Abstract
Peptide identification by tandem mass spectrometry is the dominant proteomics workflow for protein characterization in complex samples. Traditional search engines, which match peptide sequences with tandem mass spectra to identify the samples' proteins, use protein sequence databases to suggest peptide candidates for consideration. Although the acquisition of tandem mass spectra is not biased toward well-understood protein isoforms, this computational strategy is failing to identify peptides from alternative splicing and coding SNP protein isoforms despite the acquisition of good-quality tandem mass spectra. We propose, instead, that expressed sequence tags (ESTs) be searched. Ordinarily, such a strategy would be computationally infeasible due to the size of EST sequence databases; however, we show that a sophisticated sequence database compression strategy, applied to human ESTs, reduces the sequence database size approximately 35-fold. Once compressed, our EST sequence database is comparable in size to other commonly used protein sequence databases, making routine EST searching feasible. We demonstrate that our EST sequence database enables the discovery of novel peptides in a variety of public data sets.
Keywords: alternative splicing, expressed sequence tags, proteomics, sequence compression, tandem mass spectra
Introduction
Peptide identification by tandem mass spectrometry is the dominant proteomics workflow for protein characterization in complex samples. Traditional search engines, which match peptide sequences with tandem mass spectra to identify the samples' proteins, use protein sequence databases to suggest peptide candidates for consideration. This algorithmic strategy for peptide identification has been extremely successful, making peptide identification by tandem mass spectrometry one of the cornerstones of the systems biology revolution. However, only peptides contained in the protein sequence database can be identified. The Swiss-Prot protein sequence database (Apweiler et al, 2004) contains only well-characterized protein sequences and deliberately suppresses sequence variations and alternative splicing to reduce redundancy. The IPI protein sequence database (Kersey et al, 2004) preserves alternative splicing isoforms, if they are present in its source databases, but suppresses small variations in the amino-acid sequence.
An unfortunate consequence of the reliance on protein sequence databases is a computational bias against the identification of peptides from poorly understood protein isoforms, such as those from alternative splicing or coding SNP isoforms. As protein sequence databases seek to address this problem and capture more of these variants they become highly redundant, at the level of exact peptide sequence repetition. This is not only a computational inefficiency, leading to increased running times for no additional benefit, but it can also distort statistical significance estimates, in the form of E-values, computed by search engines (Edwards, 2005).
Expressed sequence tags (ESTs), single-pass sequencing reads from mature RNA transcripts, account for the majority of experimental evidence for alternative splicing in humans. To date, nearly 8 million ESTs have been generated from human samples and their sequences deposited in Genbank. ESTs, obtained from mRNA after intronic sequence is removed, provide experimental evidence for intron–exon boundaries and splicing that is used by all of the gene, transcript, and protein sequence prediction infrastructure that underlies protein sequence databases. Compared with genomic sequence, using ESTs as a source of putative peptide sequences eliminates the guesswork involved in finding coding sequence and determining its intron–exon structure, leaving only the question of translation frame unresolved.
We propose to remove the computational bias imposed by the use of protein sequence databases in peptide identification by searching species-specific EST databases instead. Although searching ESTs is not new (Yates et al, 1995; Neubauer et al, 1998), a number of issues have stymied the widespread adoption of routine EST searches.
First, the set of human ESTs from the dbEST database (Boguski et al, 1993) is large, currently consisting of more than 4 billion nucleotides in 7.6 million entries. Searched using a naive six-frame translation, the resulting sequence of more than 8 billion amino acids is more than 300 times the size of the commonly used IPI Human protein sequence database. As tandem mass spectrometry search engines typically require running time linear in the input amino-acid sequence database, this naive EST search strategy is quite impractical.
The second issue with EST sequences is that they are error-prone. EST sequences, single-pass DNA sequencing reads, have a relatively high error rate—approximately 1 in every 100 nucleotides with conventional capillary electrophoresis sequencing technology. As with genome sequencing, however, many EST sequences cover most transcribed bases, so uncorrelated sequencing errors can be corrected by the consensus.
We apply a number of computational strategies to the human EST database to make it more suitable for searching by tandem mass spectrometry search engines. First, we require that the EST sequence map to the vicinity of a known gene, as defined by the UniGene database (Pontius et al, 2003). Second, we require that peptides be contained in a 30-amino-acid open-reading-frame (ORF). This enforces a conservative filter on out-of-frame translation. Third, we require that all peptide sequences be confirmed by at least two ESTs, a conservative error correction for sequencing errors in the ESTs. Finally, we represent the peptide sequences in a way that ensures that most of the peptide sequence repetition is eliminated. This novel computational strategy compresses the human EST sequence database to less than 3% of the size of the naive six-frame translation with negligible impact on its peptide sequence content. This reduction in size makes routine searching of human ESTs feasible using existing search engines.
We demonstrate that our compressed human EST peptide sequence database makes it possible to re-search publicly available tandem mass spectra from human samples, such as that in the PeptideAtlas (Desiere et al, 2006) and the Human Proteome Organization (HUPO) Plasma Proteome Project (PPP) (Omenn et al, 2005) data repositories, to look for, and find, known coding SNPs, novel coding mutations, alternative splicing isoforms, alternative translation start sites, micro-exons, and alternative translation frames. Many of these novel peptides, which are missing from current protein sequence databases, straddle exon boundaries and therefore could not have been observed by searching the six-frame translation of the human genome directly, a strategy proposed by Fermin et al (2006) for the HUPO PPP project.
In addition to speeding up search times, our compression technique also increases search engine sensitivity, reducing the E-value estimates associated with each peptide identification. For our compressed human EST peptide database, we observe E-value estimates reduced by a factor of about 50 from their original values, increasing the number of significant peptide identifications.
Our redundancy elimination technique relies on the observation that the peptides selected for fragmentation by tandem mass spectrometry are relatively short, compared to the typical length of protein sequences. A conservative upper bound on peptide length, 30 amino acids, encompasses the vast majority of tryptic peptides reliably identified by popular LC/MS/MS workflows. In what follows, we will show that it is possible to compress any amino-acid sequence database such that the new, smaller, amino-acid sequence database is:CompleteEvery 30-mer from the original sequence database is present, andCorrectOnly 30-mers from the original sequence database are present.
This is an extension of previous work (Edwards and Lippert, 2004) that showed how to achieve the optimal complete, correct representation of the amino-acid 30-mers of a sequence database, with the additional constraint that each 30-mer appears exactly once (termed compactness). Counterintuitively, dropping the compactness constraint results in superior compression. The optimal complete, correct (C2) sequence database construction is described in the Materials and methods section.
Results and discussion
Compressed human EST peptide sequence database
Rather than construct a monolithic peptide sequence database as described previously (Edwards and Lippert, 2004), we applied our compression technique to construct 20 774 distinct gene-centric peptide sequence databases. For each UniGene gene, we extracted its EST sequences, retained ORFs of at least 30 amino acids, eliminated amino-acid 30-mers observed only once, and C2 compressed the result. As essentially all of the 30-mer repetitions are found in ESTs that map to the same gene, this gene-based EST partitioning had negligible impact on the compression achieved. Each gene's sequences were then concatenated into a single FASTA entry, separated by a special character (‘J'—molecular weight 10 kDa).
The final compressed human EST sequence database contains 223 Mb of amino acids in 20 774 FASTA entries, each labeled with the name of a human gene. The gene-centric ORFs of at least 30 amino acids represented 2 129 995 618 (597 020 903 distinct) amino-acid 30-mers. After elimination of 30-mers observed only once, 1 615 810 968 (82 836 253 distinct) amino-acid 30-mers remain.
Compared to the naive six-frame translation of the human EST database (approximately 8 billion amino acids), our compressed version represents a 35-fold compression, with negligible loss of potential peptide sequence. The compressed human EST peptide sequence database can be downloaded from the author's home page (direct URL: ftp://ftp.umiacs.umd.edu/pub/nedwards/PepSeqDB).
Faster, more sensitive, peptide identification
To benchmark the speed and sensitivity advantages of our compressed human EST peptide sequence database, we re-searched a single data file from the PeptideAtlas ‘raftflow' data set (von Haller et al, 2003) using the Mascot search engine (Perkins et al, 1999). The data file ‘raft-flow37.mzXML' contains 1994 tandem mass spectra acquired on a Thermo-Finnegan LCQ-DECA ion-trap mass spectrometer.
The first search, using Mascot's nucleotide database configuration option, which effectively searches the naive six-frame translation of the human EST sequences, took 22 h. The second, on the compressed human EST peptide sequence database, took 15 min, 1% of the time of the first search. The alternatively spliced peptide LQGSATAAEAQVGHQTAR was identified with the significant E-value of 0.0049 in the first search, and had the same score (73) in the second search, but with an improved E-value of 9.6 × 10−5. The E-values of each of the 37 identifications with Mascot score ⩾50 improved by approximately the same factor. The first, naive enumeration search made 16 significant peptide identifications of 13 distinct peptides at the 0.05 E-value threshold, whereas the second made 47 significant peptide identifications of 30 distinct peptides at the same E-value threshold.
Novel peptides in public data sets
The compressed human EST peptide sequence database has made it possible to re-search many of the tandem mass spectrometry data sets available in public repositories, using relatively modest computational infrastructure. In doing so, we have uncovered tandem mass spectra that represent peptides missing from IPI and other protein sequence databases, summarized in Table I.
Table 1. Novel peptides found in LC/MS/MS data sets from public data repositories.
| Data set | Peptide | Type | E-valued | ESTs | mRNA | SPVe | IPI | Straddle intron? | Gene |
|---|---|---|---|---|---|---|---|---|---|
| Raftflowa | LQGSATAAEAQVGHQTAR | Novel splice | <10−8 | 11 | Y | N | N | Y | LIME1 |
| Raftauga | TAGSPLCLPTPGAAPGSAGSCSHR | Novel frame | ∼10−4 | 34 | Y | N | N | N | LIME1 |
| Raftflowa | LQTASDESYKDPTNIQLSK | Micro-exon | <10−6 | 10 | N | Y | N | Y | SPTAN1 |
| A8 IPb | HEQASNVLSDISEFR | Novel start | <10−9 | 86 | Y | N | N | Y | THOC2 |
| PPP 29c | KADDTWEPFASGK | Novel mutation | <10−7 | 2 | N | N | N | Y | TTR |
| PPP 40c | DTEEEDFHVDQATTVK | Known cSNP | <10−9 | 54 | N | Y | N | N | SERPINA1 |
| PPP 40c | DTEEEDFHVDQVTTVK | Wild type | <10−9 | 337 | Y | N | Y | N | SERPINA1 |
| PPP 28c | LQHLVNELTHDIITK | Known cSNP | <10−9 | 4 | N | Y | N | N | SERPINA1 |
| PPP 28c | LQHLENELTHDIITK | Wild type | <10−6 | 351 | Y | N | Y | N | SERPINA1 |
Human lipid raft T-cell study from PeptideAtlas (von Haller et al, 2003).
Human erythroleukemia K562 cell line study from PeptideAtlas (Resing et al, 2004).
HUPO Plasma Proteome Project data set from numbered laboratory (Omenn et al, 2005).
E values computed by X!Tandem.
Swiss-Prot variant annotation.
These peptides represent only the most convincing of novel peptide identifications uncovered in our searches. Many other significant novel peptide identifications, with varying levels of spectral, genomic, and transcript sequence evidence, can be easily extracted from the search results, but they often do not stand up to careful manual scrutiny. In addition, while these public bottom-up LC/MS/MS data sets identify many proteins, they typically cover only a small percentage of each protein's sequence. Consequently, we are not guaranteed to observe peptides that confidently elucidate novel isoforms, even if they are present in the sample. A possible alternative workflow for targeting these isoforms would separate proteins before enzymatic digestion and LC/MS/MS analysis to drive sequence coverage to 80% or more.
Novel alternative splice isoform
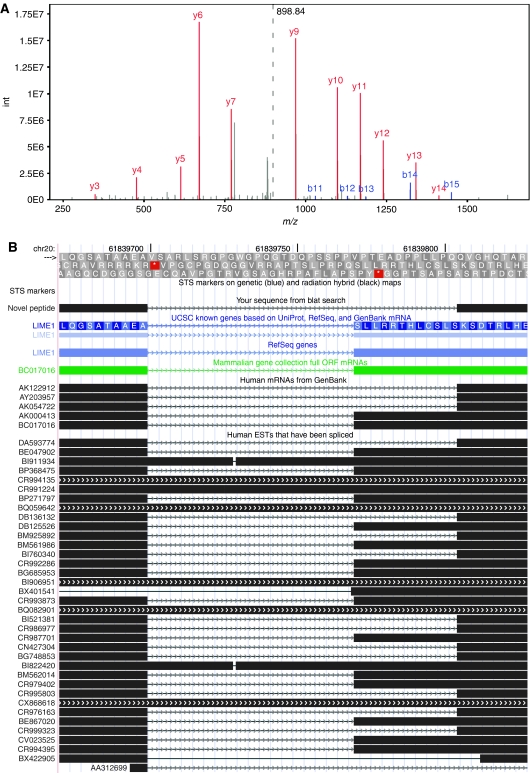
The peptide LQGSATAAEAQVGHQTAR was observed in the ‘raftflow' data set contributed to the PeptideAtlas Raw Data Repository by the Institute for Systems Biology. This data set represents the flow-through fraction of lipid rafts from human Jurkat T-cells stimulated via T-cell receptor/CD28 cross-linking and from control cells (von Haller et al, 2003). The annotated Thermo-Finnegan LCQ-DECA ion-trap MS/MS spectrum is shown in Figure 1A. This peptide identification, computed by X!Tandem, has a highly significant E-value (<10−8). This peptide sequence is found in eleven EST sequences and three mRNA from Genbank. Further investigation finds the ESTs and the peptide sequence align to an alternative splice form of the LIME1 gene on chromosome 20. This sequence straddles an intron using the same donor splice site as the primary isoform, but a different acceptor splice site. The MS/MS spectrum contains a good y-ion tag on both sides of the intron. A screen shot from the UCSC genome browser (Kent et al, 2002) in the region of this peptide is also shown in Figure 1B. This identification is supported by the weaker identification of peptide TAGSPLCLPTPGAAPGSAGSCSHR from the ICAT fraction of the same experiment, which appears in a novel frame consistent with the frame shift introduced by the alternative splicing event (Supplementary Figures 1–3). Nesvizhskii et al (2006) also identified the peptide LQGSATAAEAQVGHQTAR and other novel peptides from this LC/MS/MS data set using an early version of the compressed EST sequence database.
Figure 1.
(A) MS/MS spectrum from novel peptide LQGSATAAEAQVGHQTAR, found in PeptideAtlas data set ‘raftflow', and (B) UCSC genome browser (http://genome.ucsc.edu/) screen shot of genomic region.
Micro-exon isoform
The peptide LQTASDESYKDPTNIQLSK is also found in the PeptideAtlas ‘raftflow' data set. This peptide, from the SPTAN1 gene, is present as an isoform annotation in Swiss-Prot, but is missing from the IPI protein sequence database. Its annotated MS/MS spectrum is shown in Supplementary Figure 4. This peptide identification, computed by X!Tandem, has a highly significant E-value (<10−6). This peptide is found in about 10 ESTs and one full-length mRNA sequence. The relevant region of the human genome is shown in Supplementary Figures 5–7. The last three amino acids of this peptide are contained in a micro-exon of just five amino acids.
Novel translation start site
The peptide HEQASNVLSDISEFR was observed in the ‘A8_IP' data set contributed to the PeptideAtlas Raw Data Repository by the Ahn laboratory at the University of Colorado (Resing et al, 2004). The data set is generated from a human erythroleukemia K562 cell line using a Thermo-Finnigan LCQ classic ion-trap mass spectrometer. The annotated spectrum is shown in Supplementary Figure 8. This peptide identification, computed by X!Tandem, has a highly significant E-value (<10−9).
This peptide sequence is found in 86 ESTs and one mRNA from Genbank. Further investigation finds this sequence aligns to chromosome X, far upstream of the annotated translation and transcription start site of gene THOC2, despite full-length mRNA and significant EST evidence for transcription in this region. Screen shots from the UCSC genome browser in the region of this peptide is shown in Supplementary Figures 9 and 10. We point out that the last three amino acids of the peptide align perfectly to the first nine nucleotides of the 3′ exon defined by these EST and mRNA sequences.
Novel mutation
The peptide KADDTWEPFASGK was observed in the HUPO PPP (Omenn et al, 2005) LC/MS/MS data set from Lab 29, also downloaded from the Peptide Atlas Raw Data Repository. This peptide contains a completely novel mutation in the TTR gene. The wild-type peptide, KAADDTWEPFASGK, suggests an Ala deletion at position 2 (or 3). This peptide identification, computed by X!Tandem, has a highly significant E-value (<10−7) (Supplementary Figure 11). The corresponding three-nucleotide deletion was observed in two ESTs, derived from two clones in the same clone library. Screen shots (Supplementary Figures 12–14) of the relevant region in the UCSC genome browser shows the deleted nucleotides. Furthermore, the deleted Ala at position 2 is associated with familial amyloidotic polyneuropathy, when the Ala is changed to Pro. We stress that this peptide also straddles an intron and could not have been identified by scanning the human genome directly, even under a mutation model that permitted amino-acid insertion and deletion.
Known coding SNPs
Two different known coding SNPs and their wild-type alleles were observed for the gene SERPINA1 in HUPO PPP data sets from laboratories 28 and 40. In the HUPO PPP data set from laboratory 40, a known coding SNP was observed in the wild-type peptide DTEEEDFHVDQVTTVK, substituting Val at position 12 with Ala (Supplementary Figures 15–17), whereas in HUPO PPP data set from laboratory 28, a known coding SNP was observed in the wild-type peptide LQHLENELTHDIITK, substituting Glu at position 5 with Val. E-values in each case were <10−9, except for the wild-type version of LQHLENELTHDIITK, which had an E-value <10−6.
Materials and methods
Human EST peptide enumeration
The human EST database and the associated human UniGene index used in this work were downloaded from NCBI's ftp site in March 2006. EST sequences and the UniGene index were loaded into a relational database, which was then used to construct gene-centric EST sequence databases. Each EST sequence was translated in six frames and ORFs of at least 30 amino acids were retained. Codons containing ambiguous nucleotides, such as N, were treated as a stop codon only if the resulting amino acid was also ambiguous.
Compressed SBH graphs
The compressed sequencing-by-hybridization graph (CSBH graph) representation of a sequence database is used to select those amino-acid 30-mers that are observed at least twice in each gene-centric EST peptide enumeration. In addition, the C2 and C3 compression algorithms make use of this representation.
A sequencing-by-hybridization graph (SBH graph) contains a directed edge for each k-mer in the sequence database, from a node representing the first k−1 symbols to a node representing the last k−1 symbols. This construction is a subgraph of the de Bruijn graph (de Bruijn, 1946), which represents all possible k-mers. The de Bruijn sequence, which is the shortest string containing all possible k-mers from some alphabet, can be trivially extracted using an Eulerian path on the corresponding de Bruijn graph. We strive for a similar result for the SBH graph, which represents a specific subset of the set of all k-mers.
As described in Edwards and Lippert (2004), and by analogy with suffix trees, we suppress trivial nodes in the SBH graph—those nodes with a single edge in and out, and substitute a single edge representing the multiple adjacent k-mers. In addition, we consider the nodes representing the beginning and end of sequences to be non-trivial. We call the resulting representation of the sequence database, for a fixed mer-size k, the compressed SBH graph of the sequence database.
The CSBH graph, which we build for each of the human EST peptide enumeration databases, has a number of important properties. First, each k-mer in the original sequence database is represented by some edge of the CSBH graph. Second, all k-mers represented by some edge are found in the original sequence database, and any path of edges forms a set of overlapping k-mers, all of which are found in the original sequence database. Therefore, any set of paths on the k-mer CSBH graph that uses all of the edges generates a complete, correct sequence database of the k-mers of the original sequence database. Notice that the original sequence database, which is, of course, complete and correct, traces out a set of paths on the CSBH graph that uses all of the edges. We will show how to choose a different set of paths on the CSBH graph that minimizes the size of the resulting sequence database.
The CSBH graph construction algorithm computes, as a natural side effect, the number of times each k-mer is observed in the original sequence database. The non-trivial end-of-sequence nodes ensure that every trivial node's (k−1)-mer sequence has a k-mer to the right for every k-mer to the left. As a trivial node's left k-mers must all be the same (as trivial nodes have in-degree one) and its right k-mers must all be the same (out-degree one), the two k-mers to the left and right of each trivial node must have the same mer-count. By induction, all k-mers represented by a CSBH graph edge have the same mer-count. Consequently, collapsing and counting CSBH graph edge occurrences effectively determines mer-counts for all k-mers in the original sequence database. Labeled with edge counts, the CSBH graph representation for the original sequence database can be quickly and easily restricted to the subgraph representing k-mers that occur at least c times in the original sequence database.
For each EST peptide enumeration, we construct the 30-mer CSBH graph, and then delete all edges with count 1, thereby ensuring that all amino-acid 30-mers we output occur at least twice in the original sequence database.
Optimal complete, correct, compact (C3) compression
As outlined above, the problem of constructing a complete, correct representation of the amino-acid 30-mers of our EST peptide enumeration amounts to determining a set of paths on the 30-mer CSBH graph that use all of the edges. The additional compactness constraint, which requires each 30-mer to be represented exactly once, can be modeled by ensuring that each CSBH-graph edge is contained in exactly one path.
Edwards and Lippert (2004) showed that the optimal C3 compression of a sequence database contains Nk+k (m+B+) symbols, where Nk is the number of distinct k-mers in the original sequence database, k is the mer-size, m is the number of components of the CSBH graph with no unbalanced nodes, and B+ is the total net degree of nodes with positive net degree. The net degree of a node is the number of in-edges minus the number of out-edges; a node with zero net-degree is called balanced. The optimal C3 compression must use at least m+B+ paths, and each path incurs a cost of k symbols to start (one sequence separator, plus the k−1 symbols of the initial node of the path), plus the length of its edges.
The construction of the optimal C3compression path set proceeds by determining an Eulerian path for each component of the CSBH graph. For balanced components or those with exactly one pair of unbalanced nodes with net degree 1 and −1, this is carried out using the standard Eulerian path algorithm. For components with B+>1, B+−1 artificial restart edges are added to the component, from nodes with positive net degree to nodes with negative net degree. Each artificial restart edge ‘costs' k symbols—a sequence separator symbol, plus the k−1 symbols of the node it points to. These artificial edges make it possible to run the standard Eulerian path algorithm on the component. Once the Eulerian path is determined, guaranteeing the optimal compression length, the artificial edges are removed, leaving the correct number of paths on the original edges.
Optimal complete, correct (C2) compression
Although it might seem that dropping the compactness constraint should increase the length of the resulting compressed sequence database, in fact, the opposite is true. Suppose that instead of using an artificial restart edge, at a cost of k symbols, we can find a path in the CSBH graph component from a node of positive net degree to a node of negative net degree that costs less than k symbols. Using the edges of this path to connect two elements of the path set, rather than using an artificial restart edge, results in a shorter compressed sequence. Note that we cannot necessarily use all such shortcuts; choosing the right combination of restart edges and shortcuts requires some care.
As an aside, we note that the problem of finding a minimum cost tour that visits every edge of a directed graph at least once is known in the operations research literature as the ‘Chinese Postman Problem' (Edmonds and Johnson, 1973). The Chinese Postman Problem can be solved in polynomial time by using a matching formulation to determine which edges to reuse. Our problem differs in the respect that we can use an artificial restart edge at fixed cost instead of reusing real edges if this is more cost-effective. Similar too, is the work on DNA sequence assembly from SBH experiments by Pevzner (1989), in which a minimum cost-flow instance is used to make the SBH graph Eulerian. This work presumes that a single unique DNA assembly is desired and that missing k-mers may be added. In our formulation, we are unconcerned with the question of forming a single unique output string and insist that no new k-mers be created (correctness).
We will also use a matching style formulation to choose the optimal combination of restart edges and shortcuts for each unbalanced component. Our subproblem is easiest stated as a minimum cost network flow problem. This formulation consists of supply, demand, and transit nodes, and capacitated arcs with a cost per unit flow. Minimum cost network flow problems can be solved in polynomial time and for suitable instances, such as ours, they have integer optimal solutions (Chvátal, 1983).
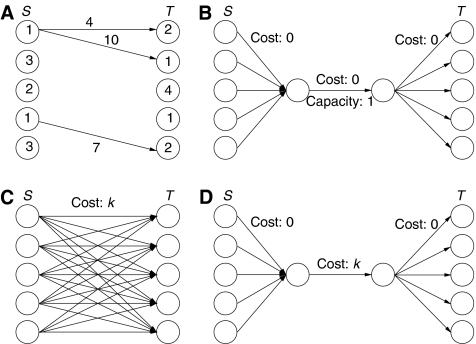
We use a minimum cost network flow instance for each component with B+>1. The instance has supply nodes S for each positive net degree node and demand nodes T for each negative net degree node. The magnitude of the supply or demand at the nodes of S∪T is the net degree of the corresponding CSBH graph node. We add an arc between a node of S and a node of T if there is a path between the corresponding nodes that uses less than k symbols. The cost of these arcs is the number of symbols on the shortcut path. Figure 2A gives an example of these elements of the formulation. We add a special widget, shown in Figure 2D, to account for the use of artificial restart edges between any pair of nodes from S and T. The use of this widget, instead of the complete bipartite graph on S and T (Figure 2C) reduces the number of edges in the formulation due to artificial restart edges from O(∣S∣ × ∣T∣) to O(∣S∣+∣T∣). Lastly, as we must leave one node with net degree 1 (and hence one other node with net degree −1) we add the widget of Figure 2B, which selects the Eulerian path start and end nodes. Unlike the widget that represents the artificial restart edges, this structure is not merely an efficiency, it is necessary to ensure that shortcut edges are not used to satisfy all supply and demand constraints, as this would represent a cycle on the CSBH graph component.
Figure 2.
(A) Basic structure of minimum cost network flow instance showing supply in the nodes of S, demand in the nodes of T, and shortcut edges; (B) graph widget to select the Eulerian path start and end nodes; (C) the dense bipartite subgraph to account for restart edges; and (D) the graph widget that replaces it.
Solved to optimality for each component using the CS2 minimum cost network flow solver (Goldberg, 1997), these minimum cost network flow instances determine the optimal use of restart edges and shortcut paths. Artificial edges representing each of the selected restart edges and shortcuts are added to the component, and an Eulerian path is determined, as before. The resulting sequence database represents the optimal C2 compression of the original sequence database.
Re-searching public LC/MS/MS data sets
Public LC/MS/MS data sets are downloaded from their respective data repositories. Sources include PeptideAtlas (Desiere et al, 2006) and HUPO PPP (Omenn et al, 2005). In all, more than 2.3 million spectra are stored locally for searching, representing 722 data files, and about 33 different laboratories or projects. Tandem mass spectra are searched using the X!Tandem (Craig and Beavis, 2004) or Mascot (Perkins et al, 1999) search engines with conservative search parameters, such as one missed cleavage, tryptic N- and C-termini, methionine oxidation and cysteine alkylation modifications only, and precursor mass tolerance of 2 Da. We use a computational grid of approximately 250 Linux processors, managed by the condor scheduling infrastructure, and the X!Tandem search engine for re-searching the public data sets. The Mascot search engine, tied to a single processor, is used for benchmarking, comparison studies, and to confirm specific identifications.
Supplementary Material
Supplementary Figures
Acknowledgments
This work is supported by NIH/NCI Grant CA126189.
References
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL (2004) UniProt: the universal protein knowledgebase. Nucl Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski M, Lowe T, Tolstoshev C (1993) dbEST—database for ‘expressed sequence tags'. Nat Genet 4: 332–333 [DOI] [PubMed] [Google Scholar]
- Chvátal V (1983) Linear Programming. W.H. Freeman and Company, New York [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- de Bruijn N (1946) A combinatorial problem. Proc Kon Ned Akad Wetensch 49: 758–764 [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R (2006) The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds J, Johnson E (1973) Matching, Euler tours and the Chinese postman. Mathematical Programming 5: 88–124 [Google Scholar]
- Edwards N, Lippert R (2004) Sequence database compression for peptide identification from tandem mass spectra. In Proceedings of the 4th International Workshop, WABI 2004, Volume 3240 of Lecture Notes in Computer Science, pages 230–241 [Google Scholar]
- Edwards NJ (2005) Faster, more sensitive peptide identification from tandem mass spectra by sequence database compression. Poster. 1st Annual US HUPO Symposium.
- Fermin D, Allen BB, Blackwell TW, Menon R, Adamski M, Xu Y, Ulintz P, Omenn GS, States DJ (2006) Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol 7: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AV (1997) An efficient implementation of a scaling minimum-cost flow algorithm. J Algorithms 22: 1–29 [Google Scholar]
- Kent W, Sugnet CW, Furey TS, Roskin K, Pringle TH, Zahler AM, Haussler D (2002) The Human Genome Browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4: 1985–1988 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, Eddes JS, Gruissem W, Baginsky S, Aebersold R (2006) Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteomics 5: 652–670 [DOI] [PubMed] [Google Scholar]
- Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet 20: 46–50 [DOI] [PubMed] [Google Scholar]
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM (2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 5: 3226–3245 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pevzner PA (1989) l-tuple DNA sequencing: Computer analysis. J Biomol Struct Dyn 7: 63–73 [DOI] [PubMed] [Google Scholar]
- Pontius J, Wagner L, Schuler G (2003) The NCBI Handbook, chapter 21, UniGene: a unified view of the transcriptome. National Center for Biotechnology Information, Bethesda, MD
- Resing K, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, Old WM, Cheung HT, Russell S, Wattawa JL, Goehle GR, Knight RD, Ahn NG (2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal Chem 76: 3556–3568 [DOI] [PubMed] [Google Scholar]
- von Haller P, Yi E, Donohoe S, Vaughn K, Keller A, Nesvizhskii AI, Eng J, Li XJ, Goodlett DR, Aebersold R, Watts JD (2003) The application of new software tools to quantitative protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry. Mol Cell Proteomics 2: 426–427 [DOI] [PubMed] [Google Scholar]
- Yates JR III, Eng JK, McCormack AL (1995) Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem 67: 3202–3210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures