Abstract
The Helicobacter pylori vacA gene encodes a secreted protein (VacA) that alters the function of gastric epithelial cells and T lymphocytes. H. pylori strains containing particular vacA alleles are associated with differential risk of disease. Because the VacA midregion may exist as one of two major types, m1 or m2, serologic responses may potentially be used to differentiate between patients colonized with vacA m1- or vacA m2-positive H. pylori strains. In this study, we examined the utility of specific antigens from the m regions of VacA as allele-specific diagnostic antigens. We report that serological responses to P44M1, an H. pylori m1-specific antigen, are observed predominantly in patients colonized with m1-positive strains, whereas responses to VacA m2 antigens, P48M2 and P55M2, are observed in patients colonized with either m1- or m2-positive strains. In an Asian-American population, serologic responses to VacA m region-specific antigens were not able to predict the risk of development of gastric cancer.
Helicobacter pylori, a gram-negative microaerophilic bacterium that persistently colonizes the gastric mucosa of human hosts (5), is an important risk factor for the development of peptic ulcer disease and gastric cancer (36). More than 50% of the world's population is colonized by H. pylori, but only a relatively small percentage develops the associated diseases (17). The spectrum of consequences may depend on differences in host responses and/or on the H. pylori genotype. Of the bacterial factors implicated in pathogenesis, the vacuolating cytotoxin (VacA) and CagA, a marker for the pathogenicity island that encodes a type IV secretion system, have been the most extensively studied (7, 43).
In the 3,864-bp vacA open reading frame, which encodes a secreted product, VacA (7), the ∼700-bp midregion exists as two major allelic types, m1 and m2 (3). The m1 and m2 forms of the mature VacA protein differ in the capacity to bind to gastric epithelial cells (24, 33) and differ in functional activity. The m1 and m2 regions are <60% identical in amino acid sequence, while the other regions of the mature protein show >90% identity across a variety of strains (2, 11). There also is diversity in the 5′ vacA region, which encodes a signal sequence and the amino terminus of the secreted toxin (s1 and s2 alleles); vacA s1 H. pylori strains are associated with both gastric cancer and peptic ulcer disease (2, 14). In other investigations, vacA m1 strains have been associated with a higher gastric cancer risk than m2 strains (1, 15, 16, 26).
In prior studies, about 50% of H. pylori-positive patients had detectable serum immunoglobulin G (IgG) antibodies that recognize purified VacA from s1/m1 H. pylori strain 60190 (9, 39); similar seropositivity rates have been reported when whole purified VacA or whole recombinant VacA has been used (13, 48, 49). We previously developed enzyme-linked immunosorbent assays (ELISAs) that use purified VacA antigens from s1/m1-positive or s2/m2-positive strains to distinguish patients colonized with strains of the different VacA subtypes (39). However, the serologic differences were not marked and cross-reactivity was high, presumably reflecting the extensive sequence conservation among the VacA subtypes across the entire secreted protein. Since identifying the VacA subtype in a particular patient currently requires endoscopic biopsies, followed by H. pylori culture and PCR amplification (12, 38), there is a need for noninvasive, simpler assays to diagnose the presence of more-virulent strains.
In this study, we identified and cloned specific vacA sequences that are present in either vacA m1- or vacA m2-positive strains, expressed their recombinant products, and assessed human serologic recognition of these antigens as a means toward the development of serological tests to differentiate between patients colonized with vacA m1 or vacA m2 strains. We also report the use of VacA m region-specific antigens for predicting gastric cancer development in an Asian-American population.
MATERIALS AND METHODS
Bacterial strains and sequences.
H. pylori strain 60190 (ATCC 49503) (vacA s1/m1) was used as a source of DNA encoding m1-specific VacA antigens, and H. pylori strain Tx30a (ATCC 51932) (vacA s2/m2) was used as a source of DNA encoding m2-specific VacA antigens (8, 10, 22). Bacterial strains were grown, and genomic DNA was extracted by the phenol-chloroform method as previously described (47). Previously published nucleotide and amino acid sequences from 22 vacA m1-type strains and 6 vacA m2-type strains were aligned with CLUSTAL_X and GeneDoc, and areas within the m1 and m2 regions specific for the allelic types were identified (27, 41).
Synthesis of VacA peptides.
Synthetic peptides corresponding to allele-specific regions of VacA m1 (VacA479M1, a 15-amino-acid peptide [TNKLAFGPQGSPWGT]) and VacA m2 (VacA632M2, an 18-amino-acid peptide [NIYLGKSTNLRVNGHSAH]) were obtained from Alpha Diagnostics International (San Antonio, TX) (23, 35, 46). The peptides were conjugated in a 2:1 ratio with bovine serum albumin (28) to improve binding to polystyrene plates.
Expression of VacA m1 and m2 antigens.
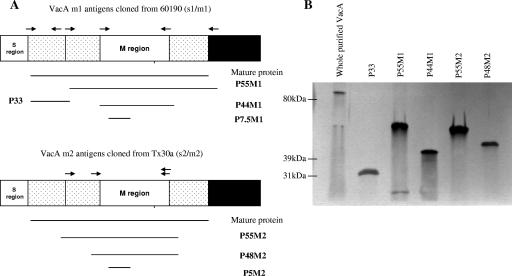
Regions unique to the vacA m1 and m2 alleles of H. pylori were PCR amplified (for the primers used, see Table S1 in the supplemental material). Short segments of the vacA m1 region of strain 60190 P7.5M1 (encoding a 7.5-kDa fragment, corresponding to amino acids F513 to F581) and the vacA m2 region of strain Tx30a P5M2 (encoding a 5-kDa fragment corresponding to amino acids V457 to F509), each of which includes domains of high antigenicity, corresponding to the synthetic peptides described above, were cloned into the XhoI site in pET15b and expressed in Escherichia coli strain BL21DE3 (Novagen, Madison, WI) (Fig. 1; see Table S1 in the supplemental material). Several other putatively antigenic regions encoded by the vacA variants were each cloned by a similar approach, i.e., P44M1 (encoding amino acids V507 to I825 in strain 60190), P48M2 (encoding amino acids V457 to I851 in strain Tx30a), P55M2 (encoding amino acids N390 to I851 in strain Tx30a), P33 (encoding amino acids A1 to A312 in strain 60190), and P55M1 (encoding amino acids A312 to Y821 in strain 60190) (Fig. 1A) (25). In brief, the region corresponding to P33 was amplified by PCR from plasmid pMM592 (including the A1M substitution), digested with SpeI and PstI, and then ligated to XbaI- and PstI-digested pET-41b to create pVT27 (25, 42). The region corresponding to the P55M1 fragment was amplified by PCR from pMM592, digested with XbaI and SalI, and ligated to XbaI- and SalI-digested pET-41b to create pVT30b (New England BioLabs, Beverly, MA) (25, 42). All recombinant antigens included a histidine (His) tag, which allowed purification on Ni-nitrilotriacetic acid agarose resin columns (QIAGEN, Chatsworth, CA). Plasmids pVT27 and pVT30b were transformed into E. coli ER2566 (New England BioLabs), and proteins were expressed in inclusion bodies by inducing the cultures with isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C. Insoluble proteins were then subjected to the Ni-nitrilotriacetic acid protocol for purification of denatured proteins as described by the manufacturer (QIAexpressionist; QIAGEN, Chatsworth, CA). Immunoblotting of the recombinant proteins with rabbit polyclonal anti-VacA s1/m1 antiserum at a 1:5,000 dilution and an anti-rabbit alkaline phosphatase conjugate confirmed expression of VacA proteins of the expected sizes (18). As a control, oligomeric VacA, comprising 88-kDa monomers (amino acids 1 to 821), was purified from H. pylori strain 60190 by gel filtration chromatography as previously described (10).
FIG. 1.
Cloning and expression of VacA antigens. (A) Schematic representation of VacA antigens from vacA s1/m1 strain 60190 and vacA s2/m2 strain Tx30a. From strain 60190, antigens P55M1, P44M1, P33M1, and P7.5M1 were cloned, and from strain Tx30a, antigens P55M2, P48M2, and P5M2 were cloned. (B) Western blot assay of proteins separated on a 10% polyacrylamide gel by using rabbit hyperimmune serum to detect expression of the VacA antigens.
ELISAs for detection of VacA antibodies in serum from rabbits.
To assess the specificity and antigenicity of the recombinant VacA proteins, ELISAs were performed with rabbit hyperimmune serum and 100 ng/well VacA recombinant protein P44M1, P48M2, or P55M2. Sera from adult New Zealand White rabbits immunized with heat-killed or formalin-treated whole cell extracts of H. pylori strains 84-180, 84-182, 84-183, and 85-456 (37) were used at a 1:100 dilution. Hyperimmune serum from adult New Zealand White rabbits immunized with whole purified VacA from strain 60190 (vacA s1/m1), J133 (vacA s1/m2), 86-338 (vacA s2/m2), or Tx30a (vacA s2/m2) (7) and control sera from four nonimmunized rabbits were diluted 1:1,000 and then twofold for use in the antigen-specific ELISAs.
Patient populations.
We studied 174 patients who underwent upper gastrointestinal endoscopy at the New York Harbor VA Medical Center (n = 54) or New York University (NYU) Downtown Hospital (n = 45), both in Manhattan, NY, or at the VA Medical Center in Nashville, TN (n = 75). Of these patients, 77 (44%) were confirmed to be H. pylori negative at endoscopy by rapid urea test, histology, culture, and serology and were used as the source of serum from H. pylori-negative persons. The other 97 patients were H. pylori culture positive and were used as the source of serum to study the utility of the VacA antigens for serodiagnosis. For each of these patients, the vacA s and m types of the isolate were determined by PCR as previously described (3, 44). vacA m1-type H. pylori was present in 32 patients, and a vacA m2-type strain was found in 65 patients. None of these patients had PCR evidence of mixed colonization with vacA m1-type and vacA m2-type strains. Serum samples were collected from another group of 40 patients from the VA Medical Center in Nashville who were H. pylori negative by rapid urea test, histology, culture, and serology. These patients served as a VacA-seronegative reference group (referred to as the reference group). All of the patients included in these studies provided written informed consent, and the protocols used were approved by the Vanderbilt University and NYU School of Medicine Institutional Review Boards.
ELISAs for detecting antibodies to VacA with antigens specific for the m1 and m2 regions.
To assess the diagnostic value of the VacA antigens of interest, we used antigen-specific ELISAs to measure the presence of serum IgG antibodies as previously described (39). The optimal concentration of the VacA antigens and working serum dilutions were determined by checkerboard titration with sera from H. pylori-negative persons and from persons from whom H. pylori strains with a particular vacA m type was isolated. An antigen concentration of 100 ng/well and a 1:100 serum dilution were considered to be optimal in all cases, and each ELISA was done in triplicate.
To establish cutoff values to define seropositivity, we assessed sera from the 40 reference group H. pylori-negative patients. The threshold for seropositivity was defined as an optical density (OD) that was ≥2 standard deviations above the mean OD value for the 40 reference group patients. For each assay, sera from patients who were known to be colonized with a vacA m1-strain (n = 3) or a vacA m2-strain (n = 2) or known to be H. pylori negative (n = 1) were included as additional controls.
Prospective value of VacA serological testing.
To determine whether colonization with H. pylori strains of particular vacA m subtypes is associated with an increased risk of development of gastric cancer, we tested sera from Japanese-American men living in Hawaii who developed gastric cancer and from their matched controls from a nested case-control study that involved 28 years of follow-up (28, 30). The study population consists of 7,498 men born between 1900 and 1919, who were examined between 1967 and 1970 and have been part of the Honolulu Heart study (32); serum samples obtained from 5,924 men (based on an 80% random sample) were stored at −20°C. Between 1971 and 1975, 6,860 of the 7,498 men returned for a second examination and serum samples were collected from 6,813 men. Between the two examinations, serum samples were available from 7,429 individuals. An additional 2,534 men were recruited who were born between 1889 and 1938 and who were identified as brothers of the group who had returned for the second exam. Of the 9,963 men with available blood samples, 28 had received a diagnosis of gastric cancer before their initial exam and were excluded from the study. Of the remaining 9,935 men, 279 developed gastric cancer over the course of a 28-year follow-up to 1996; there were 18 with cancer involving the gastric cardia who were excluded. Among the 261 remaining cases, there were 205 with intestinal gastric cancer, 49 with diffuse cancer, and 7 with unknown types (31). Serum samples were available for the present study from 141 men who developed gastric cancer (114 with intestinal and 27 with diffuse-type gastric cancer). Each case was matched by age at examination and date of serum collection with a control subject from the cohort.
Statistical analysis.
Student's t-test analysis assuming equal variance with Bonferroni's correction was performed; P < 0.05 was considered significant in all comparisons. Receiver operator characteristic (ROC) curves were generated with SPSS software (SPSS Inc., Chicago, IL) (20). From the ROC curves, the area under the curve (AUC) and the sensitivities and specificities associated with using each of the antigens at different cutoff values were calculated (20). A chi-square analysis was performed with the 141 pairs of gastric cancer samples and matched controls to assess the relationships of H. pylori and CagA status with the risk of development of gastric cancer, in comparison with studies using the complete (261-pair) data set (31). Subsequent chi-square analyses compared all gastric cancer patients with all controls, as well as each patient with his matched control. Associations of VacA m types with diffuse and intestinal gastric cancer also were analyzed by chi-square analysis.
RESULTS
Identification of m1- and m2-specific regions in vacA in silico.
In silico analysis of vacA sequences from 22 vacA s1/m1 and 6 vacA s2/m2 H. pylori strains allowed identification of m1-specific and m2-specific regions which are predicted to have hydrophilicity, antigenicity, and surface accessibility properties characteristic of robust antigens (23, 35, 46). In the translated VacA m1 sequences, a 15-amino-acid peptide (TNKLAFGPQGSPWGT) starting at amino acid 479 of the mature VacA m1 protein was identified and designated VacA479m1. The VacA479m1 sequence was present in 18 of the 22 vacA m1-positive strains analyzed. For the VacA m2 strains, an 18-amino-acid peptide (NIYLGKSTNLRVNGHSAH) deduced to start at amino acid 632 of the mature VacA m2 protein was identified and designated VacA632m2. This VacA632m2 sequence was present in all six vacA m2-positive strains. Since BLAST analysis of each peptide only revealed other known m1 and m2 sequences, respectively (e values of 0.065 for VacA479m1 and 0.007 for VacA632m2), these peptides were synthesized as described in Materials and Methods.
Using parallel in silico analyses, we also identified longer m1-specific and m2-specific regions of vacA. The two regions were cloned, and the encoded proteins (P7.5M1 and P5M2), which include the sequences of the two synthetic peptides described previously (Fig. 1A), were expressed. To optimize appropriate protein folding and to favor antigen-antibody binding, we also expressed several larger antigens (recombinant P33, P44M1, P55M1, P48M2, and P55M2) (Fig. 1A).
Assessment of rabbit serum IgG recognition of VacA antigens.
Antigenicity of the VacA recombinant proteins was examined with serum from a rabbit that had been immunized with whole purified VacA from strain 60190 (Fig. 1B) (7). In immunoblot assays, the rabbit serum recognized the VacA m1 and VacA m2 recombinant proteins of the expected sizes (data not shown for P7.5M1 and P5M2). To determine the specificity of antigen recognition, we examined sera from rabbits immunized with H. pylori whole cells. Since VacA is a secreted protein present in relatively small quantities in whole cell preparations of H. pylori, none of the sera from rabbits immunized with H. pylori whole cells recognized the VacA recombinant proteins (data not shown). Next, we assessed antigen recognition with sera from rabbits immunized with whole purified VacA from strains expressing different VacA subtypes. Serum from the rabbit immunized with VacA from strain 60190 (s1/m1) had the strongest reactivity to P44M1, even at a 1:8,000 dilution (see Fig. S2A in the supplemental material). Rabbits immunized with an m2-specific VacA molecule also recognized P44M1, confirming the cross-reactivity observed in the immunoblot experiment (Fig. 1B). Normal rabbit sera did not recognize P44M1.
Sera from rabbits immunized with whole purified VacA from H. pylori m2-positive strains had the strongest reactivity to the two VacA m2 recombinant proteins examined, whereas serum from a VacA m1-immunized rabbit also had reactivity (see Fig. S2B and C in the supplemental material). These results indicate that the VacA proteins produced are antigenic. Although there is extensive cross-reactivity, in general, the homologous m type-specific recognition was stronger than the heterologous recognition.
Human serum IgG responses to synthetic peptides VacA479m1 and VacA632m2 and to recombinant antigens P7.5M1 and P5M2.
We first analyzed the human immune recognition of the synthetic peptides VacA479m1 and VacA632m2 with control serum from H. pylori-negative patients (n = 10) and from patients colonized with vacA m1-positive strains (n = 7) or vacA m2-positive strains (n = 6) (see Fig. S3A in the supplemental material). There was little recognition of the synthetic peptides, compared to that for the whole purified VacA, even at 1 μg/well, indicating the lack of utility of these antigens to assess immune responses to VacA. Next, we analyzed the recognition of the P7.5M1 and P5M2 recombinant antigens, as well as pET15b as a control antigen (see Fig. S3B in the supplemental material). In contrast to whole purified VacA, these small recombinant antigens did not distinguish sera from the different patient groups.
Assessment of human serum IgG response to recombinant VacA antigens P55M1, P44M1, P33, P55M2, and P48M2.
We next examined the immune response to the larger P33 and P55M1 antigens. P55M1 was differentially recognized by sera from H. pylori-negative persons versus H. pylori-positive persons, although the recognition was not as strong as for whole purified VacA (see Fig. S3C in the supplemental material). P33 failed to differentiate between H. pylori-negative and H. pylori-colonized patients. Consequently, we focused subsequent analyses on the p55 region of VacA, which contains the VacA m region.
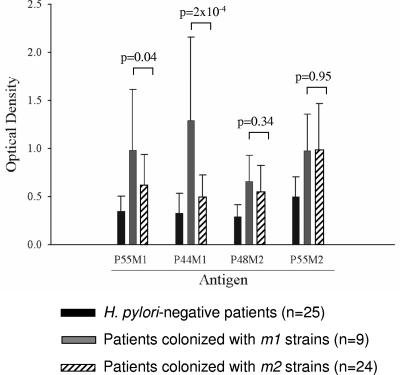
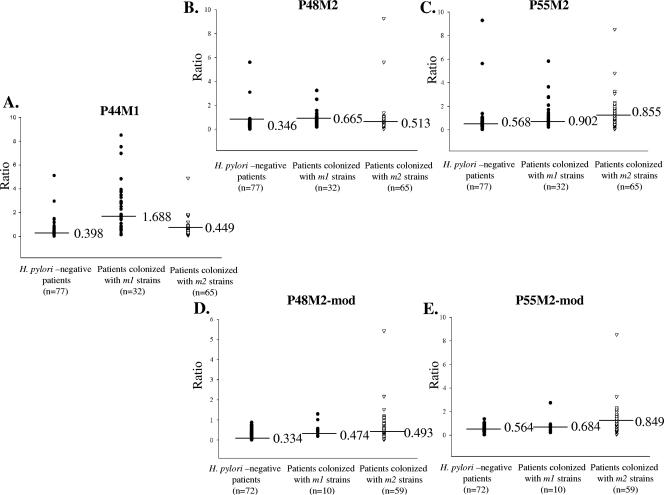
After cloning recombinant proteins P44M1, P48M2, and P55M2 (Fig. 1), we performed ELISAs with P55M1 and these antigens with sera from 25 H. pylori-negative persons and from patients colonized with vacA m1 (n = 9) and vacA m2 (n = 24) strains (Fig. 2). Each of these VacA antigens differentiated between the H. pylori-positive and -negative persons. Since P44M1 was superior to P55M1 in differentiating between patients colonized with vacA m1 and vacA m2 strains, P55M1 was not further studied. For the P48M2 and P55M2 recombinant VacA m2-type proteins, the specificity for patients colonized with m2 versus m1 strains was much lower than for the parallel m1 antigen. To further examine the three possible antigen candidates (P44M1, P48M2, and P55M2), we increased the number of patients whose serum samples were studied to 174 (Fig. 3; Table 1). The P44M1 antigen was able to significantly differentiate patients who were colonized with vacA m1 strains from those who were H. pylori negative (P = 2.0 × 10−10) or who were colonized with vacA m2 strains (P = 8.0 × 10−9) (Fig. 3A and Table 1). However, increasing the sample size did not significantly change the low specificity of the P48M2 and P55M2 antigens for distinguishing between persons with m1 or m2 strains (P = 0.503 and P = 0.791, respectively) (Fig. 3B and C and Table 1).
FIG. 2.
Assessment of serum IgG responses to four recombinant VacA antigens. The antigens examined were P55M1, P44M1, P48M2, and P55M2, and they were evaluated in 58 patients with known H. pylori VacA statuses, as measured by antigen-specific ELISAs.
FIG. 3.
Assessment of serum IgG responses to VacA antigens in 174 patients with known H. pylori/VacA statuses. On the ordinate is shown the ratio of the mean OD value of the sample in relation to the seropositivity threshold. Seropositivity is defined as the mean OD from H. pylori-negative patients plus 2 standard deviations. The abscissa divides the population into H. pylori-negative patients and those colonized with m1 or m2 strains. The median value for each group is indicated by a horizontal bar in the scatter plots. (A to C) Assessment of responses of 174 patients (77 H. pylori negative, 32 m1 colonized, and 65 m2 colonized) to cloned VacA antigens P44M1 (A), P48M2 (B), and P55M2 (C). (D, E) Assessment of responses of 141 patients (72 H. pylori negative, 10 m1 colonized, and 59 m2 colonized) after modification to exclude serum samples that have a positive serological response to cloned VacA m1 antigen P44M1. An assessment of modified responses to VacA antigens P48M2 (D) and P55M2 (E) is shown.
TABLE 1.
Analysis of serologic responses to VacA m1- and m2-specific antigens in 174 patients, according to H. pylori status
| Serum sample group and antigen | OD for samples from patients who were:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
H. pylori negative
|
H. pylori positive
|
||||||||
| m1 colonized
|
m2 colonized
|
||||||||
| Range | Mean | Median | Range | Mean | Median | Range | Mean | Median | |
| Unmodifieda | |||||||||
| P44M1 | 0.028-5.108 | 0.524c | 0.398 | 0.140-8.507 | 2.409c,d | 1.688 | 0.094-4.896 | 0.598d | 0.449 |
| P48M2 | 0.069-5.599 | 0.478e,f | 0.346 | 0.188-3.249 | 0.839e,g | 0.665 | 0.028-5.432 | 0.729f,g | 0.513 |
| P55M2 | 0.023-9.293 | 0.759h | 0.568 | 0.222-5.832 | 1.233i | 0.902 | 0.072-8.546 | 1.642h,i | 0.855 |
| Selected samplesb | |||||||||
| P48M2-mod | 0.009-0.878 | 0.372j,k | 0.334 | 0.188-1.302 | 0.551j,l | 0.474 | 0.028-5.432 | 0.652k,l | 0.493 |
| P55M2-mod | 0.023-1.382 | 0.574m | 0.546 | 0.222-2.754 | 0.800n | 0.684 | 0.072-8.546 | 1.065m,n | 0.849 |
Includes all serum samples from 77 H. pylori-negative patients and 32 m1-colonized and 65 m2-colonized, H. pylori-positive patients.
Excludes samples with a positive serologic response to P44M1 antigen; samples were from 72 H. pylori-negative patients and 10 m1-colonized and 59 m2-colonized, H. pylori-positive patients.
P = 2.0 × 10−10.
P = 8.0 × 10−9.
P = 0.012.
P = 0.047.
P = 0.503.
P = 0.002.
P = 0.791.
P = 0.046.
P = 0.013.
P = 0.669.
P = 0.006.
P = 0.486.
From the experiments performed with rabbit hyperimmune sera specific to a particular VacA subtype, we observed substantial cross-reactivity directed toward the VacA recombinant proteins (see Fig. S2 in the supplemental material), which correlates with the limited accuracy of the diagnostic tests for distinguishing between m1- and m2-specific responses in humans. In particular, serological responses to the VacA m2 antigens were observed in patients colonized with either m1- or m2-positive strains. Therefore, we also examined a reclassification of the serological results based on a new definition; serum samples reacting with the P44M1 antigen were considered to reflect carriage of H. pylori vacA m1-positive strains, regardless of reactivity to an m2-based antigen. By this definition, serum samples that showed a positive response to either of the two VacA m2 antigens but a negative response to P44M1 were considered to reflect carriage of H. pylori vacA m2-positive strains. With this modification, we excluded the results from 33 patients whose samples showed a positive response to the P44M1 antigen, leaving 141 serum samples to be analyzed (Fig. 3D and E). These included 72 from H. pylori-negative patients, 10 from H. pylori vacA m1-positive patients, and 59 from H. pylori vacA m2-positive patients (Table 1). With this modification, the diagnostic value of the VacA m2 antigens improved somewhat, with P55M2 discriminating better than P48M2 (Table 1; Fig. 3D and E). This analysis indicates that serologic responses to VacA antigens occur in some persons colonized with vacA m2-positive strains and that such serologic responses can be detected by assays using VacA m2 antigens.
Analysis of diagnostic accuracy.
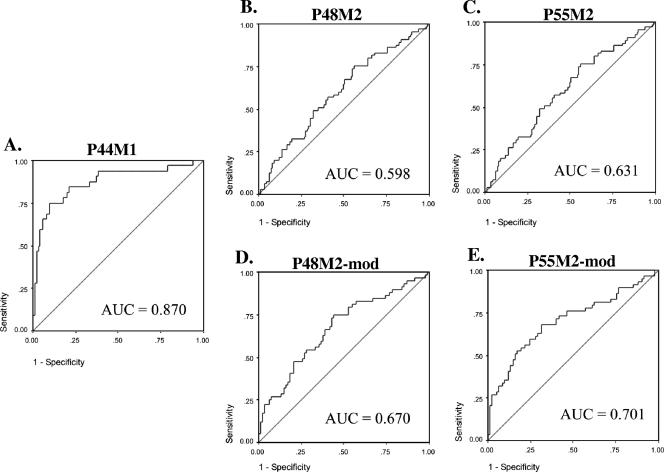
The diagnostic accuracy of the ELISAs using the P44M1, P48M2, and P55M2 antigens was examined by ROC analyses (20). For the P44M1 antigen, the AUC was 0.87, whereas for the P48M2 and P55M2 antigens, the AUCs were 0.60 and 0.63, respectively (Fig. 4A, B, and C). Thus, the P44M1 antigen has the greatest diagnostic value in differentiating between individuals colonized with H. pylori m1-positive strains and those who are H. pylori negative or colonized with H. pylori m2-positive strains. The m2-based antigens P48M2 and P55M2 did not perform as well in differentiating persons with m2 strains from others.
FIG. 4.
ROCs of VacA antigen-specific ELISAs. For each ROC, the AUC is shown. Sensitivity is shown on the ordinate, and (1 − specificity) is shown on the abscissa. The ROC curves in panels A to C are based on 174 patients, and those in panels D and E are based on 141 patients with the modification of excluding H. pylori-positive persons with responses to the P44M1 antigen. The antigens used in the ELISAs were P44M1 (A), P48M2 (B and D), and P55M2 (C and E). Panels D and E show AUCs for the modified analyses.
The ROC analyses permitted examination of the sensitivity and specificity of the three antigens. With a cutoff of 1.0 for the P44M1 antigen, the sensitivity and specificity for the antigen were 69% and 92%, respectively (Table 2). For the P48M2 and P55M2 antigens, the diagnostic accuracy with a predetermined threshold (0.6) was lower, with sensitivities of 35% and 74%, respectively, and specificities of 73% and 47%, respectively (Table 2). Excluding the subjects positive for anti-m1 antibodies, the AUCs for the P48M2 and P55M2 antigens were 0.67 and 0.70, respectively (Fig. 4C and D). When the subjects positive for anti-m1 antibodies were excluded, the sensitivities of the assays for detecting antibody responses to VacA m2-based antigens decreased slightly (from 35 to 31% for P48M2 and from 74 to 72% for P55M2), but the specificity of the assays improved to a greater extent (from 73 to 85% for P48M2 and from 47 to 59% for P55M2).
TABLE 2.
Diagnostic accuracy of VacA antigens at predetermined cutoff values
| VacA antigen and analysis | Sample characteristics | AUCc | Cutoff valued | % Sensitivity | % Specificity |
|---|---|---|---|---|---|
| Unmodifieda | |||||
| P44M1 | 32 m1 colonized, 142 not m1 colonized | 0.870 | 0.8 | 75 | 89 |
| 1.0 | 69 | 92 | |||
| P48M2 | 65 m2 colonized, 109 not m2 colonized | 0.598 | 0.6 | 35 | 73 |
| 0.8 | 26 | 86 | |||
| P55M2 | 65 m2 colonized, 109 not m2 colonized | 0.631 | 0.6 | 74 | 47 |
| 0.8 | 54 | 70 | |||
| Modifiedb | |||||
| P48M2-mod | 59 m2 colonized, 82 not m2 colonized | 0.670 | 0.6 | 31 | 85 |
| 0.8 | 22 | 96 | |||
| P55M2-mod | 59 m2 colonized, 82 not m2 colonized | 0.701 | 0.6 | 72 | 59 |
| 0.8 | 52 | 82 |
Includes all serum samples from 174 patients.
Excludes samples with a positive serologic response to P44M1 antigen; samples were from 141 patients.
AUC in ROC analysis (19).
The cutoff value is the ratio of the OD value to the seropositivity cutoff value. Values greater than or equal to the cutoff are considered to indicate immune responses to the specific antigen.
Analysis of the prevalence of VacA m subtypes by the VacA m-specific ELISAs in gastric cancer patients and controls.
In a prior study of 261 gastric cancer patients and controls, the odds ratio for the development of gastric cancer to the presence of H. pylori was 2.4 (95% confidence interval, 1.1 to 4.0). For the present analysis, for the 141 of these pairs that were available, the odds ratio observed for the development of gastric cancer associated with the presence of H. pylori was 2.14 (95% confidence interval, 1.15 to 4.05) (31), indicating that this group could be used to study the utility of anti-VacA antibodies to understand cancer risk. Analysis based on the 141 paired gastric cancer patients and controls showed no association of cancer risk and the measured anti-VacA responses (Table 3). For example, among the 141 pairs of patients and controls, 42% of the controls and 47% of the patients had positive serologic responses to P44M1. For the two VacA m2 antigens, there also was no association of gastric cancer and a measured VacA m2 response. Paired analysis based on cancer subtypes, either intestinal or diffuse, did not show any association with the VacA m-specific antigens (see Table S2 in the supplemental material).
TABLE 3.
Prevalence of vacA m subtypes by vacA m-specific ELISAs in serum obtained from gastric cancer patients and matched controls
| VacA antigen and analysis and study subject characteristic(s)a | No. of pairs | % Positivec
|
|
|---|---|---|---|
| Controls | Patients | ||
| P44M1 | |||
| All | 141 | 42 | 47 |
| All H. pylori positive | 87 | 52 | 48 |
| All H. pylori or CagA positive | 90 | 51 | 48 |
| All H. pylori positive and CagA positive | 47 | 53 | 53 |
| P44M2-modifiedb | |||
| All | 45 | 20 | 15 |
| All H. pylori positive | 23 | 26 | 30 |
| All H. pylori or CagA positive | 24 | 25 | 29 |
| All H. pylori positive and CagA positive | 11 | 18 | 27 |
| P55M2-modifiedb | |||
| All | 45 | 47 | 47 |
| All H. pylori positive | 23 | 61 | 61 |
| All H. pylori or CagA positive | 24 | 62 | 62 |
| All H. pylori positive and CagA positive | 11 | 54 | 54 |
Characteristics based on prior analyses (30).
Excludes samples with a positive serologic response to P44M1 antigen.
All P > 0.5.
DISCUSSION
Among diverse H. pylori strains, the peptides encoded by the vacA m1 and m2 regions have less than 60% amino acid sequence identity, whereas the rest of the deduced protein has more than 90% identity (7). In this study, we have exploited this variation to develop serological assays with the aim of differentiating between patients colonized with either vacA m1 and those colonized with vacA m2 H. pylori.
We first performed ELISAs with small synthetic or recombinant peptides (P7.5M1 and P5M2) that contain VacA m regions of the highest predicted antigenicity so as to differentiate between the two groups. The lack of sensitivity and specificity of these peptides suggests that they are too small for proper folding of the protein to reveal specific epitopes or that too few epitopes are present.
With a larger antigen, the P44M1 protein, whose coding sequence spans the vacA m1 region, there was better differentiation of those with m1 strains from persons who were H. pylori negative and or were colonized with an m2 strain (specificity = 92%). The sensitivity of 69% was higher than the reported sensitivity of whole purified VacA from strain 60190 (39), and the AUC of 0.87 suggests that an ELISA based on P44M1 may be a useful diagnostic tool. Although all H. pylori strains contain vacA, the VacA product may not be expressed because of point mutations, frameshifts, or insertions and deletions within the gene (21). Therefore, among patients colonized with vacA m1 strains, some may be carrying strains that do not actually express VacA; therefore, the sensitivity of the assay in relation to VacA m1-expressing strains may be greater than indicated. Multiple H. pylori strains expressing different vacA alleles might colonize an individual host. This would not affect the sensitivity or specificity of the VacA P44M1 assay since chimeric vacA alleles also exist (3, 34, 40, 45). Substantial sequence diversity in these vacA chimeras from the classical m1 and m2 forms might lead to a lack of serologic responses to the prototypic m1 and m2 antigens. The serological responses to the P44M1 antigen vary greatly (Fig. 3), as reported for antibodies to the whole VacA protein (8), representing underlying biological variation.
To improve upon the initial results obtained when the vacA m2-based antigens P48M2 and P55M2 were used, we excluded serum samples that were m1 positive. Studies with immunized rabbits (see Fig. S2 in the supplemental material) showed that, despite the m1/m2 sequence differences, VacA proteins possess conserved epitopes that can explain the apparent cross-reactivity of the VacA m1- and m2-based antigens. With the m1 exclusion, the diagnostic value of the VacA m2 antigens improved somewhat, with P55M2 discriminating better than P48M2. However, the current assays using vacA m2-based antigens do not have sufficient accuracy to be used for an m type-specific diagnosis in patients. It is possible that the accuracy of a VacA m2-based ELISA could be improved by using native m2 antigens rather than the recombinant antigens described in this report.
Multiple H. pylori strains expressing different vacA m-specific alleles may colonize an individual host, a phenomenon that appears especially common in populations from developing countries (19). A stand-alone diagnostic tool such as the VacA P44M1-specific ELISA would be able to identify persons colonized with vacA m1-specific strains whether or not they are also colonized with vacA m2-specific strains. In contrast, the current assays using vacA m2-based antigens do not have sufficient accuracy to be used for an m type-specific diagnosis in patients colonized with either a single vacA m2-positive strain or multiple strains with different m types.
H. pylori colonization has been associated with gastric cancer in both cohort and case-control studies (29, 31). Most studies that have reported an association between H. pylori and gastric cancer have used serological assays with pools of antigenic preparations from the bacteria. Since studies based on H. pylori culture show an association of the vacA m1 genotype with gastric cancer, development of an accurate serologic test would aid in diagnosis (26). A cohort of Japanese-American men, a group at high risk for gastric cancer, has been used to address questions about the association of H. pylori and its virulence factors with a risk for developing upper gastrointestinal diseases (29-32). The several advantages of studying this group include that serum was collected from individuals before they were diagnosed with gastric cancer, and it is a large, well-studied, and homogeneous group, which diminishes confounding factors. However, our inability to observe correlations between vacA m1 and m2 types and gastric cancer in this group may not be surprising. The patients in the cohort studied originate from Japan, mostly from Hiroshima Province, where the circulating H. pylori strains are mostly cagA positive and vacA s1/m1 positive (39, 40). Thus, there likely was not sufficient power to detect m1 type-specific associations with disease. An analysis with VacA antigens cloned from a strain isolated from a person of Japanese origin also might clarify our results, which are based on antigens cloned from a Western strain (60190). Several other studies in Japan also have not correlated vacA subtypes with gastrointestinal diseases, possibly for similar reasons (4, 6). Studies with populations where there are high proportions of both m1 and m2 strains (such as among Caucasians in Europe or in the United States) in circulation might be better able to address whether m1-type vacA strains are associated with an enhanced gastric cancer risk.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01GM63270, R01AI39657, RO3 CA099512), the Medical Research Service of the Department of Veterans Affairs, the Institute for Urban and Global Health (NYU), the Thrasher Foundation, and the Diane Belfer Program for Human Microbial Ecology in Health and Disease.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Ashour, A. A., P. P. Magalhaes, E. N. Mendes, G. B. Collares, V. R. de Gusmao, D. M. Queiroz, A. M. Nogueira, G. A. Rocha, and C. A. de Oliveira. 2002. Distribution of vacA genotypes in Helicobacter pylori strains isolated from Brazilian adult patients with gastritis, duodenal ulcer or gastric carcinoma. FEMS Immunol. Med. Microbiol. 33:173-178. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., P. M. Sharp, T. L. Cover, G. Gonzalez-Valencia, R. M. Peek, Jr., S. A. Thompson, C. J. Hawkey, and M. J. Blaser. 1999. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr. Microbiol. 39:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuma, T., S. Kato, W. Zhou, S. Yamazaki, A. Yamakawa, M. Ohtani, S. Fujiwara, T. Minoura, K. Iinuma, and T. Kato. 2004. Diversity of vacA and cagA genes of Helicobacter pylori in Japanese children. Aliment. Pharmacol. Ther. 20(Suppl. 1):7-12. [DOI] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J., K. Kobayashi, T. L. Cover, P. Cao, I. D. Feurer, and G. I. Perez-Perez. 1993. Helicobacter pylori infection in Japanese patients with adenocarcinoma of the stomach. Int. J. Cancer 55:799-802. [DOI] [PubMed] [Google Scholar]
- 7.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 8.Cover, T. L., P. Cao, C. D. Lind, K. T. Tham, and M. J. Blaser. 1993. Correlation between vacuolating cytotoxin production by Helicobacter pylori isolates in vitro and in vivo. Infect. Immun. 61:5008-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cover, T. L., P. Cao, U. K. Murthy, M. S. Sipple, and M. J. Blaser. 1992. Serum neutralizing antibody response to the vacuolating cytotoxin of Helicobacter pylori. J. Clin. Investig. 90:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover, T. L., P. I. Hanson, and J. E. Heuser. 1997. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J. Cell Biol. 138:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cover, T. L., M. K. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566-10573. [PubMed] [Google Scholar]
- 12.Cutler, A. F., S. Havstad, C. K. Ma, M. J. Blaser, G. I. Perez-Perez, and T. T. Schubert. 1995. Accuracy of invasive and noninvasive tests to diagnose Helicobacter pylori infection. Gastroenterology 109:136-141. [DOI] [PubMed] [Google Scholar]
- 13.Donati, M., S. Moreno, E. Storni, A. Tucci, L. Poli, C. Mazzoni, O. Varoli, V. Sambri, A. Farencena, and R. Cevenini. 1997. Detection of serum antibodies to CagA and VacA and of serum neutralizing activity for vacuolating cytotoxin in patients with Helicobacter pylori-induced gastritis. Clin. Diagn. Lab. Immunol. 4:478-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, K. A., T. L. Cover, M. K. Tummuru, M. J. Blaser, and S. Krakowka. 1997. Role of vacuolating cytotoxin in gastritis due to Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 65:3462-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figueiredo, C., W. Quint, N. Nouhan, H. van den Munckhof, P. Herbrink, J. Scherpenisse, W. de Boer, P. Schneeberger, G. Perez-Perez, M. J. Blaser, and L. J. van Doorn. 2001. Assessment of Helicobacter pylori vacA and cagA genotypes and host serological response. J. Clin. Microbiol. 39:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueiredo, C., L. J. Van Doorn, C. Nogueira, J. M. Soares, C. Pinho, P. Figueira, W. G. Quint, and F. Carneiro. 2001. Helicobacter pylori genotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand. J. Gastroenterol. 36:128-135. [DOI] [PubMed] [Google Scholar]
- 17.Frenck, R. W., Jr., and J. Clemens. 2003. Helicobacter in the developing world. Microbes Infect. 5:705-713. [DOI] [PubMed] [Google Scholar]
- 18.Garner, J. A., and T. L. Cover. 1996. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect. Immun. 64:4197-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghose, C., G. I. Perez-Perez, L. J. van Doorn, M. G. Dominguez-Bello, and M. J. Blaser. 2005. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J. Clin. Microbiol. 43:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greiner, M., D. Pfeiffer, and R. D. Smith. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 45:23-41. [DOI] [PubMed] [Google Scholar]
- 21.Ito, Y., T. Azuma, S. Ito, H. Suto, H. Miyaji, Y. Yamazaki, Y. Kohli, and M. Kuriyama. 1998. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J. Infect. Dis. 178:1391-1398. [DOI] [PubMed] [Google Scholar]
- 22.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 23.Lo Conte, L., C. Chothia, and J. Janin. 1999. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285:2177-2198. [DOI] [PubMed] [Google Scholar]
- 24.Massari, P., R. Manetti, D. Burroni, S. Nuti, N. Norais, R. Rappuoli, and J. L. Telford. 1998. Binding of the Helicobacter pylori vacuolating cytotoxin to target cells. Infect. Immun. 66:3981-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClain, M. S., and T. L. Cover. 2003. Expression of Helicobacter pylori vacuolating toxin in Escherichia coli. Infect. Immun. 71:2266-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miehlke, S., C. Kirsch, K. Agha-Amiri, T. Gunther, N. Lehn, P. Malfertheiner, M. Stolte, G. Ehninger, and E. Bayerdorffer. 2000. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer. 87:322-327. [PubMed] [Google Scholar]
- 27.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet. News 4:14. [Google Scholar]
- 28.Nieto, A., A. Gaya, C. Moreno, M. Jansa, and J. Vives. 1986. Adsorption-desorption of antigen to polystyrene plates used in ELISA. Ann. Inst. Pasteur Immunol. 137C:161-172. [DOI] [PubMed] [Google Scholar]
- 29.Nomura, A., G. N. Stemmermann, P. H. Chyou, I. Kato, G. I. Perez-Perez, and M. J. Blaser. 1991. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 325:1132-1136. [DOI] [PubMed] [Google Scholar]
- 30.Nomura, A., G. N. Stemmermann, P. H. Chyou, G. I. Perez-Perez, and M. J. Blaser. 1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med. 120:977-981. [DOI] [PubMed] [Google Scholar]
- 31.Nomura, A. M., J. Lee, G. N. Stemmermann, R. Y. Nomura, G. I. Perez-Perez, and M. J. Blaser. 2002. Helicobacter pylori CagA seropositivity and gastric carcinoma risk in a Japanese American population. J. Infect. Dis. 186:1138-1144. [DOI] [PubMed] [Google Scholar]
- 32.Nomura, A. M., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser. 2002. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155:1054-1059. [DOI] [PubMed] [Google Scholar]
- 33.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. USA 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 35.Parker, J. M., D. Guo, and R. S. Hodges. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425-5432. [DOI] [PubMed] [Google Scholar]
- 36.Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Pérez, G. I., and M. J. Blaser. 1987. Conservation and diversity of Campylobacter pyloridis major antigens. Infect. Immun. 55:1256-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Pérez, G. I., A. F. Cutler, and M. J. Blaser. 1997. Value of serology as a noninvasive method for evaluating the efficacy of treatment of Helicobacter pylori infection. Clin. Infect. Dis. 25:1038-1043. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Pérez, G. I., R. M. Peek, Jr., J. C. Atherton, M. J. Blaser, and T. L. Cover. 1999. Detection of anti-VacA antibody responses in serum and gastric juice samples using type s1/m1 and s2/m2 Helicobacter pylori VacA antigens. Clin. Diagn. Lab. Immunol. 6:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strobel, S., S. Bereswill, P. Balig, P. Allgaier, H. G. Sonntag, and M. Kist. 1998. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J. Clin. Microbiol. 36:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres, V. J., S. E. Ivie, M. S. McClain, and T. L. Cover. 2005. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J. Biol. Chem. 280:21107-21114. [DOI] [PubMed] [Google Scholar]
- 43.Tummuru, M. K., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61:1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H. J., C. H. Kuo, A. A. Yeh, P. C. Chang, and W. C. Wang. 1998. Vacuolating toxin production in clinical isolates of Helicobacter pylori with different vacA genotypes. J. Infect. Dis. 178:207-212. [DOI] [PubMed] [Google Scholar]
- 46.Welling, G. W., W. J. Weijer, R. van der Zee, and S. Welling-Wester. 1985. Prediction of sequential antigenic regions in proteins. FEBS Lett. 188:215-218. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, K. 1995. Preparation of genomic DNA from bacteria, p. 2.4.1.-2.4.5. In R. B. F. M. Ausubel, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 48.Yan, J., and Y. F. Mao. 2004. Construction of a prokaryotic expression system of vacA gene and detection of vacA gene, VacA protein in Helicobacter pylori isolates and anti-VacA antibody in patients' sera. World J. Gastroenterol. 10:985-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, J., Y. F. Mao, and Z. X. Shao. 2005. Frequencies of the expression of main protein antigens from Helicobacter pylori isolates and production of specific serum antibodies in infected patients. World J. Gastroenterol. 11:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.