Abstract
Imprinted genes are expressed in a parent-of-origin manner and are located in clusters throughout the genome. Aberrations in the expression of imprinted genes on human Chromosome 7 have been suggested to play a role in the etiologies of Russell-Silver Syndrome and autism. We describe the imprinting of KLF14, an intronless member of the Krüppel-like family of transcription factors located at Chromosome 7q32. We show that it has monoallelic maternal expression in all embryonic and extra-embryonic tissues studied, in both human and mouse. We examine epigenetic modifications in the KLF14 CpG island in both species and find this region to be hypomethylated. In addition, we perform chromatin immunoprecipitation and find that the murine Klf14 CpG island lacks allele-specific histone modifications. Despite the absence of these defining features, our analysis of Klf14 in offspring from DNA methyltransferase 3a conditional knockout mice reveals that the gene's expression is dependent upon a maternally methylated region. Due to the intronless nature of Klf14 and its homology to Klf16, we suggest that the gene is an ancient retrotransposed copy of Klf16. By sequence analysis of numerous species, we place the timing of this event after the divergence of Marsupialia, yet prior to the divergence of the Xenarthra superclade. We identify a large number of sequence variants in KLF14 and, using several measures of diversity, we determine that there is greater variability in the human lineage with a significantly increased number of nonsynonymous changes, suggesting human-specific accelerated evolution. Thus, KLF14 may be the first example of an imprinted transcript undergoing accelerated evolution in the human lineage.
Author Summary
Imprinted genes are expressed in a parent-of-origin manner, where one of the two inherited copies of the imprinted gene is silenced. Aberrations in the expression of these genes, which generally regulate growth, are associated with various developmental disorders, emphasizing the importance of their discovery and analysis. In this study, we identify a novel imprinted gene, named KLF14, on human Chromosome 7. It is predicted to bind DNA and regulate transcription and was shown to be expressed from the maternally inherited chromosome in all human and mouse tissues examined. Surprisingly, we did not identify molecular signatures generally associated with imprinted regions, such as DNA methylation. Additionally, the identification of numerous DNA sequence variants led to an in-depth analysis of the gene's evolution. It was determined that there is greater variability in KLF14 in the human lineage, when compared to other primates, with a significantly increased number of polymorphisms encoding for changes at the protein level, suggesting human-specific accelerated evolution. As the first example of an imprinted transcript undergoing accelerated evolution in the human lineage, we propose that the accumulation of polymorphisms in KLF14 may be aided by the silencing of the inactive allele, allowing for stronger selection.
Introduction
Genomic imprinting is an epigenetic phenomenon characterized by the expression of alleles in a parent-of-origin manner, giving rise to monoallelic or heavily biased gene expression. Imprinted genes are generally located in clusters that often contain maternally and paternally expressed protein-coding genes, as well as imprinted noncoding transcripts [1]. Aberrations in the expression of imprinted genes have been associated with various developmental and behavioral disorders, such as Prader-Willi syndrome and Beckwith-Wiedemann syndrome.
Imprinted genes on human Chromosome 7 have been suggested to underlie several disorders that show parent-of-origin effects, including Russell-Silver Syndrome (RSS). This genetically heterogeneous disorder, which is characterized by intrauterine and postnatal growth restriction as well as dysmorphic facial features, has been associated with numerous chromosomal rearrangements and anomalies. Most recently, hypomethylation of the imprinting control region (ICR) at Chromosome 11p15 has been associated with RSS [2]. However, various reports have suggested a possible role for human Chromosome 7 in the etiology of this disorder, based on evidence indicating that 10% of affected individuals have maternal uniparental disomy for this chromosome [3,4]. A causative gene for the Chromosome-7 form of RSS has not been found, but absence of a paternally inherited FOXP2 gene might explain the verbal dyspraxia phenotype usually observed in this subtype [5]. A recent study has also shown evidence for the existence of a parent-of-origin effect in autism linked to Chromosome 7 [6]. As a consequence, studies that have attempted to identify imprinted genes associated with these disorders have concentrated on Chromosome 7 [7,8].
To date, three distinct imprinted loci have been identified on human Chromosome 7. The first, located at 7p12.2, contains the growth factor receptor-bound protein 10 gene (GRB10) [9]. The second locus, at 7q21.3, includes the retrotransposon derived PEG10 [10] and ɛ-sarcoglycan (SGCE) [11]. The third cluster, located at 7q32.3, includes the genes encoding carboxypeptidase-A4 (CPA4) [12] and mesoderm specific transcript homolog (MEST) [13] (Figure 1). Hannula and colleagues described a patient with RSS with partial maternal uniparental disomy for 7q31-qter, highlighting the genes in this third region as candidates for the syndrome [5,14]. However, analyses of imprinted transcripts in this interval have only excluded them as candidates for RSS [15,16]. Several studies have attempted to identify additional imprinted transcripts in the region, but have not found parent-of-origin specific expression in tissues analyzed [17,18].
Figure 1. Human and Murine KLF14 Structure and Expression.
(A) Human KLF14 and (B) murine Klf14 structure are shown. Genes in 7q32.3 region in the upper panel with maternally and paternally expressed genes are depicted in grey and black, respectively. Striped patterns represent genes with tissue specific imprinting. Arrows indicate transcriptional direction. Lower panels show the gene structure of KLF14/Klf14, including results from RACE, the ORF, CpG island, and primers used in various analyses (represented by thin black bars). The grey block representing AK030435 denotes the fact that evidence of splicing was not identified in our experiments. (C) Expression of Klf14 in murine embryonic and extra-embryonic and (D) brain tissues is shown. Samples lacking reverse transcriptase are indicated by -. Results indicate higher levels of expression in extra-embryonic tissues. (E) Expression of KLF14 in human tissues. Human expression results concur with those of mouse expression, in that there is higher expression in prenatal stages of development.
In this paper, we describe the identification of a novel maternally expressed imprinted transcript located at 7q32.3, telomeric to TSGA13. This gene, named KLF14 (Krüppel-like factor 14), is an intronless molecule and a member of the Sp/KLF family of transcription factors. These proteins are characterized by three highly conserved C2H2-type zinc-fingers at the carboxy-terminal end joined to each other by linker sequences, known as Krüppel-links [19]. In contrast, the N terminus is highly variable between KLF paralogues and has lower levels of conservation between orthologues [20]. Members of the KLF family are known to act as transcriptional activators, repressors, or both [21].
We show that KLF14 has monoallelic maternal expression in a variety of embryonic and extra-embryonic tissues in human and mouse. In addition, we determine that KLF14 has undergone accelerated evolution in the human lineage with numerous amino acid substitutions identified in different populations and demonstrate that this variability is increased in the human lineage.
Results
Imprinting Analyses
Maternal specific expression of human and murine KLF14 in embryonic and extra-embryonic murine tissues.
To determine the allelic expression of murine Klf14, reciprocal crosses of C57BL/6J (BL6) and JF1/Ms (JF1) were carried out and cDNA was extracted from embryonic and extra-embryonic tissues at 15.5 days post coitum (dpc). To distinguish the two parental alleles, a G/A polymorphism corresponding to nucleotide 451 of AK030435 was identified. Equal peak heights of the two alleles were observed in sequencing electropherograms from PCR-amplified genomic DNA of BL6 × JF1 F 1 hybrids, indicating lack of amplification bias (Figure 2A). cDNA from the F 1 hybrids was amplified, the products were sequenced, and allelic expression was analyzed in an intronless PCR fragment amplified by primers AK030435F/AK030435R. Due to the intronless nature of the amplicon, samples without reverse transcriptase were also prepared to account for the possibility of genomic DNA contamination. By use of reciprocal crosses of hybrid mice, monoallelic expression of the maternal allele was identified in all tissues examined, as noted by the expression of a single peak at the position corresponding to the G/A polymorphism (Figure 2A). In addition, Klf14 was found to be imprinted in tissues extracted from 9.5 dpc embryos and neonates (unpublished data), indicating that the imprinted expression of Klf14 is not developmental-stage specific and is an imprint established early in development.
Figure 2. Imprinting Analysis of Human and Murine KLF14 .
(A) Imprinted expression of murine Klf14 is shown. Sequence analysis of genomic DNA and RT-PCR products from 15.5-dpc hybrid mice are shown in the left and right panels, respectively. Genomic sequencing results indicate the genotype for JF1 (G) at the polymorphism. RT-PCR sequencing results show the expression of the JF1 allele in all tissues where JF1 is the maternal allele (upper row in right panel) and expression of the BL6 allele in the reciprocal cross (lower row in right panel), indicating maternal expression.
(B) Imprinted expression of human KLF14. The first column of panels shows genomic sequencing electropherograms for three fetal samples (rows) heterozygous for a polymorphism in KLF14. The second column presents the genotype for the corresponding maternal samples (maternal DNA was not available for fetus number 62). The third column shows sequencing results of RT-PCR products indicating the monoallelic expression of various tissues, as indicated on the right of the column. Results from fetus number 66, which is informative for parental origin, indicate that KLF14 is maternally expressed. *, sequencing of tongue, stomach, eye, kidney, and intestine cDNA from fetus number 62 showed monoallelic expression.
(C) Maternal expression of human KLF14 in somatic cell hybrids. RT-PCR was performed for three independent maternal or paternal monochromosomal hybrid cell lines for human Chromosome 7. Results confirm the maternal expression of KLF14, as seen in (B). The expression of the paternally expressed MEST and mouse A9 cell line, which lacks human Chromosome 7, are also shown.
To distinguish parental alleles of human KLF14, DNA derived from fetal samples was genotyped for a C/T polymorphism at nucleotide 336 of NM_138693, and three fetuses heterozygous for the polymorphism were identified (Figure 2B). cDNA corresponding to KLF14 was sequenced, and the expression of the alleles was noted. Monoallelic expression of KLF14 was observed in lung (fetus number 66), heart (fetus number 65), tongue, stomach, eye, intestine, and placental samples (fetus number 62). One informative fetus-mother DNA pair indicated monoallelic expression of the maternal allele (fetus number 66).
We examined the expression of KLF14 in cDNA extracted from somatic cell hybrid lines containing a maternal or paternal human Chromosome 7. These lines have been shown to maintain the monoallelic expression of MEST-isoform-1 and MESTIT1 [22]. PCR was carried out, amplifying a fragment specific to KLF14. The absence of a PCR product in a cell line not containing the human chromosome (A9) indicated that the amplification was specific to a human Chromosome-7 transcript. Amplification of KLF14 was observed exclusively in cell lines containing a maternal human Chromosome 7 (Figure 2C), indicating maternal specific expression.
Histone modifications in Klf14 and Mest CpG islands.
The allele-specific modification of histones has been shown to be a hallmark of promoters and ICRs of imprinted loci [23,24]. Though such modifications are a feature of preferential expression when found in the promoter, they are integral at the level of ICRs. The unmethylated allele of ICRs is associated with “open chromatin” marks, such as the acetylation of histones and dimethylation of lysine 4 on histone 3 (H3K4me2). In contrast, the methylated allele of ICRs is characterized by heterochromatic features, such as hypoacetylation of lysine 9 on histone 3 (H3K9ac), trimethylation of lysine 9 on histone 3 (H3K9me3), and trimethylation of lysine 20 on histone 4 (H4K20me3) [25,26]. Thus, by performing chromatin immunoprecipitation (ChIP), we investigated allele-specific covalent histone modifications that have been shown to be characteristic of maternally and paternally methylated ICRs [23,24,27,28] (P. Arnaud and R. Feil, personal communication).
We performed ChIP by formaldehyde fixation using murine fibroblasts from the F 1 hybrid offspring of BL6 × JF1. We previously determined that Klf14 is imprinted in these cells (Figure 2A), and therefore epigenetic modifications associated with the gene would be present. To distinguish each allele, we identified single nucleotide polymorphisms (SNPs) in the CpG islands of murine Mest and Klf14, which were also restriction fragment length polymorphisms of MaeI and SacII (Figure 3A). Although histone 3 acetylation (at lysines 9 and 14), histone 4 acetylation (at lysines 5, 8, 12, and 16), and H3K4me2 were exclusively enriched in nucleosomes of the unmethylated paternal allele of Mest, no differences were observed between the parental alleles at Klf14 (Figure 3B).
Figure 3. Epigenetic Modifications of Murine Klf14 .
(A) The location and distribution of regions analyzed in Mest and Klf14. The CpG islands overlapping Mest exon 1 and Klf14 are depicted by grey bars (row 1). The regions examined in the methylation analysis and ChIP assay are indicated in rows 2 and 3, respectively. The restriction enzymes used in the ChIP assay and the polymorphisms identified in BL6 and JF1 strains are also shown.
(B) Analysis of histone modifications by ChIP in fibroblast cells of BL6 × JF1 and JF1 × BL6 hybrids. ChIP was performed using formaldehyde fixed chromatin. Antibodies against histone 3 acetylated at lysines 9 and 14 (H3K9acK14ac), histone 4 acetylated at lysines 5, 8, 12, and 16 (H4ac), and H3K4me2 were used in the ChIP assay. Precipitated DNAs were PCR amplified using primers specific to the CpG islands of Mest and Klf14 and subsequently digested as shown in (A). DNA before immunoprecipitation (input) and the product obtained with no antibody (N.C.) were also included in the analysis. The difference in band intensities between the precipitated products and input DNA reveals that there is preferential precipitation of H3K9acK14ac, H4ac, and H3K4me2 on the paternal allele of the Mest CpG island, but no allelic differences were detected at the Klf14 region.
(C) Analysis of histone modifications by native-ChIP in whole embryos of BL6 × JF1 hybrids. Chromatin was immunoprecipitated using antibodies against H3K9ac, H3K4me2, H3K9me3, and H4K20me3. Anti-chicken was used as a nonspecific antibody (mock). Input DNA is denoted by I. Antibody-bound and unbound fractions of the precipitate are denoted by B and U, respectively. Precipitated DNA was PCR amplified using the same primers as in (B). The amplified DNA was analyzed by single strand conformation polymorphism. The results show differences in histone modifications between the two parental alleles in the Mest CpG island, but allelic enrichments were not observed for Klf14.
(D) Expression of Klf14 in offspring of Dnmt3a conditional knockout mice. The expression of Mest and Klf14 was examined in two embryos (e1–2) and corresponding extra-embryonic tissues (e1-2ex) from the offspring of female Dnmt3a conditional knockout mice, as well as a wild-type (wt) embryo. Klf14 expression is lost in the knockout mice, suggesting that its expression is dependent upon a maternally methylated region.
(E) Model of Klf14 expression in wild-type (wt) and Dnmt3a conditional knockout mice. In wild-type mice (upper panel), the maternally methylated CpG island in Mest (black circle) silences the expression of the gene from the maternal allele (M), while Klf14 is actively transcribed on this allele. The opposite pattern of expression is seen on the paternal strand (P), because of the unmethylated CpG island (white circle). In Dnmt3a −/+ embryos (lower panel), maternal methylation of the Mest CpG island is lost, causing increased expression of Mest and loss of expression of Klf14.
(F) Bisulfite sequencing results from 12.5-dpc whole embryos of BL6 × JF1 hybrids. Each block corresponds to a separate region analyzed in the Klf14 CpG island, as shown in (A) (Mest methylation analysis is not shown). Hollow circles and black circles indicate unmethylated and methylated CpG dinucleotides, respectively (N, could not be determined). Each row of circles represents CpGs in an individual PCR product clone. In each block, the top section and bottom sections correspond to clones from the maternal allele (BL6) and paternal (JF1) alleles, respectively, as determined by use of polymorphisms. The three regions analyzed indicate that the Klf14 CpG island is hypomethylated on both alleles.
We subsequently performed native ChIP, which precipitates a higher quantity of chromatin in comparison to formaldehyde fixed chromatin [29], to determine if a more sensitive assay would identify differences in histone modifications in the CpG island of Klf14. For this analysis we used DNA from the 13.5-dpc whole embryos of hybrid BL6 × JF1 mice. Precipitated DNA was PCR amplified and analyzed by the single strand conformation polymorphism method (Figure 3C). Again, we determined that H3K9ac and H3K4me2 were associated with the unmethylated paternal allele in the Mest promoter, and that H3K9me3 and H4K20me3 were associated with the methylated maternal allele. However, we did not observe differences between the alleles of Klf14.
Methylation analysis and Klf14 expression in DNA methyltransferase 3a knockout mice.
The establishment of a differentially methylated region (DMR) in the germ-line is the primary hallmark of imprinted loci and has been shown to be necessary for the proper imprinting of many regions (for review, see [30]). The study of these regions has been greatly enhanced by DNA methyltransferase 3a (Dnmt3a) conditional knockout mice. These mice have Dnmt3a disrupted specifically in germ cells, while somatic cells express the wild-type protein, allowing conditional knockouts to be viable and enabling the study of their offspring. The progeny of female conditional knockouts die in utero at approximately 10.5 dpc and have been shown to be hypomethylated at maternally methylated DMRs, while methylation patterns at paternally methylated regions and repetitive regions remain unaltered [31]. Correspondingly, a 1.6-fold increase in the expression of Mest has been measured in these Dnmt3a− /wt embryos indicating a relaxation of the imprinted expression of the gene. We examined the expression of Mest and Klf14 in the offspring of female Dnmt3a conditional knockout mice. Our results indicate a substantial decrease in Klf14 expression, in both embryonic and extra-embryonic tissues in Dnmt3a− /wt embryos (Figure 3D). This suggests that Klf14 expression is dependent upon the establishment of maternal imprints in oocytes (Figure 3E).
We examined the methylation status of the CpG island located in Klf14. Using three different PCR primers spanning the CpG island (Figure 3A), we subcloned PCR-amplified bisulfite-treated DNA extracted from 12.5-dpc BL6 × JF1 hybrids. We observed hypomethylation of both alleles throughout the CpG island (Figure 3F). Similar results were obtained from the bisulfite sequencing of fibroblast DNA (unpublished data). Using the same materials, we examined the methylation of Mest and observed enrichment of methylated CpGs on the maternal allele, as previously described [32] (unpublished data).
The methylation status of 94 CpG dinucleotides located in the open reading frame (ORF) and 5′ UTR regions of human KLF14 were examined by bisulfite sequencing of fibroblast DNA. Extensive hypomethylation was again observed in the subcloned fragments (unpublished data).
Klf14 Structure and Function
Characterization of the human and murine KLF14 transcripts.
An in silico analysis of human KLF14 was performed to identify the full-length transcript of the gene by EST assembly. A single intronless transcript of approximately 1.4 Kb was found in the Chromosome 7 annotation database (http://www.chr7.org) [33]. This reference sequence, derived from mRNA AF490374, contains an ORF of 972 nucleotides, as well as an in-frame stop codon. Rapid amplification of cDNA ends (RACE) was performed, and single band of approximately 1.6 Kb was amplified and directly sequenced (Figure 1A). The unspliced fragment contained a poly-A tail and poly-A signal. We performed 3′ RACE again using primers located closer to the new 3′ end, and a second fragment, also containing a poly-A tail, was identified.
In silico analysis was performed to identify the full length of murine Klf14. The spliced transcript AK030435, whose putative second exon was originally used to determine the imprinted expression of the gene, contains a poly-A signal. Reverse transcriptase-PCR (RT-PCR) was performed to confirm the expression of the spliced transcript, yet such attempts only succeeded in amplifying cDNA intronic to AK030435 and did not find evidence of splicing. An ORF of 978 nucleotides was identified, partially located in the intron of AK030435, corresponding to Klf14 (Figure 1B). We performed 3′ RACE, and the results did not extend the transcript beyond the 3′ end of AK030435, though they identified a poly-A tail.
Expression of human and murine KLF14.
To determine the expression of KLF14 in human tissues, RT-PCR was carried out using numerous adult and fetal RNA samples. In general, the transcript was found to have low levels of expression in both human and mouse. The transcript was found to be expressed in many tissues (Figure 1E), but its expression was absent in liver and lymphoblast (unpublished data). It was found to have higher levels of expression in fetal tissues and placenta than in adult tissues.
RT-PCR was performed on murine cDNA, where higher levels of Klf14 expression were observed in embryonic and extra-embryonic tissues with respect to adult tissues (Figure 1C). We cultured glia and neurons from mouse embryos, as previously described [34], and observed much higher level of expression in the latter (Figure 1D).
Syntenic analysis of KLF14.
The intronless nature of KLF14 suggested that the gene may have arisen through retrotransposition [35]. Phylogenetic studies of the KLF family have revealed that the KLF14 protein is most closely related to KLF16, encoded on human Chromosome 19 [36]. An amino acid alignment of these two proteins (BLASTP) demonstrated that they are 58% identical and bear most similarity in the first 26 amino acids (N terminus) and the zinc-finger domains (C-terminal end). Thus, it is plausible that this gene is an ancient retrotransposon-derived duplication of KLF16.
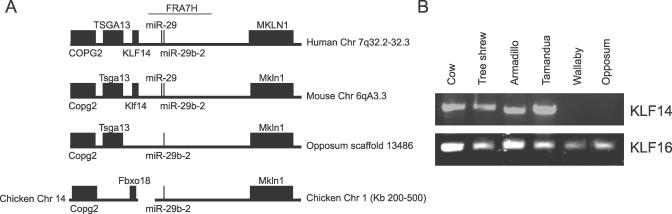
To determine the timing of the retrotransposition event during vertebrae evolution, we examined the synteny of the region encompassing Klf14 in human, mouse, opossum, and chicken using the genome browser at University of California, Santa Cruz (http://genome.ucsc.edu). We used COPG2/TSGA13 and MKLN1 as reference points, which are located centromeric and telomeric to human KLF14, respectively. Two highly conserved segments corresponding to microRNAs (miR-29 and miR-29b-2) were also used as anchors. Synteny with all these elements, including Klf14, was maintained in mouse (Figure 4A). However, Klf14 and miR-29 were absent in the opossum. A break in synteny was observed in chicken, where Copg2 and Mkln1 were located on different chromosome. This analysis suggested that Klf14 was retrotransposed after the divergence of marsupials from eutherian mammals.
Figure 4. Retrotransposition of KLF14 and Mammalian Evolution.
(A) Genomic distribution of KLF14 and flanking genes is shown. KLF14 is flanked by MKLN1 and TSGA13 in human and mouse. These two genes are present in opossum, yet KLF14 is absent. In chicken, there is a syntenic break in the region, placing genes on different chromosomes. Two microRNAs (miR-29 and miR-29b-2), which lie in the fragile site adjacent to KLF14 (FRA7H), are conserved.
(B) Presence of KLF14 and KLF16 in distant mammals is shown. The lower panel shows PCR amplification of KLF16 from mammals of diverse clades, and the upper panel shows the amplification of KLF14. The mammals shown are (L-R) cow (Bos taurus), tree shrew (Tupaia glis), nine-banded armadillo (Dasypus novemcinctus), tamandua (Tamandua tetradactyla), red-necked wallaby (M. rufogriseus), and red-legged short-tailed opossum (M. brevicaudata). It indicates that KLF14 is present in eutherian, but not marsupial mammals, and shows that KLF16 is more ancient than KLF14.
A more precise timing of this retrotransposition event in eutherian evolution was examined by amplifying Klf14 in organisms from each of the supraordinal clades: Afrotheria, Xenarthra, Euarchontoglires, and Laurasiatheria [37]. Using a variety of different PCR conditions, Klf14 was amplified from numerous species and its sequence was confirmed by direct sequencing of products. Several primers were designed, located in the conserved zinc-finger domains of the gene. Multiple attempts using increasingly more permissive conditions were used to amplify DNA from red-legged short-tailed opossum (Monodelphis brevicaudata), red-necked wallaby (Macropus rufogriseus), and echidna (Tachyglossus aculeatus), yet bands corresponding to a Klf14 homologue were not obtained. However, Klf16 was amplified in both eutherian and marsupial mammals (Figure 4B).
The syntenic analysis together with the amplification of Klf14 in each of the superclades, most notably in members of the Xenarthra order (represented by the armadillo and tamandua), suggest that the gene is present in all eutherian mammals, yet absent in monotremes and marsupials. Based on estimates of mammalian evolution, this would place the retrotransposition event, which gave rise to KLF14, between 130 and 170 million years ago (i.e., prior to the divergence of Xenarthra and after the divergence of Marsupialia) [38]. At the same time, the presence of Klf16 in marsupials indicates that it is more ancient than KLF14 and supports our hypothesis that the latter gene may be a retrotransposed copy of KLF16.
KLF14 Variation and Selection
Sequence of KLF14 in RSS and autistic individuals.
As previously mentioned, imprinted genes on Chromosome 7 are hypothesized to play a role in the etiologies of RSS and autism. The KLF14 ORF was sequenced in 60 RSS patients and 160 autistic individuals to test for mutations. Although, we did not identify mutations specific to these affected groups, we identified numerous nonsynonymous base-pair substitutions in the N-terminal region (Figure S1). All polymorphic changes were found at equal frequencies in controls, suggesting that they are not disease-associated.
The KLF14 ORF was sequenced in a total of 704 chromosomes, representing both patients and controls, the latter of which included an ethnically diverse panel of 61 individuals. We found eight haplotypes (Figure S1): two were specific to the Japanese population (haplotypes 7 and 8), and one was found predominantly in individuals of African descent (haplotype 6) (Figure 5A). The frequency of these haplotypes varied greatly in different populations, suggesting that the gene may have recently undergone relaxed selection or accelerated evolution.
Figure 5. Haplotype Frequencies of KLF14 ORF in the Human Population.
(A) The frequency of eight KLF14 haplotypes in various ethnic populations is shown. The frequency of each haplotype, as defined in Figure S1, identified in ethnic populations is shown (n = number of chromosomes genotyped).
(B) KLF14 primate species and human haplotype tree is represented. This tree was created manually by parsimony, with the inferred number of changes shown on each branch (thick lines represent nonsynonymous changes, while thin lines represent synonymous changes). The tree is rooted using two macaque sequences (M. mulatta and M. nemestrina). In orang-utan, gorilla, and chimpanzee, only a single haplotype is represented (where polymorphisms were present, the ancestral allele was used; when descendent alleles were used for polymorphisms, the results did not differ significantly). MRCAs, manually inferred by parsimony, are represented by shaded circles at four nodes in the tree. dN/dS values were calculated for each lineage, including the fixed human sequence (HH-MRCA) and each of the human haplotypes. For the human haplotypes, the dN/dS value is calculated to the human-chimpanzee MRCA, and not to the HH-MRCA. Two methods were utilized: maximum likelihood pairwise comparison (top) and Nei and Gojobori comparison (bottom). Where dS = 0, these methods give values −1.0000.
Lineage-specific accelerated evolution of KLF14.
To analyze evolutionary features of KLF14, we sequenced its ORF in three gorillas, two orang-utans, two macaques, two bonobos, and 20 chimpanzees (for genotypes, see Tables S4 and S5). The ratio of nonsynonymous to synonymous substitutions (dN/dS) was calculated in phylogenetic analysis by maximum likelihood (PAML) software [39] using a two-ratio maximum-likelihood analysis between the human haplotype's most recent common ancestor (HH-MRCA) and the human-chimpanzee MRCA (Figure 5B). This measurement, which only evaluates fixed changes, is used to determine selective pressures in a coding region, where values >1 suggest positive selection, while values <1 indicate purifying selection. This dN/dS value was found to be −1.0000 (or infinity), because no synonymous changes occurred in this lineage.
We also calculated dN/dS values for each of the eight human haplotypes from the human-chimpanzee MRCA (Figure 5B). In each of these haplotypes, the dN/dS value was >1, with the two most common haplotypes (1 and 2) having values of 2.25 and −1.0000 (or infinity), respectively. Such values suggest that positive selection has occurred in the human lineage. Consequently, we used the likelihood ratio test within PAML [39] for detecting positive selection and found that the dN/dS from the HH-MRCA was not significantly >1 (p = 0.1345) (Table S1). Therefore, we could not reject the hypothesis of neutral evolution in the human lineage. This conclusion was confirmed by analyzing the dN/dS values for each of the human haplotypes. Only one haplotype (7) had a dN/dS value significantly >1 (p < 0.05) before Bonferroni multiple test correction. However, this is a rare haplotype, and after Bonferroni correction its significance is lost. The common haplotypes (1 and 2) did not have dN/dS values significantly >1. Similar results were obtained when all human haplotypes were simultaneously used to identify significant elevation in the human dN/dS (Text S1).
By sequencing regions flanking the ORF of KLF14, we attempted to find evidence of positive selection in the form of a recent selective sweep. We sequenced 9,880 bps, including the ORF, in 18 individuals from our diversity panel. A chimpanzee was also sequenced to identify the ancestral allele for each variant. In total, 42 SNPs and two polymorphic insertion/deletions were identified, 13 of which were singletons, and 29 of which were parsimony-informative sites. Using DnaSP 4.0 [40], we performed various analyses including Tajima's D-test [41], Fu and Li's D-test [42], Fu and Li's F* [42], Fu's Fs statistic [43], and Fay and Wu's H test statistic [44]. These tests are designed to detect recent selective sweeps by looking for skews in frequency spectrums or recent mutations relative to ancient mutations. Departures from neutrality detected by these tests could indicate that positive selection has occurred. However, none of these analyses gave a significant result.
The McDonald-Kreitman test [45], which examines ratios of synonymous and nonsynonymous nucleotide substitutions within and between species, was performed using the human ORF sequences compared to those from the 20 chimpanzee and two bonobo samples. Again, this test did not give a significant result. These findings, together with the results from the previous analyses, indicate that we cannot reject the null hypothesis of neutrality in KLF14. Although there is evidence suggesting that the gene may be undergoing positive selection, such as the presence of three fixed nonsynonymous changes (bps 194, 545, and 559) and two nonsynonymous SNPs (bps 140 and 172) for which the derived allele is more frequent than the ancestral allele, our results may also be explained by accelerated neutral evolution.
To establish whether accelerated evolution has ocurred, dN/dS values were calculated for the sequenced primates with respect to their MRCAs. In each case, the dN/dS values were <1, indicating purifying selection (Figure 5B). Subsequently, we examined whether the dN/dS values observed in the human lineage were significantly elevated relative to the primate background dN/dS ratio by using the likelihood ratio test [39]. For this analysis, the dN/dS value for HH-MRCA showed significance (p = 0.0139). Additionally, all haplotypes with the exception of haplotype 3 had significantly increased dN/dS values (p < 0.05) before Bonferroni correction. After Bonferroni correction, only haplotypes 2 and 7 remained significant (see Table S1). We confirmed the significantly higher dN/dS values in the human lineage relative to the primate background using the haplotype tree (see Text S1). Significant results were obtained for the human lineage using both the two- and three-ratio tests in PAML. These results suggest accelerated evolution in the human lineage.
We used a binomial distribution [46] to determine if there is an increased rate of amino acid substitutions in the human lineage relative to the ape lineages (orang-utan, gorilla, and chimpanzee). The results indicated that seven of the eight haplotypes were significant before and after Bonferroni correction (p < 0.05), with the exception of haplotype 3 (p = 0.0540) (Table S2). Yet when this test was applied to the HH-MRCA, a significant result was not obtained (p = 0.0540). However, the significance observed in the seven haplotypes suggests that, although there is no significant evidence for positive selection, there is human lineage-specific accelerated protein evolution of KLF14.
Sequence variability in KLF14.
To determine if the sequence variability observed in KLF14 was significantly greater than average, we used information available from the Seattle SNP (http://pga.gs.washington.edu) and Environmental Genome (http://www.niehs.nih.gov/envgenom/home.htm) projects to extract polymorphism data from 826 genes [47,48]. These genes were sequenced in >96 haploid genomes of ethnically diverse individuals, allowing for the identification of 99.9% of SNPs with minor allele frequencies ≥5% [49]. Using the allele frequencies of identified polymorphisms, we determined π values for each of the sequenced genes and for KLF14. π is the measure of nucleotide diversity or the average number of pairwise nucleotide differences between aligned sequences divided by the sequence length [50]. We found that KLF14 has a higher π value than 798 of the 827 total genes (96.5%). We then calculated π values at nonsynonymous and synonymous SNP sites separately and determined that KLF14 had a higher π value than 779 of the 827 genes (94.2%) within each of these sets, revealing that the gene's significant enrichment in SNPs is due to elevated numbers of both synonymous and nonsynonymous polymorphisms.
To determine if the degree of variability observed is unique to human KLF14 or is common in other primates, polymorphisms in the ORF were identified in 20 chimpanzees. Only three SNPs were observed, all of which were synonymous singleton changes. We found two of these variations in a single chimpanzee of the subspecies Pan troglodytes schweinfurthii. In addition, sequence comparison between two bonobos (P. paniscus) and the chimpanzees did not identify any polymorphisms. To look for increased diversity in the human lineage, we calculated diversity statistics (π and θ) for both synonymous and nonsynonymous sites using non-patient human samples (264 chromosomes) and chimpanzees (40 chromosomes) (Table S3). θ is a measure of genetic diversity within populations that describes the amount of variation expected at each nucleotide site [50]. In general, π and θ values were higher in the human gene than in the chimpanzee's, suggesting greater diversity. This increased diversity appears to be mainly due to a higher number of nonsynonymous SNPs, since the chimpanzee's synonymous θ value is higher than that of human's. These results lend support to an increased level of divergence, especially at nonsynonymous sites, occurring in the human lineage. However, such a disparity between these species is seen often, due to the recent population explosion in humans.
From these results we conclude that KLF14 is a highly variable gene relative to other genes in the genome, and its variability is not common to chimpanzees. This high level of variability lends support to the finding that KLF14 has undergone, and may be continuing to undergo, accelerated evolution in the human lineage.
Discussion
Imprinting of KLF14
We have identified a novel imprinted gene, KLF14, and show that it is maternally expressed in every tissue examined in both human and mouse. The gene encodes a putative Krüppel-like transcription factor, containing three C2H2 zinc-fingers joined together by a characteristic linker sequence [19]. The function of KLF14 is currently unknown. Previous studies have shown that it is similar to KLF16 [36], and an alignment of the two proteins identified a conserved α-helical repression motif at the N terminus, which has been shown to act as a transcriptional repressor by directly interacting with mSin3A-histone deacetylase [51,52], suggesting that KLF14 may also share this regulatory activity.
The relatively greater expression level of KLF14 in embryonic and extra-embryonic tissues compared to adult tissues suggests a role for the transcript in development. Indeed, many imprinted genes regulate growth and embryonic development (for a review, see [30]). This observation has led to the proposal of a conflict hypothesis, where maternally expressed genes suppress fetal growth, allowing nutrient supply to be available for future pregnancies and increasing the mother's survival rate. In contrast, paternally expressed transcripts enhance fetal growth to ensure survival of their genetic offspring [53]. Under this hypothesis, the maternally expressed KLF14 is predicted to suppress embryonic growth. This, together with its predicted function as a transcriptional repressor, suggests that the protein may suppress the expression of genes that enhance fetal or placental development.
Our analysis of the Klf14 CpG island indicates that the region is hypomethylated. The identification of an unmethylated CpG-rich region in an imprinted cluster has been previously described, such as the promoter region of Dlk1 [54], the promoter and first exon of Gsα [55], and Ascl2 [56]. Our ChIP results, performed using antibodies specific to various histone modifications, did not identify differences between the two alleles of Klf14. However, clear allele-specific precipitation of histone modifications was observed at the DMR of Mest. This is the first description of allele-specific histone modifications associated with the Mest germ-line DMR. Despite the lack of these trademark epigenetic features at the Klf14 CpG island, its imprinting is maintained, and its expression is dependent on the function of Dnmt3a in female germ cells. To date, the only DMR identified in this region is the maternally methylated CpG island at the 5′ end of MEST/Mest, which has been shown to be established in gametes [32]. As such, we propose that this DMR may regulate the imprinted expression of KLF14, possibly through long-range chromatin regulatory interactions and may act as an ICR for the entire locus, spanning carboxypeptidase-A4 (CPA4) to KLF14 (Figure 3E). Further studies are necessary to determine if the expression of these genes is regulated by long-range insulator or enhancer elements, as has been shown for genes in the H19/Igf2 locus and KCNQ1 region [57].
Evolution of KLF14
Our analysis suggests that KLF14 arose through the retrotransposition of KLF16. Thus, KLF14 is the ninth imprinted retrotransposed gene identified to date and the first protein-coding maternally expressed retrotransposed gene identified in mouse, adding further support to the hypothesis that imprinting serves as a mechanism for regulating increased gene dosage [1]. We postulate that KLF14 acquired imprinting through cis- and trans-acting elements associated with the more ancient MEST, which is known to be imprinted in marsupials [58]. This would allow the host eutherian mammal to adapt to the increased gene dosage caused by the retrotransposition of KLF16. By analyzing the expression and epigenetic modifications of KLF14 and other retrotransposed genes in eutherian mammals, it may be possible to elucidate the mechanism whereby these genes have acquired imprinted expression and the control elements upon which their imprinting depend.
KLF14, unlike its closest relatives in the KLF family of genes, has a large CpG island spanning the vast majority of its ORF. At the same time, the gene is enriched for proline and arginine amino acids, which are encoded by the codons CCN and CGN, respectively. Hence, it is plausible that, upon the retrotransposition of KLF16, the fusion of the proline/arginine-rich exons created a CG-rich region, which is predicted to be a CpG island, yet lacks the biological functions generally associated with such regions. Such an occurrence would also account for the absence of epigenetic modifications observed in the CpG island. Thus, we cannot exclude the existence of a CG-neutral differentially methylated promoter or additional exons upstream of the KLF14 ORF. However, attempts to identify the 5′′ end of the gene in both human and mouse were fruitless (unpublished data).
The variability observed in the KLF14 ORF in humans, particularly the three fixed nonsynonymous changes and two nonsynonymous SNPs with higher frequencies of the derived allele, are suggestive of positive selection. A recent investigation of selection in the human and chimpanzee genomes identified KLF14 as a gene that has undergone adaptive evolution in the human genome [59]. However, our detailed analyses of this gene failed to provide significant results in tests for positive selection, even when the gene was divided into the variable N terminus and the conserved C-terminal zinc-finger domains (unpublished data). Since the null hypothesis of neutrality could not be rejected, the evolution observed in KLF14 could be due to relaxed functional constraint. The lack of significance observed may be caused by limitations in the analyses due to the gene's small size or by ongoing positive selection, as observed in previous studies [60]. Evidence for the latter lies in the numerous polymorphisms that are not fixed in the human lineage. Further experiments, such as the Extended Haplotype Homozygosity method [60] are needed to elucidate whether selection is active at this locus.
While we cannot reject neutrality or provide conclusive evidence that KLF14 is under positive selection, we demonstrate that this gene has undergone accelerated evolution in the human lineage. It is important to note that although the dN/dS values corresponding to the human lineage are >1, this accelerated evolution may in fact be caused by a relaxation of purifying selection. Although such a relaxation is unlikely, due to KLF14's conservation throughout mammalian evolution, the gene's functional significance may have changed or decreased within the human lineage, allowing for multiple mutations to accumulate in nonsynonymous sites.
Numerous studies have focused their efforts on the identification of genes under positive selection or accelerated evolution in the human species [61–63] and have found that several functional categories, including transcription factors, are enriched for rapidly evolving transcripts [62]. Since KLF14 had not been fully sequenced in the chimpanzee, and possibly due to the gene's size, these studies did not detect positive selection in the transcript. Thus, our analysis of KLF14 adds to the mounting body of literature suggesting that transcription factors are often evolving rapidly or with positive selection. However, KLF14 stands alone in this category due to its imprinting, which may have played a role in the evolution of this transcript. Previous papers have also failed to find evidence of positive selection across imprinted genes [64,65].
Our data suggest that the KLF14 sequence is highly variable, specifically in the human species. We propose that this variability may be due to the transcript's monoallelic expression, which allows for the accumulation of mutations on the silenced allele. Maternal inheritance of mutated alleles would give rise to their expression and deleterious mutations would face strong purifying selection due to haploinsufficiency. In contrast, beneficial mutations would undergo stronger and more rapid positive selection since their impact would be greater due to the gene's monoallelic expression. The latter phenomenon has been implicated in the increased rate of nonsynonymous substitution on the X chromosome [66]. Consequently, the inherited variations seen in KLF14 should be nondeleterious and possibly advantageous. This is supported by the fact that the haplotypes carrying rare alleles are transmitted from both mothers and fathers and are consequently expressed in healthy individuals, as evidenced in the unaffected siblings of RSS and autism patients. All of these sequence variations, with the exception of a synonymous polymorphism observed in haplotype 6 (Figure S1), occur in the N-terminal end of the putative protein that has low sequence conservation, suggesting that variation in the C-terminal end of the protein may not be tolerated.
In 2004, Dorus and colleagues examined the evolutionary rates of nervous system-related genes between primates and rodents, thereby identifying genes under accelerated evolution in primates. They proposed that these genes may have played important roles in human speciation by developing human behavior and brain size [61]. Consequently, the disruption of genes under accelerated evolution or positive selection in the human lineage has been associated with disease. For example, mutations in FOXP2 underlie severe language and speech impairment/developmental verbal dyspraxia [5,67], while Microcephalin and ASPM are associated with microcephaly [68,69]. Due to KLF14's increased expression in neuronal cells, as well as the accelerated evolution observed in the human lineage, this gene may have played a role in the acquisition of human-specific traits. Such a function would agree with our hypothesis postulated above, describing the role of imprinting in the variability of the gene. Despite being imprinted in ancestral mammals, we observe that the evolutionary aspects of KLF14 are unique to the human lineage. This observation could be due to selective pressures unique to this species, and possibly unique to demographic populations within the human population, which have accelerated the evolution of the gene. Consequently, the variations seen in KLF14 may be beneficial to humans or subpopulations of the human species, particularly the variations that are fixed or are going towards fixation. However, further studies are required to determine the gene's function in the brain, particularly in neurons, to assess its contribution to human speciation and its putative role in cognitive disease.
Although, the sequencing of the KLF14 ORF in autistic individuals and RSS patients did not identify mutations unique to these populations, it does not rule out the involvement of KLF14 in these phenotypes since mutations may be present in regulatory regions causing changes in expression levels, loss of imprinting, or transcript instability. Due to the lack of KLF14 expression in lymphoblasts, we were unable to quantify transcript levels in patients, nor were we able to verify imprinted expression in these patients. Future studies may ascertain the involvement of KLF14 in these disorders by obtaining fibroblast cells from patients, thereby elucidating the role of both imprinted genes and positively selected genes on development and cognitive function, respectively.
Materials and Methods
RT-PCR using RNA from somatic cell hybrids, human tissues, and murine embryonic samples.
cDNA was obtained and amplified from somatic cell hybrids–cell lines and human tissues [22]. The primers used for amplification of KLF14 are forward (5′-CCACCCAACCTATCATCCAG-3′) and reverse (5′-GTACCTCCCCAGAGTCCACA-′). Reciprocal hybrid crosses were performed between C57BL/6J and JF1/Ms, and tissues were obtained from the F 1 generations. Glia and neurons were cultured as previously described [34], and cDNA was obtained [22]. To identify genomic SNP in Klf14, an 851-bp fragment was amplified using primers AK030435F (5′-TGGACACCCTCTCCAAAGTC-3′) and AK030435R (5′-AAGCGACATCAGTGCTCCTT-3′), and a SNP corresponding to bp 451 of AK030435 was found. Amplified DNA and cDNA fragments were purified and directly sequenced.
Methylation analysis.
Genomic DNA was extracted from BL6 × JF1 12.5-dpc whole embryos and fibroblasts. Bisulfite treatment was performed using the EZ methylation protocol (Zymo Research, http://www.zymoresearch.com). One microliter of the eluted DNA was used for PCR using the following primers: F 1, 5′-TGGTTGTAATAAGGTTTATTATAAGT-3′ and R 1, 5′-AAACCAAAACTTTCCACCATAACTA-3′; F 2, 5′-TGGAGGATTGGGGGTATTTATA-3′ and R 2 5′-CAAACAAATAATTTCCCAAACTACTAA-3′; F 3 5′-TTTGGGGTTATTTTTTATTTGAGTT-3′ and R 3 5′-TCAAACAAAATCCTAAAAACTTTTT-3′. PCR products were subcloned using the pGEM-T easy system (Promega, http://www.promega.com), and transformants were sequenced.
RACE to determine full-length sequence of KLF14.
Marathon ready cDNA from 11-day embryo and placenta (Clontech, http://www.clontech.com) were used for RACE to determine the full length of KLF14 in mouse and human, respectively. The manufacturer's protocol was followed using the following primers for 3′ RACE: human GSP1 (5′- GAAGGGATGAACTCCCGTACTCTCCA-3′), human GSP1-nested (5′-AACCAGGGATGTGAAACTGG-3′), mouse GSP1 (5′-GGGTGTTGTGATCTCATGGAGTTG-3′), and mouse GSP1-nested (5′-TGCTAAGTTTCTGCCAAGAGC-3′). The amplified cDNA fragments were purified using microCLEAN (Microzone, http://www.microzone.co.uk) and directly sequenced.
ChIP and analysis of histone modification.
Formaldehyde fixed ChIP assay was carried out using Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology, http://www.upstate.com) as previously described [70]. The antisera used were against H3K9acK14ac (Upstate Biotechnology, 06–599), H4ac (Upstate Biotechnology, 06–866), and H3K4me2 (Upstate Biotechnology, 07–030). DNA obtained from the precipitated fractions was amplified using primers for Mest (MCF1: 5′-AGGGGGTAGCGGGTCAATAC-3′ and MCR1: 5′-ATGTGCTGGTGGCCGAAGCAG-3′) and Klf14 (KCF1: 5′-TTGGAGCCAGACGAGCTGGAAG-3′ and KCR1: 5′-AGGCTGCTGGGAATGCCATAGC-3′). Alleles between BL6 and JF1 were distinguished by MaeI and SacII polymorphisms. We determined the dominant alleles by analyzing band intensities.
At the same time, ChIP was performed using nonfixed chromatin from BL6 × JF1 13.5-dpc hybrid embryos, and precipitated DNA was analyzed by PCR single strand conformation polymorphism as previously described [71]. The antibodies used for native ChIP were directed against H3K9ac (Upstate Biotechnology, 06–942), H3K4me2 (Upstate Biotechnology, 07–030), H3K9me3 (Upstate Biotechnology, 07–442), and H4K20me3 (Upstate Biotechnology, 07–463).
Amplification of KLF14 and KLF16 in mammalian species.
The KLF14 ORF was sequenced in 352 individuals (704 chromosomes), representing both patients (60 RSS and 160 autistic) and controls (78 individuals of Western European descent from the human variation panel and 54 individuals from an ethnically diverse panel containing nine African American, four Arabian, four Armenian, four Chinese, three Greek, four Indo-Pakistani, four Italian, four Iranian, eight Japanese, and 10 Somalian individuals). Haplotypes were determined manually, and when necessary, samples were subcloned to determine the phase of polymorphisms. The KLF14 ORF was also sequenced from three gorillas, two orang-utans, two macaques, two bonobos, and 20 chimpanzees. In the latter, at least one individual from three chimpanzee subspecies was included.
Primers used to amplify KLF16 were: F 1 (5′-CCCGCCACCACCGGAC-3′) or F 2 (5′-CCCGGCACTACCGGAC-3′) and R (5′-TGCAGGGCAGCGAGTCG-3′). KLF14 was amplified using the following primers and combinations thereof: F 1 (5′-AACTTCTTGTCGCAGTCGAG-3′), F 2 (5′-ACTTCTTGTCGCAGTCGA-3′), R 1 (5′-CGTGCCTGGACTACTTCGC-3′), R 2 (5′-GCCCCACCTGCTGGCT-3′), R 3 (5′-GCCCCACCTGCTGGC-3′).
Evolution analyses of KLF14.
Primate and human haplotype sequences were manually aligned and used to create a parsimony tree (Figure 5B). This tree does not represent the evolution of the gene, since haplotypes evolve through recombination as well as mutation making phylogeny difficult to reconstruct. Instead, the tree represents the similarity between the sequences. A neighbor-joining tree was also created from this alignment using MEGA3 [72]. We created nine other joining-joining primate trees, with each one using a single human haplotype or the HH-MRCA, created manually by parsimony, as the human sequence. These trees were used to test the neutral model of molecular evolution within the primate lineages. Lineage-specific dN/dS ratios were computed using codon-based maximum likelihood methods. These dN/dS tests were performed using the CodeML program within the PAML software package [73], which uses the codon substitution model of Goldman and Yang [74]. In these models we allowed for unequal codon frequencies and unequal transition and transversion substitution rates.
The lineage-specific dN/dS ratios for each of the nine single human haplotype trees were calculated using the maximum likelihood free-ratio model, as well as the Nei-Gojobori method (Figure 5B) [75]. The likelihood values and dN/dS ratios for each tree were also calculated, using the one-ratio model, the two-ratio model, where the ratio of the human lineage is allowed to differ from the ratio of the other primate lineages, and the two-ratio model, with dN/dS of the human lineage set equal to one. To test the null hypothesis that KLF14 is evolving neutrally in the human lineage, the likelihood ratio test was used (Table S1) [39] to assess whether KLF14's human dN/dS ratio is significantly greater than one or if the human dN/dS ratio is significantly greater than the primate background.
Additionally, 9,880 bps (including the ORF) encompassing KLF14 were sequenced in 18 individuals from the diversity panel and in one chimpanzee (to deduce the ancestral allele). Haplotypes were inferred using the PHASE program [76], and DnaSP 4.0 [40] was used to test for positive selection using the Tajima's D-test [41], Fu and Li's D-test [42], Fu and Li's F* [42], Fu's Fs statistic [43], and Fay and Wu's H test statistic [44].
To detect positive selection, the McDonald-Kreitman test [45] was performed using the ORF sequences from the 20 chimpanzee and two bonobo samples. DnaSP 4.0 [40] was then used to look for a significant departure from neutrality. A binomial distribution test [46] was also used to look for an increased rate of amino acid substitutions in the human lineage relative to the ape lineages, which would suggest that accelerated amino acid evolution has occurred in the human lineage.
Sequence variability in KLF14.
Polymorphism data were extracted on 826 genes sequenced in the Seattle SNP and Environmental Genome (EG) projects. π values were calculated for each of the sequenced genes and for KLF14 using the allele frequencies for each identified polymorphism. Values of π were calculated using SNPs at all sites, at sites with synonymous polymorphisms, and at sites with nonsynonymous polymorphisms. Diversity statistics π and θ were also calculated for both the human and chimpanzee KLF14 gene to compare the variability between species.
Supporting Information
Shown are eight different haplotypes (1–6, 7J, and 8J) identified in diverse human populations, as well as the sequence for the KLF14 ORF in the chimpanzee and gorilla (C and G, respectively). Synonymous polymorphisms and nonsynonymous polymorphisms are denoted by gray and black boxes, respectively. The corresponding amino acid substitution is noted above each polymorphism. The identity of the ancestral allele was identified by comparison to orang-utan (Pongo pygmaeus) and macaque (Macaca mulatta). The sequence corresponding to zinc-finger domains are enclosed, and the sequence between the boxes comprises the conserved Krüppel-link. Haplotypes specific to the Japanese population are indicated by J.
(563 KB PDF)
(52 KB DOC)
(35 KB DOC)
(25 KB DOC)
(53KB DOC)
(27 KB DOC)
(31 KB DOC)
Accession Numbers
The Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) numbers discussed in this paper are Prader-Willi syndrome, MIM:176270; Beckwith-Wiedemann syndrome, MIM:130650; and RSS, MIM:180860. The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) accession numbers for the genes and gene products discussed in this paper are KLF14, NM_138693; Klf14, AK030435 and DQ534758; and KLF16, NP_114124.
Acknowledgments
We thank The Centre for Applied Genetics and Jennifer Skaug for technical assistance and Matthew Hurles and Lars Feuk for advice on evolutionary analyses. We thank the following institutions, research facilities, and zoological parks for the primate samples used in this study: Riverside Zoo (http://www.riversidezoo.org), New Iberia Research Center (http://nirc.louisiana.edu) (supported by National Institutes of Health National Center for Research Resources Grant 3 U42 RR015087-05S1), Primate Foundation of Arizona (Mesa, Arizona, United States), Southwest Foundation for Biomedical Research (http://www.sfbr.org), Coriell Institute for Medical Research (http://www.coriell.org), and the Integrated Primate Biomaterials and Information Resource (http://www.coriell.org). We thank the Royal Ontario Museum (http://www.rom.on.ca) for providing opossum and tamandua tissues.
Abbreviations
- ChIP
chromatin immunoprecipitation
- DMR
differentially methylated region
- dN/dS
ratio of nonsynonymous to synonymous substitutions
- Dnmt3a
DNA methyltransferase 3a
- dpc
days post coitum
- H3K4me2
dimethylation of lysine 4 on histone 3
- H3K9ac
acetylation of lysine 9 on histone 3
- H3K9me3
trimethylation of lysine 9 on histone 3
- H4K20me3
trimethylation of lysine 20 on histone 4
- HH-MRCA
human haplotype's most recent common ancestor
- ICR
imprinting control region
- KLF14
Krüppel-like factor 14
- MEST
mesoderm specific transcript homolog
- MRCA
most recent common ancestor
- ORF
open reading frame
- PAML
phylogenetic analysis by maximum likelihood
- RACE
rapid amplification of cDNA ends
- RSS
Russell-Silver Syndrome
- RT-PCR
reverse transcriptase-PCR
- SNP
single nucleotide polymorphism
Footnotes
¤a Current address: Department of Obstetrics and Gynaecology, Hokkaido University Graduate School of Medicine, Sapporo, Japan
¤b Current address: Department of Maternal-Fetal Biology, National Center for Child Health and Development, Setagaya, Tokyo, Japan
¤c Current address: Wellcome Trust/Cancer Research UK Gurdon Institute of Cancer and Developmental Biology, University of Cambridge, Cambridge, United Kingdom
Competing interests. The authors have declared that no competing interests exist.
A previous version of this article appeared as an Early Online Release on March 12, 2007 (doi:10.1371/journal.pgen.0030065.eor).
Author contributions. LPK, ARC, TY, HS, KN, and SWS conceived and designed the experiments. LPK, TY, PA, SNAA, GHP, MMH, and KN performed the experiments. LPK, ARC, TY, PA, RF, GHP, KN, and SWS analyzed the data. LPK, TY, SNAA, GEM, MK, GHP, ACS, CL, HS, KK, and KN contributed reagents/materials/analysis tools. LPK, ARC, and RF wrote the paper.
Funding. LPK and ARC are supported by Natural Sciences and Engineering Research Council of Canada Postgraduate Studentship and Canada Graduate Scholarship, respectively. SAA is a WellBeing for Women Post-doctoral Fellow. TY is supported by the Hospital of Sick Children's Research Training Competition. Grant support is from Genome Canada/Ontario Genomics Institute to SWS. SWS is an Investigator of the Canadian Institutes of Health Research and an International Scholar of the Howard Hughes Medical Institute.
References
- Walter J, Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum Mol Genet. 2003;12(Spec No 2):R 215–R220. doi: 10.1093/hmg/ddg296. [DOI] [PubMed] [Google Scholar]
- Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, et al. Epimutation of the telomeric imprinting center region on Chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- Eggermann T, Wollmann HA, Kuner R, Eggermann K, Enders H, et al. Molecular studies in 37 Silver-Russell syndrome patients: Frequency and etiology of uniparental disomy. Hum Genet. 1997;100:415–419. doi: 10.1007/s004390050526. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Fernandez BA, Teshima I, Shuman C, Proud VK, et al. Molecular genetic studies of human Chromosome 7 in Russell-Silver syndrome. Genomics. 2002;79:186–196. doi: 10.1006/geno.2002.6695. [DOI] [PubMed] [Google Scholar]
- Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Barnby G, Bonora E, Sykes N, Bacchelli E, et al. Analysis of IMGSAC autism susceptibility loci: Evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Kayashima T, Yamasaki K, Ohta T, Yoshiura K-i, et al. The gene TSGA14, adjacent to the imprinted gene MEST, escapes genomic imprinting. Gene. 2002;288:57–63. doi: 10.1016/s0378-1119(02)00428-6. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Hayashida S, Miura K, Masuzaki H, Ishimaru T, et al. The novel gene, gamma2-COP (COPG2), in the 7q32 imprinted domain escapes genomic imprinting. Genomics. 2000;68:330–335. doi: 10.1006/geno.2000.6265. [DOI] [PubMed] [Google Scholar]
- Blagitko N, Mergenthaler S, Schulz U, Wollmann HA, Craigen W, et al. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum Mol Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- Ono R, Kobayashi S, Wagatusuma H, Aisaka K, Kohda T, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- Piras G, El Kharroubi A, Kozlov S, Escalante-Alcalde D, Hernandez L, et al. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for varepsilon-sarcoglycan are maternally imprinted genes: Identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol. 2000;20:3308–3315. doi: 10.1128/mcb.20.9.3308-3315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley L, Nakabayashi K, Monk D, Beechey C, Peters J, et al. The imprinted region on human Chromosome 7q32 extends to the carboxypeptidase A gene cluster: An imprinted candidate for Silver-Russell syndrome. J Med Genet. 2003;40:249–256. doi: 10.1136/jmg.40.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Kohda T, Miyoshi N, Kuroiwa Y, Aisaka K, et al. Human PEG1/MEST, an imprinted gene on Chromosome 7. Hum Mol Genet. 1997;6:781–786. doi: 10.1093/hmg/6.5.781. [DOI] [PubMed] [Google Scholar]
- Hannula K, Lipsanen-Nyman M, Kontiokari T, Kere J. A narrow segment of maternal uniparental disomy of Chromosome 7q31-qter in Silver-Russell syndrome delimits a candidate gene region. Am J Hum Genet. 2001;68:247–253. doi: 10.1086/316937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesewijk AM, Blagitko N, Schinzel AA, Hu L, Schulz U, et al. Evidence against a major role of PEG1/MEST in Silver-Russell syndrome. Eur J Hum Genet. 1998;6:114–120. doi: 10.1038/sj.ejhg.5200164. [DOI] [PubMed] [Google Scholar]
- Meyer E, Wollmann HA, Eggermann T. Searching for genomic variants in the MESTIT1 transcripts in Silver-Russell syndrome patients. J Med Genet. 2003;40:e65. doi: 10.1136/jmg.40.5.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Mitsuya K, Kayashima T, Yamasaki K, Ohta T, et al. Imprinting analysis of 10 genes and/or transcripts in a 1.5 Mb MEST-flanking region at human Chromosome 7q32. Genomics. 2004;83:402–412. doi: 10.1016/j.ygeno.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Bonora E, Bacchelli E, Levy ER, Blasi F, Marlow A, et al. Mutation screening and imprinting analysis of four candidate genes for autism in the 7q32 region. Mol Psychiatry. 2002;7:289–301. doi: 10.1038/sj.mp.4001004. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. Mammalian Krüppel-like transcription factors: More than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, et al. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Bentley L, Hitchins MP, Mitsuya K, Meguro M, et al. Identification and characterization of an imprinted antisense RNA (MESTIT1) in the human MEST locus on Chromosome 7q32. Hum Mol Genet. 2002;11:1743–1756. doi: 10.1093/hmg/11.15.1743. [DOI] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. Embo J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, et al. Imprinting along the Kcnq1 domain on mouse Chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. Embo J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Tsai TF, Beaudet AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20:2859–2870. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Gomez M, Brockdorff N, Feil R. Differential patterns of histone methylation and acetylation distinguish active and repressed alleles at X-linked genes. Cytogenet Genome Res. 2002;99:66–74. doi: 10.1159/000071576. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kerjean A, Dupont J-M, Vasseur C, Le Tessier D, Cuisset L, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Cheung J, MacDonald JR, Osborne LR, Nakabayashi K, et al. Human Chromosome 7: DNA sequence and biology. Science. 2003;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet. 2004;36:339–341. doi: 10.1038/ng1327. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Kitano T, Liu YH, Ueda S, Saitou N. Human-specific amino acid changes found in 103 protein-coding genes. Mol Biol Evol. 2004;21:936–944. doi: 10.1093/molbev/msh100. [DOI] [PubMed] [Google Scholar]
- Livingston RJ, von Niederhausern A, Jegga AG, Crawford DC, Carlson CS, et al. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, et al. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- Page RDM, Holmes EC. Molecular evolution: A phylogenetic approach. Malden (Massachusetts): Blackwell Science; 1998. 346 [Google Scholar]
- Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, et al. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Zhang JS, Ellenrieder V, Conley A, Duenes T, et al. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J Biol Chem. 2001;276:36749–36756. doi: 10.1074/jbc.M105831200. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, et al. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse Chromosome 12. Curr Biol. 2000;10:1135–1138. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal Chromosome 7 gene cluster. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Beatty LG, Zhou W, Lew J, Schoenherr C, et al. Insulator and silencer sequences in the imprinted region of human Chromosome 11p15.5. Hum Mol Genet. 2003;12:1927–1939. doi: 10.1093/hmg/ddg194. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Renfree MB, Pask AJ, Shaw G, Kobayashi S, et al. Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech Dev. 2005;122:213–222. doi: 10.1016/j.mod.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Arbiza L, Dopazo J, Dopazo H. Positive selection, relaxation, and acceleration in the evolution of the human and chimp genome. PLoS Comput Biol. 2006;2:e38. doi: 10.1371/journal.pcbi.0020038. doi: 10.1371/journal.pcbi.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, et al. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GT, Hurst LD. Molecular evolution of imprinted genes: No evidence for antagonistic coevolution. Proc Biol Sci. 1997;264:739–746. doi: 10.1098/rspb.1997.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Hurst LD. Molecular evolution of an imprinted gene: Repeatability of patterns of evolution within the mammalian insulin-like growth factor type II receptor. Genetics. 1998;150:823–833. doi: 10.1093/genetics/150.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu CI. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc Natl Acad Sci U S A. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li T, Vu TH, Ulaner GA, Hu J-F, et al. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology. 2003;144:5658–5670. doi: 10.1210/en.2003-0798. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nei M. Positive darwinian selection observed at the variable-region genes of immunoglobulins. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are eight different haplotypes (1–6, 7J, and 8J) identified in diverse human populations, as well as the sequence for the KLF14 ORF in the chimpanzee and gorilla (C and G, respectively). Synonymous polymorphisms and nonsynonymous polymorphisms are denoted by gray and black boxes, respectively. The corresponding amino acid substitution is noted above each polymorphism. The identity of the ancestral allele was identified by comparison to orang-utan (Pongo pygmaeus) and macaque (Macaca mulatta). The sequence corresponding to zinc-finger domains are enclosed, and the sequence between the boxes comprises the conserved Krüppel-link. Haplotypes specific to the Japanese population are indicated by J.
(563 KB PDF)
(52 KB DOC)
(35 KB DOC)
(25 KB DOC)
(53KB DOC)
(27 KB DOC)
(31 KB DOC)