH locus directly. (A) RT–PCR to detect the mRNA levels of molecules involved in early B-cell development in sorted pro-B cells from heterozygous and KO mice. (B) RT–PCR to detect the IgH germline transcript. (A,B) Input cDNA was normalized by the amplification of the HPRT mRNA (Fig. 3D). (C) Quantitative ChIP–PCR. ChIP assays showing YY1 binding to the Eiμ enhancer, but not several other selective regions of the IgH locus. Experiments were performed with rabbit polyclonal anti-YY1 or normal rabbit IgG as control. Real-time PCR was carried out using the Roche 480 LightCycler. Data are presented as “fold enrichment” of anti-YY1 signal relative to that of IgG. (JH and DFL16.1) J and D gene regions; (7183aRSS) the most proximal VH gene family; (S107bp) the middle region of the VH gene; (J588aP and J558aRSS) the proximal VHJ588 genes, (J558cRSS) the distal regions of the VHJ588 genes; (Intergenic D) intergenic regions between members of the VHJ588 family. RPL30 and Actin-B are positive and negative controls, respectively. Each DNA sample was repeated two to three times by real-time PCR for each primer pair, and a total of eight to 12 independent ChIP preparations were performed.
YY1 binds to Eiμ within the IgH locus
The direct model predicts YY1 binding at the IgH locus. Previous in vitro studies identified a potential YY1-binding site at the μE1 site of the IgH intronic enhancer (Eiμ) (Park and Atchison 1991). Using quantitative ChIP assays, we confirmed binding of YY1 to Eiμ in ex vivo cultured pro-B cells (Fig. 5C), consistent with a potentially important role of this YY1 site. We also tested a few selected areas in the VH region, but found no significant YY1 binding. (Fig. 5C). Since the selected regions only represent a small percentage of the total VH regions, further systematic analysis (ChIP–chip) is necessary to determine whether YY1 binds to other VH regions in addition to Eiμ.
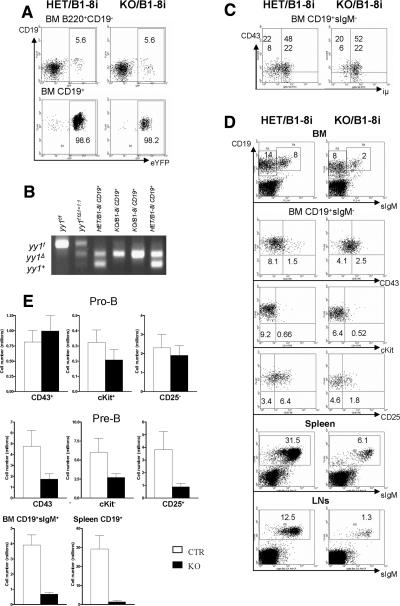
A prerecombined IgH transgene partially rescues pro-B-cell differentiation defect in the yy1 KO mice
To further investigate whether YY1 controls pro-B-cell differentiation mainly through regulation of VHDHJH recombination, we introduced a prerearranged VHDHJH segment (B1-8i) into the YY1 B-cell-specific mb1-Cre YY1 KO mice. The B1-8i IgH transgene is inserted into the endogenous IgH locus (Sonoda et al. 1997). As shown in Figure 6A,B, yy1f alleles were deleted efficiently in CD19+ BM B cells from both HET/B1-8i and KO/B1-8i mice, as reflected by the percentage of eYFP+ cells and PCR analysis.
Figure 6.
The prerearranged IgH transgene (B1-8i) partially rescued the pro-B/pre-B-cell differentiation defect in the KO mice. (A) Visualization of Cre activities by detecting green-fluorescing cells at different stages of BM B-cell development with the R26eYFP Cre reporter. Results are representative of three experiments. (B) PCR detection of deletion efficiency in purified CD19+ BM B cells from HET/B1-8i and KO/B1-8i mice. A sample of yy1f:fΔ1 = 1:1 was used to show similar amplifying efficiency of the yy1f and yy1Δ alleles with the primer mixture of 1, 2, and 4. (C) Detection of iμ-chain expression in pro-B and pre-B cells from CTR and KO mice. Results are representative of three experiments. (D) FACs analysis of CTR/B1-8i and KO/B1-8i mice. The numbers shown on the dot plot are an average percentage of each subpopulation in the total cells of BM, spleen, or LNs from five to 10 experiments. (E) Total cell number of each B-cell subpopulation in KO/B1-8i and CTR/ B1-8i mice (Mean ± SE). Pro-B cells were identified as CD19+CD43+sIgM−, CD19+cKit+sIgM−, or CD19+CD25−sIgM−, and pre-B cells as CD19+CD43−sIgM−, CD19+cKit−sIgM−, or CD19+CD25+sIgM−. Cells from two femurs and two tibias were counted. A total of five to 10 mice were analyzed for each genotype.
As shown in Figure 6C, both CTR/B1-8i and KO/B1-8i mice had a similar percentage of iμ-chain-positive cells among the CD19+sIgM− B-cell population, indicating that YY1 was not required for the transcription of the recombined IgH gene. We found very few cKit+CD19+ pro-B cells in both CTR/B1-8i and KO/B1-8i BM (Fig. 6D,E), indicating that expression of the B1-8i transgene bypassed the requirement for YY1 and successfully transduced the necessary signal to down-regulate surface expression of c-Kit to facilitate the transition of pro-B to pre-B cell. Compared with the yy1 KO mice, yy1 KO/B-8i mice displayed reduced pro-B-cell number, and increased numbers of pre-B, immature and mature B cells in BM, spleen, and LNs (Fig. 6D,E), indicating that regulation of VH to DHJH recombination is an important function of YY1 in pro-B-cell differentiation. However, the pre-B, immature, and mature B-cell populations were still significantly reduced in yy1 KO/B1-8i compared with those in CTR/B1-8i mice (Fig. 6D,E). In addition, the incomplete down-regulation of CD43 and up-regulation of CD25 upon expression of the prerecombined IgH transgene in the YY1-depleted pre-B cells, point to other functions for YY1 in pre-B-cell expansion and differentiation.
Discussion
Pro-B-cell differentiation requires successful recombination and expression of a single IgH allele. In this report, we have demonstrated that the ubiquitously expressed YY1 transcription factor is important for VH to DHJH recombination and pro-B-cell differentiation. Furthermore, we have provided evidence supporting a role for YY1 in regulating V(D)J recombination by controling IgH locus contraction.
In pro-B cells, the spatially separated VH, DH and JH gene segments are brought together by means of DNA looping and IgH locus contraction to allow for RAG recombinase-mediated VHDHJH recombination (Kosak et al. 2002; Roldan et al. 2005; Sayegh et al. 2005). Pax5 is the first protein identified to play a role in IgH locus contraction (Fuxa et al. 2004). Pax5-binding sites within the IgH locus and direct interaction between Pax5 and IgH locus were described in recent reports (Pawlitzky et al. 2006; Zhang et al. 2006), suggesting that Pax5 may participate in DNA loop formation within the IgH locus. Here we show that loss of YY1 interfered with IgH locus contraction, but did not affect IgH locus relocation. The normal expression of Pax5 in the pro-B cells lacking YY1 (Fig. 5A) indicates that YY1 does not control IgH locus contraction by regulating transcription of Pax5. It is likely that YY1 and Pax5 play nonredundant roles in IgH locus contraction. For instance, YY1 and Pax5 may require each other for efficient binding to the IgH locus and DNA loop formation. YY1 and Pax5 may also work together to induce histone modifications and alterations in chromatin structures that may facilitate IgH locus contraction and recombination.
The direct interaction between YY1 and Eiμ as shown by ChIP, together with a lack of requirement for YY1 in the transcription of a number of genes whose products are important for IgH recombination (Fig. 5A), support a direct involvement of YY1 at the IgH locus in regulating IgH locus contraction. Thus far, cis-elements required for IgH DNA looping and locus contraction remain unknown. The importance of the core Eiμ in VHDHJH recombination and its short distance to the DHJH region makes it a good candidate as a site that participates in the regulation of DNA looping (Sakai et al. 1999; Perlot et al. 2005). Future studies on the status of IgH locus contraction with pro-B cells derived from Eiμ knockout mice should provide insights into the role of Eiμ in IgH DNA looping and locus contraction. The Eiμ enhancer contains binding sites for multiple transcription factors including YY1, E2A, and Pu.1 (Park and Atchison 1991; Ernst and Smale 1995). Transgenic mutant mice studies suggested that each individual transcription factor-binding site within the Eiμ may play distinct role in different aspects of VHDJH recombination (Fernex et al. 1994, 1995) Our analysis of the published data suggests that a 38-base-pair (bp) fragment in the most 5′ region of the core Eiμ may play an important role in VH to DJH recombination (Fernex et al. 1995; Sakai et al. 1999; Perlot et al. 2005). This 38-bp cis-acting element contains the μE1–YY1-binding sites and μE5–E2A-binding sites. A recent study showed that expression of E2A and binding of E2A to μE5 is not required for VHDHJH recombination if a sufficient amount of the B-cell-specific EBF protein is present in the pro-B cells (Seet et al. 2004), suggesting that the YY1/μE1 interaction is functionally important for the VH to DHJH recombination. DNA looping was originally suggested to be a mechanism of transcriptional regulation mediated by long-range cis-elements, such as distal enhancers and locus control regions within the same or different chromosomes (Tolhuis et al. 2002; de Laat and Grosveld 2003; Spilianakis and Flavell 2004; Spilianakis et al. 2005). Therefore, understanding YY1-mediated IgH locus contraction will have important implications for understanding mechanisms that control communications among noncontiguous chromosomal DNA elements regulating both transcription and recombination.
An alternative, but not mutually exclusive possibility is that YY1 regulates IgH locus contraction indirectly by recruiting histone modifiers to change local histone modifications and chromatin structure, which, in turn, could influence the local chromatin accessibility for proteins directly involved in DNA loop formation. Histone acetylation, while a marker for an accessible IgH locus and critical for IgH germline transcription (Chowdhury and Sen 2001; Johnson et al. 2003), is neither required for nor dependent on IgH locus contraction as suggested by studies on Pax5 and Stat5 knockout pro-B cells (Fuxa et al. 2004; Bertolino et al. 2005). The normal levels of IgH germline transcripts in the YY1−/− pro-B cells suggests that loss of YY1 may not change the histone acetylation status of the IgH locus. The fact that Pax5 controls both loss of histone H3K9 methylation at the IgH locus and IgH locus contraction in pro-B cells (Fuxa et al. 2004; Johnson et al. 2004) suggests a possible link between methylation and IgH locus contraction. Consistent with this, EZH2-mediated methylation of histone H3 Lys 27 (K27) is believed to be important for VH to DHJH rearrangement and IgH locus contraction (Su et al. 2003; A. Tarakhovsky, pers. comm.). YY1 interacts with the EED/EZH2 PcG complex and was shown to be required for recruiting EZH2 to DNA during muscle differentiation (Satijn et al. 2001; Caretti et al. 2004). Whether YY1 is required for recruiting EZH2 specifically to the IgH locus remains to be determined.
Both YY1KO and interleukin-7 receptor α (IL-7Rα) KO pro-B cells have a more severe defect in distal than proximal VH to DHJH recombination (Corcoran et al. 1998). Although we cannot completely exclude an interplay between YY1 and IL-7-mediated signals, the loss of YY1 has no obvious effects on a number of IL-7-dependent processes, including expression of distal VH gene germline transcription and histone acetylation of the distal VH gene segments, as well as expression of Pax5 and its target genes (Corcoran et al. 1998; Chowdhury and Sen 2001).
Lastly, it is worth pointing out that our phenotypic analysis of the B-cell-specific YY1 KO mice suggests that YY1 plays roles in B-cell development in addition to pro-B-cell differentiation. In the YY1KO mice, ∼10% of the pro-B cells expressed iμ chain (Fig. 3A). If YY1 were dispensable for B-cell development beyond pro-B-cell differentiation, these μ-chain-expressing B cells would have differentiated into pre-B, immature, and mature B cells and accumulated in the peripheral lymphoid organs. In contrast, we observed an almost complete absence of immature and mature B cells in the YY1KO mice, suggesting that YY1 is also required at later stages of B-cell development following VHDHJH recombination and μ-chain expression. This hypothesis is further supported by the incomplete rescue of the YY1KO phenotype by a prerearranged IgH transgene. The partial rescue was reflected by the significantly reduced pre-B-cell number, and the incomplete down-regulation of CD43 and up-regulation of CD25 upon expression of the prerecombined IgH transgene in the YY1-deficient pre-B cells (Fig. 6D,E). A much lower percentage of pre-B cells from the YY1KO/B1-8i mice contain intracellular κ-chain expression compared with that of CTR/B1-8i mice, suggesting an additional role of YY1 in light-chain recombination (H. Liu and Y. Shi, unpubl.). Collectively, our results suggest that YY1 is likely to play critical roles at multiple stages of B-cell development.
In summary, our study has identified a novel lineage-specific role for YY1 in early B-cell development. Our findings not only provide new insights into the molecular mechanisms underlying VH-DHJH recombination and locus contraction, but also shed significant light on the role and mechanism of action of YY1 in living organisms. The findings here also highlight YY1 as a potential regulator, which may facilitate communications among noncontiguous DNA elements in the genome.
Materials and methods
Generation of the floxed YY1 conditional knockout mice
The generation of the loxP-flanked yy1 allele (yy1f) was described previously (Affar et al. 2006). Generation of the B-cell-specific mb1-Cre transgenic mice will be described elsewhere (Hobeika et al. 2006). The Rosa26eYFP cre reporter mice were kindly provided by Dr. Frank Costantini (Srinivas et al. 2001). Generation of the IgH transgenic B1-8i mice was described previously (Sonoda et al. 1997). All mice were bred and maintained under specific pathogen-free conditions at the animal facility of Harvard Medical School. All mouse protocols were approved by the Harvard Medical School IACUC. Mice are maintained on a mixed background of 129SvEvXC57BL/6. Analyzed animals range from 2 to 14 wk old, including both males and females. The observed phenotype is consistent at different ages and both sexes in the KO mice. Mutant mice were genotyped by PCR (for primer sequences, see Supplementary Table 2).
FACS analysis and cell sorting
Single-cell suspension prepared from BM, spleen, and LNs were stained with antibodies conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein (PerCP), allophycocyanin (APC), or biotin. For FACs analysis, the following antibodies were purchased from BD biosciences: PerCP- or APC-conjugated anti-B220 (RA3-6B2), FITC- or PE-conjugated anti-CD43 (S7), PE- or APC-conjugated anti-CD19(1D3), and PerCP-conjugated-streptavidin. The following antibodies were purchased from eBiosciences: PE- and APC-anti-cKit, and PE- and APC-anti-CD25. FITC- or biotin-conjugated anti-μ (M41) were prepared from the corresponding hybridoma. Flow cytometric acquisition was conducted with a FACscalibur (BD Biosciences) and data was analyzed with WinMDI2.8. For biochemical analysis, CD19+ B lymphocytes were enriched with the EasySep B-cell enrichment kit (StemCell Technology) from freshly prepared BM cell suspension and purified according to the manufacturer’s protocol. Enriched cells were then stained with anti-B220, anti-CD43, and anti-μ antibodies. Pro-B cells were sorted as B220loCD19+CD43+sIgM− and pre-B cells were sorted as B220medCD19+CD43−sIgM−. For 3D FISH, freshly prepared BM cell suspension was stained directly with anti-CD19-PE and anti-cKit-APC and sorted as CD19+cKit+ pro-B cells with an Aria (BD Biosciences). The purity of sorted cells was verified by re-sorting. Sorted cells for further experiments were at least 95% pure.
PCR detection of deletion efficiency and recombination efficiency
Sorted pro-B cells and pre-B cells or total thymocytes (1 × 105) from heterozygous mb1-Cre yy1f/+ and homozygous mb1-Cre yy1f/f mice were dissolved in 80 μL of 50 mM NaOH, heated for 5 min at 95°C, and vortexed to dissolve the cell pellets. NaOH was neutralized with 20 μL of 1 M Tris.HCl (pH 6.8). The resulting DNA solution was serially diluted at a 1:5 ratio. About 1 μL of solution was used for each PCR reaction to detect either the deletion efficiency of the floxed yy1 allele in pro-B and pre-B cells or the VH(D)JH recombination efficiency in pro-B cells using primers described previously (Fuxa et al. 2004). PCR products were separated on 2% agarose gel and visualized by ethidium-bromide staining. For primer sequence, see Supplementary Table 2.
RT–PCR analysis
Total RNA from 0.5 × 105 to 1 × 105 sorted pro-B cells were extracted with Trizol (Invitrogen) followed by RNase-free DNase (Promega) digestion for 1 h at 37°C. RNA was then purified with phenol-chloroform-ethanol precipitation and dissolved in DEPC-treated H2O. cDNA was synthesized using the ReverseIt first-strand synthesis kit according to manufacturer’s protocol (ABGENE) using oligo-dT primer, random hexamer, or gene-specific primers. Total cDNA were serially diluted at a 1:5 ratio and ∼1 μL of diluted cDNA was used for each PCR reaction. Most primer sequences for RT–PCR analysis were described previously and are provided in Supplementary Table 2 (DeKoter et al. 2002; Bolland et al. 2004; Fuxa et al. 2004).
Pro-B-cell culture
Total BM suspension was prepared from 2- to 3-wk-old pups and seeded on mitomycin C-treated S17 stromal cells at a density of 2–4 million per well of six-well tissue culture plates with Iscove’s modified Dulbecco’s medium, supplemented with 2% fetal bovine serum (FBS), 0.03% (w/v) Primatone RL (P8388, Sigma), 2.5% conditioned supernatant of rIL-7 (recombinant interleukin 7) secreting J558L cells, 1 mM glutamine, 50 μM β mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptamycin. After 7–10 d, the culture contains >90% B220+CD43+sIgM− pro-B cells, as shown by FACs analysis.
ChIP
ChIP assay was essentially conducted according to the protocol of Upstate Biotechnology with some modifications. About 5 × 106 to 10 × 106 cultured pro-B cells from wild-type animals were used for each immunoprecipitation and rabbit anti-mouse YY1 antibody (H414, Santa Cruz Biotechnology) was used at 1:50 dilution followed by protein G bead precipitation. Control for ChIP was conducted with a twofold amount of normal rabbit IgG. Following reverse linking and proteinase K digestion, DNA was purified with phenol-chloroform extraction and ethanol precipitation. DNA samples were analyzed by real-time PCR using the Roche 480 LightCycler and LightCycler 480 SYBR Green 1 Master (Roche) with primer sets specific for different regions of the mouse IgH gene. Results are presented as fold changes enriched by anti-YY1 antibody relative to normal rabbit IgG control, calcaulated as 2^(Ct CTR − Ct YY1), where Ct is the cycle threshold. The mouse actin-B promoter region serves as negative control, while the mouse RPL30 promoter region serves as positive control. Most primer information was described previously and is available in Supplementary Table 2 (Johnson et al. 2004).
3D DNA FISH and confocal analysis
Three-color 3D DNA FISH was carried out using sorted CD19+cKit+ pro-B cells from control and YY1KO mice as previously described in detail (Kosak et al. 2002; Fuxa et al. 2004). Cells were analyzed by confocal microscopy on a Leica SP2 AOBS (Acoustica Opical Beam Splitter) system. Optical Z sections were collected at 0.3-μm steps through individual nuclei. Only cells containing signals of both IgH alleles were evaluated. The locus-specific DNA probes were prepared from the bacterial artificial chromosomes (BACs) 526A21 (VHJ558), 243G9 (VH15), 167C1 (VH7183), and C34H6 (CH) by nick translation and directly labeled with dUTP Cy5, dUTP Cy3, and dUTP fluorogreen (Amersham Pharmacia). The distance separating the signals of the different IgH gene probes in the nucleus was measured on individual confocal images. A distance of <0.3 μm was defined as colocalized, 0.3–0.5 μm was defined as “apart,” and a distance of 0.5–1.5 μm was referred to as “far apart.” Calculation of P value was performed by applying the χ2 test to observed and expected frequencies (Supplementary Table 1).
Acknowledgments
We thank Dr. Frank Costantini for the Rosa26eYFP Cre reporter mice and Dr. Alexander Tarakhovsky for sharing unpublished information. We thank Drs. Changchun Xiao, Stefano Casola, David Lombard, and Cesar Cobaleda for advice and reagents in establishing the pro-B cell culture; Dr. Marjorie A. Oettinger for her advice; and Dr. Katrina B. Morshead for the primer sequence information to detect the recombination efficiency of VH(D)JH recombination. We also thank Dr. Hui Hu for discussion and for providing RT–PCR primers and antibodies for FACS analysis, Victoria Drier and Sara Linehan for assistance in transferring the mb1-Cre and the B1-8i mice, and Ken Ketman for cell sorting. We thank Dr. Anne Corcoran and Andrew Wood for helpful discussion of the manuscript and bioinformatic analysis of the IgH locus. This work was supported by grants from the NIH (GM53874 to Y.S. and AI057947 to K.R.), a Wellcome Trust project grant (J.S.), and the DFG through SFB 620 (M.R.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1529307
References
- Affar El B., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Gay F., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Shi Y., Liu H., Huarte M., Wu S., Collins T., Li E., Liu H., Huarte M., Wu S., Collins T., Li E., Huarte M., Wu S., Collins T., Li E., Wu S., Collins T., Li E., Collins T., Li E., Li E. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol. Cell. Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F.W., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Yancopoulos G.D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Blackwell T.K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Coffman R., Rosenberg N., Tonegawa S., Baltimore D., Rosenberg N., Tonegawa S., Baltimore D., Tonegawa S., Baltimore D., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L., Ghias A., Wilkinson F., Bonini N., Atchison M.L., Ghias A., Wilkinson F., Bonini N., Atchison M.L., Wilkinson F., Bonini N., Atchison M.L., Bonini N., Atchison M.L., Atchison M.L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E., Reddy K., Medina K.L., Parganas E., Ihle J., Singh H., Reddy K., Medina K.L., Parganas E., Ihle J., Singh H., Medina K.L., Parganas E., Ihle J., Singh H., Parganas E., Ihle J., Singh H., Ihle J., Singh H., Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangeents by transcription factor STAT5. Nat. Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- Bolland D.J., Wood A.L., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E., Wood A.L., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E., Morgan G., Chakalova L., Fraser P.J., Corcoran A.E., Chakalova L., Fraser P.J., Corcoran A.E., Fraser P.J., Corcoran A.E., Corcoran A.E. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Mucci D., Whiteley M., Dirksen M.L., Kassis J.A., Mucci D., Whiteley M., Dirksen M.L., Kassis J.A., Whiteley M., Dirksen M.L., Kassis J.A., Dirksen M.L., Kassis J.A., Kassis J.A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Fritsch C., Mueller J., Kassis J.A., Fritsch C., Mueller J., Kassis J.A., Mueller J., Kassis J.A., Kassis J.A. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional control of early B cell development. Annu. Rev. Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Cancro M.P. Peripheral B-cell maturation: The intersection of selection and homeostasis. Immunol. Rev. 2004;197:89–101. doi: 10.1111/j.0105-2896.2004.0099.x. [DOI] [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V., Di Padova M., Micales B., Lyons G.E., Sartorelli V., Micales B., Lyons G.E., Sartorelli V., Lyons G.E., Sartorelli V., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes & Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Sen R., Sen R. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Sen R., Sen R. Regulation of immunoglobulin heavy-chain gene rearrangements. Immunol. Rev. 2004;200:182–196. doi: 10.1111/j.0105-2896.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- Corcoran A.E., Riddell A., Krooshoop D., Venkitaraman A.R., Riddell A., Krooshoop D., Venkitaraman A.R., Krooshoop D., Venkitaraman A.R., Venkitaraman A.R. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Hasbold J., Dietrich W., Hawkins E., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Hasbold J., Dietrich W., Hawkins E., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Dietrich W., Hawkins E., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Hawkins E., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Kallies A., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Nutt S.L., Tarlinton D.M., Matthias P., Hodgkin P.D., Tarlinton D.M., Matthias P., Hodgkin P.D., Matthias P., Hodgkin P.D., Hodgkin P.D. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J. Exp. Med. 2005;201:1385–1396. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W., Grosveld F., Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Lee H.J., Singh H., Lee H.J., Singh H., Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- Donohoe M.E., Zhang X., McGinnis L., Biggers J., Li E., Shi Y., Zhang X., McGinnis L., Biggers J., Li E., Shi Y., McGinnis L., Biggers J., Li E., Shi Y., Biggers J., Li E., Shi Y., Li E., Shi Y., Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P., Smale S.T., Smale S.T. Combinatorial regulation of transcription II: The immunoglobulin mu heavy chain gene. Immunity. 1995;2:427–438. doi: 10.1016/1074-7613(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Fernex C., Caillol D., Capone M., Krippl B., Ferrier P., Caillol D., Capone M., Krippl B., Ferrier P., Capone M., Krippl B., Ferrier P., Krippl B., Ferrier P., Ferrier P. Sequences affecting the V(D)J recombinational activity of the IgH intronic enhancer in a transgenic substrate. Nucleic Acids Res. 1994;22:792–798. doi: 10.1093/nar/22.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernex C., Capone M., Ferrier P., Capone M., Ferrier P., Ferrier P. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol. Cell. Biol. 1995;15:3217–3226. doi: 10.1128/mcb.15.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming H.E., Paige C.J., Paige C.J. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity. 2001;15:521–531. doi: 10.1016/s1074-7613(01)00216-3. [DOI] [PubMed] [Google Scholar]
- Fleming H.E., Paige C.J., Paige C.J. Cooperation between IL-7 and the pre-B cell receptor: A key to B cell selection. Semin. Immunol. 2002;14:423–430. doi: 10.1016/s1044532302000775. [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M., Souabni A., Salvagiotto G., Roldan E., Busslinger M., Salvagiotto G., Roldan E., Busslinger M., Roldan E., Busslinger M., Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.J., Saleque S., Birshtein B.K., Saleque S., Birshtein B.K., Birshtein B.K. Yin Yang 1 is a lipopolysaccharide-inducible activator of the murine 3′ Igh enhancer, hs3. J. Immunol. 2003;170:5549–5557. doi: 10.4049/jimmunol.170.11.5549. [DOI] [PubMed] [Google Scholar]
- Gronroos E., Terentiev A.A., Punga T., Ericsson J., Terentiev A.A., Punga T., Ericsson J., Punga T., Ericsson J., Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R. B cell ontogeny and B cell subsets. Curr. Opin. Immunol. 1989;2:189–198. doi: 10.1016/0952-7915(89)90187-8. [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K., Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Li Y.S., Allman D., Asano M., Gui M., Hayakawa K., Li Y.S., Allman D., Asano M., Gui M., Hayakawa K., Allman D., Asano M., Gui M., Hayakawa K., Asano M., Gui M., Hayakawa K., Gui M., Hayakawa K., Hayakawa K. B-cell commitment, development and selection. Immunol. Rev. 2000;175:23–32. [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M., Jumaa H., Nielsen P.J., Pelanda R., Reth M., Nielsen P.J., Pelanda R., Reth M., Pelanda R., Reth M., Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Angelin-Duclos C., Park S., Calame K.L., Angelin-Duclos C., Park S., Calame K.L., Park S., Calame K.L., Calame K.L. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Pflugh D.L., Yu D., Hesslein D.G., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Pflugh D.L., Yu D., Hesslein D.G., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Yu D., Hesslein D.G., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Hesslein D.G., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Lin K.I., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Bothwell A.L., Thomas-Tikhonenko A., Schatz D.G., Calame K., Thomas-Tikhonenko A., Schatz D.G., Calame K., Schatz D.G., Calame K., Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat. Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C.M., Wood A.L., Bolland D.J., Corcoran A.E., Wood A.L., Bolland D.J., Corcoran A.E., Bolland D.J., Corcoran A.E., Corcoran A.E. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- Jung D., Alt F.W., Alt F.W. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H., Riblet R., Le Beau M.M., Fisher A.G., Singh H., Le Beau M.M., Fisher A.G., Singh H., Fisher A.G., Singh H., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kwon H.J., Chung H.M., Chung H.M. Yin Yang 1, a vertebrate polycomb group gene, regulates antero–posterior neural patterning. Biochem. Biophys. Res. Commun. 2003;306:1008–1013. doi: 10.1016/s0006-291x(03)01071-4. [DOI] [PubMed] [Google Scholar]
- Liu H., Shi Y., Shi Y. Yin Yang 1. In: Iuchi S., Kuldell N., Kuldell N., editors. Zinc finger proteins: From atomic contact to cellular function. Landes Bioscience/ Eurekah.com, Kluwer Academic/Plenum Publishers; Georgetown, TX: 2005. pp. 182–194. Chapter 25. [Google Scholar]
- Morgan M.J., Woltering J.M., Inderrieden P.M., Durston A.J., Thiery J.P., Woltering J.M., Inderrieden P.M., Durston A.J., Thiery J.P., Inderrieden P.M., Durston A.J., Thiery J.P., Durston A.J., Thiery J.P., Thiery J.P. YY1 regulates the neural crest-associated slug gene in Xenopus laevis. J. Biol. Chem. 2004;279:46826–46834. doi: 10.1074/jbc.M406140200. [DOI] [PubMed] [Google Scholar]
- Morshead K.B., Ciccone D.N., Taverna S.D., Allis C.D., Oettinger M.A., Ciccone D.N., Taverna S.D., Allis C.D., Oettinger M.A., Taverna S.D., Allis C.D., Oettinger M.A., Allis C.D., Oettinger M.A., Oettinger M.A. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K., Atchison M.L., Atchison M.L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain μ E1 site. Proc. Natl. Acad. Sci. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrone L., Henson S.E., Wall R., Malone C.S., Henson S.E., Wall R., Malone C.S., Wall R., Malone C.S., Malone C.S. A conserved sequence upstream of the B29 (Ig β, CD79b) gene interacts with YY1. Mol. Biol. Rep. 2004;31:1–11. doi: 10.1023/b:mole.0000013489.04734.5e. [DOI] [PubMed] [Google Scholar]
- Pawlitzky I., Angeles C.V., Siegel A.M., Stanton M.L., Riblet R., Brodeur P.H., Angeles C.V., Siegel A.M., Stanton M.L., Riblet R., Brodeur P.H., Siegel A.M., Stanton M.L., Riblet R., Brodeur P.H., Stanton M.L., Riblet R., Brodeur P.H., Riblet R., Brodeur P.H., Brodeur P.H. Identification of a candidate regulatory element within the 5′ flanking region of the mouse Igh locus defined by pro-B cell-specific hypersensitivity associated with binding of PU.1, Pax5, and E2A. J. Immunol. 2006;176:6839–6851. doi: 10.4049/jimmunol.176.11.6839. [DOI] [PubMed] [Google Scholar]
- Perlot T., Alt F.W., Bassing C.H., Suh H., Pinaud E., Alt F.W., Bassing C.H., Suh H., Pinaud E., Bassing C.H., Suh H., Pinaud E., Suh H., Pinaud E., Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A., Martinez D., Novatchkova M., Busslinger M., Skok J.A., Novatchkova M., Busslinger M., Skok J.A., Busslinger M., Skok J.A., Skok J.A. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai E., Bottaro A., Davidson L., Sleckman B.P., Alt F.W., Bottaro A., Davidson L., Sleckman B.P., Alt F.W., Davidson L., Sleckman B.P., Alt F.W., Sleckman B.P., Alt F.W., Alt F.W. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl. Acad. Sci. 1999;96:1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn D.P., Hamer K.M., den Blaauwen J., Otte A.P., Hamer K.M., den Blaauwen J., Otte A.P., den Blaauwen J., Otte A.P., Otte A.P. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh C., Jhunjhunwala S., Riblet R., Murre C., Jhunjhunwala S., Riblet R., Murre C., Riblet R., Murre C., Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes & Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet C.S., Brumbaugh R.L., Kee B.L., Brumbaugh R.L., Kee B.L., Kee B.L. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman J.R., Alt F.W., Gorman J.R., Alt F.W., Alt F.W. Accessibility control of antigen-receptor variable-region gene assembly: Role of cis-acting elements. Annu. Rev. Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., Rajewsky K., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., Rajewsky K., Schwers S., Taki S., Jung S., Eilat D., Rajewsky K., Taki S., Jung S., Eilat D., Rajewsky K., Jung S., Eilat D., Rajewsky K., Eilat D., Rajewsky K., Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Flavell R.A., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A., Lalioti M.D., Town T., Lee G.R., Flavell R.A., Town T., Lee G.R., Flavell R.A., Lee G.R., Flavell R.A., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F., William C.M., Tanabe Y., Jessell T.M., Costantini F., Tanabe Y., Jessell T.M., Costantini F., Jessell T.M., Costantini F., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L., Atchison M.L., Atchison M.L. YY1 DNA binding and PcG recruitment requires CtBP. Genes & Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A., Ullrich A., Chait B.T., Tarakhovsky A., Chait B.T., Tarakhovsky A., Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Su K., Li X., Edberg J.C., Wu J., Ferguson P., Kimberly R.P., Li X., Edberg J.C., Wu J., Ferguson P., Kimberly R.P., Edberg J.C., Wu J., Ferguson P., Kimberly R.P., Wu J., Ferguson P., Kimberly R.P., Ferguson P., Kimberly R.P., Kimberly R.P. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcγRIIb alters receptor expression and associates with autoimmunity. II. Differential binding of GATA4 and Yin-Yang1 transcription factors and correlated receptor expression and function. J. Immunol. 2004;172:7192–7199. doi: 10.4049/jimmunol.172.11.7192. [DOI] [PubMed] [Google Scholar]
- Sui G., El Bachir A., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., El Bachir A., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Shi Y., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Brignone C., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Wall N.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Yin P., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Donohoe M., Luke M.P., Calvo D., Grossman S.R., Luke M.P., Calvo D., Grossman S.R., Calvo D., Grossman S.R., Grossman S.R. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Thomas M.J., Seto E., Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W., Palstra R.J., Splinter E., Grosveld F., de Laat W., Splinter E., Grosveld F., de Laat W., Grosveld F., de Laat W., de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Alt F.W., Alt F.W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Espinoza C.R., Yu Z., Stephan R., He T., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Espinoza C.R., Yu Z., Stephan R., He T., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Yu Z., Stephan R., He T., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Stephan R., He T., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., He T., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Williams G.S., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Burrows P.D., Hagman J., Feeney A.J., Cooper M.D., Hagman J., Feeney A.J., Cooper M.D., Feeney A.J., Cooper M.D., Cooper M.D. Transcription factor Pax5 (BSAP) transactivates the RAG-mediated V(H)-to-DJ(H) rearrangement of immunoglobulin genes. Nat. Immunol. 2006;7:616–624. doi: 10.1038/ni1339. [DOI] [PubMed] [Google Scholar]