Abstract
Polyketide synthases (PKSs) and/or nonribosomal peptide synthetases (NRPSs) are central components of secondary metabolism in bacteria, plants, and fungi. In filamentous fungi, diverse PKSs and NRPSs participate in the biosynthesis of secondary metabolites such as pigments, antibiotics, siderophores, and mycotoxins. However, many secondary metabolites as well as the enzymes involved in their production are yet to be discovered. Both PKSs and NRPSs require activation by enzyme members of the 4′-phosphopantetheinyl transferase (PPTase) family. Here, we report the isolation and characterization of Aspergillus nidulans strains carrying conditional (cfwA2) and null (ΔcfwA) mutant alleles of the cfwA gene, encoding an essential PPTase. We identify the polyketides shamixanthone, emericellin, and dehydroaustinol as well as the sterols ergosterol, peroxiergosterol, and cerevisterol in extracts from A. nidulans large-scale cultures. The PPTase CfwA/NpgA was required for the production of these polyketide compounds but dispensable for ergosterol and cerevisterol and for fatty acid biosynthesis. The asexual sporulation defects of cfwA, ΔfluG, and ΔtmpA mutants were not rescued by the cfwA-dependent compounds identified here. However, a cfwA2 mutation enhanced the sporulation defects of both ΔtmpA and ΔfluG single mutants, suggesting that unidentified CfwA-dependent PKSs and/or NRPSs are involved in the production of hitherto-unknown compounds required for sporulation. Our results expand the number of known and predicted secondary metabolites requiring CfwA/NpgA for their biosynthesis and, together with the phylogenetic analysis of fungal PPTases, suggest that a single PPTase is responsible for the activation of all PKSs and NRPSs in A. nidulans.
Filamentous fungi represent a rich and diverse source of bioactive compounds derived from secondary metabolism. Indeed, many positive and negative effects that fungi have on human activity are mediated by secondary metabolites such as antibiotics and other pharmaceutical drugs, mycotoxins, or pathogen virulence factors (22, 30, 54-56, 60, 66). In contrast to primary metabolism, common to most living organisms, secondary metabolism is not essential for the immediate survival of the producing organism. Furthermore, different taxonomic groups produce different types of secondary metabolites.
Despite their large chemical diversity, secondary metabolites can be grouped according to their primary metabolism precursors. Acetyl coenzyme A, shikimate, and amino acids are major secondary metabolite building units. Acetyl coenzyme A is used to produce terpenoids, steroids, carotenoids, and polyketides. Shikimate is a precursor of aromatic compounds, while diverse peptides are derived from amino acids (30, 56).
Most polyketide and peptide secondary metabolites are produced by complex enzymes called polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively (15, 29). Although there are many different types of PKSs and NRPSs, they all require a posttranslational modification to become active and therefore share a common point of regulation. This modification consists of the covalent attachment of the 4′-phosphopantetheine moiety of coenzyme A to a serine conserved in all acyl carrier and peptidyl carrier domains present in PKSs and NRPSs, respectively. This enzymatic activation is carried out by members of the 4′-phosphopantetheinyl transferase (PPTase) family (19, 34, 47, 63).
Three major groups of PPTases have been defined according to their primary sequences and substrate specificities. The first group includes small (120 to 140 amino acids) bacterial AcpS-type PPTases showing narrow protein substrate specificity usually associated with primary metabolism. A second group contains eukaryotic PPTases that are integral domains of type I fatty acid synthases. The third class consists of the Sfp-type PPTases, about twice the molecular mass of AcpS-type enzymes, which show broader substrate preferences and have been associated with secondary metabolism in bacteria (34, 41). Three PPTases in Saccharomyces cerevisiae and Candida albicans have been characterized. While one enzyme is part of a cytoplasmic fatty acid synthase, a second one is related to a mitochondrial acyl carrier protein. A third PPTase is essential for activating the enzyme α-aminoadipate reductase (AAR) and is thus indispensable for lysine biosynthesis (12, 14).
The fungus Aspergillus nidulans has been established as a model system to study different aspects of eukaryotic biology, including development and secondary metabolism (1, 9, 30, 36, 52, 57, 66). A. nidulans is closely related to species of economical or pathological importance, e.g., Aspergillus niger, Aspergillus oryzae, Aspergillus terreus, and Aspergillus fumigatus (18). In addition, pathways involved in the production of pigments (17, 40), penicillin, and the mycotoxin sterigmatocystin (ST), the immediate precursor of aflatoxins, have been well studied in this fungus (24, 30, 66). Notably, it has been shown that asexual development (conidiation) is linked to secondary metabolism, as FadA, the α-subunit of a heterotrimeric G protein, negatively regulates conidiation and ST biosynthesis (24).
A. nidulans asexual development depends on the activity of the transcription factor BrlA and is characterized by the production of multicellular structures called conidiophores, which form large chains of asexual spores or conidia (1, 9, 57). The characterization of “fluffy” mutants showing delayed conidiation and brlA expression has led to the study of fluG (37, 38, 50, 64) and tmpA (52), two genes that are independently required for conidiation. fluG encodes a protein showing similarity to prokaryote-type glutamine synthetases, whereas tmpA encodes a member of a family of putative membrane flavoproteins conserved in plants and fungi. The overexpression of either protein induces conidiophore development under conditions that normally repress this process, and the conidiation defects of null mutants in either gene can be rescued by growing them next to wild-type or other developmental mutants. This has led to the proposal that both genes are required for the production of different extracellular sporulation-inducing compounds (37, 38, 50, 52).
Based on the genome sequence, it has been estimated that A. nidulans contains 27 PKSs and 14 NRPSs (18, 30), but the functions and products of most of these enzymes are still unknown. It has been reported that cfwA2 partial-function mutants are impaired in the biosynthesis of the PKS product sterigmatocystin (21), the NRPS products penicillin (31) and siderophores, as well as the amino acid lysine (44). cfwA is an allele (31; this work) of the npgA gene (23, 32), which has been shown to encode a PPTase (42).
Here, we report the isolation of the conditional cfwA2 mutant and the generation of complete-lack-of-function (ΔcfwA) mutants. We use these mutants, together with a detailed chemical analysis, to demonstrate that the A. nidulans PPTase CfwA/NpgA is involved in a wide spectrum of processes, including the production of the metabolites shamixantone and dehydroaustinol, not reported previously for this fungus. Our results demonstrate a novel role of this PPTase in the regulation of asexual reproduction and suggest that all PKSs and NRPSs are activated by a single PPTase in A. nidulans and probably also in other fungi. Furthermore, our analysis suggests that a similar mutant approach can be used as a novel and powerful tool to study the roles of secondary metabolism in fungi of medical, agricultural, or biotechnological importance.
MATERIALS AND METHODS
cfwA2 cloning and sequencing and cfwA deletion.
Primers cfwA1 (CCTTCCACAAGCCTTACC) and cfwA2 (TCAATGCAATCAGTTCGC) were used to amplify a 1,146-bp cfwA/npgA PCR fragment using genomic DNA from wild-type FGSC26 or cfwA2 CRO1 mutant strains as a template. The PCR products include the entire cfwA/npgA open reading frame and were cloned into plasmid pCR2.1-TOPO (Invitrogen, Carlsbad, CA). Two independent clones from each template were sequenced by automatic fluorescence dideoxy sequencing using an ABI Prism 310 sequencer from Perkin-Elmer (Wellesley, MA). To delete the cfwA gene, a replacement construct was generated by double-joint PCR (65) using genomic DNA as a template. First, a 1,516-bp 5′ cfwA fragment was amplified with primers 1cfwA (GGCGGTTACGGGCGAGCATACG) and 2cfwA (ATTTTAATCCCATGTGATCAAACGAGCCAGATGACTAAAGAGAAATGGTAAGG). Second, a 1,703-bp 3′ cfwA fragment was amplified with primers 3cfwA (CGTCACACTCATGTAACGGTTCTGCAGCGCAGAATAGATGACGTTAAAGCTGC) and 4cfwA (AACCAAAAAGATTCAAGACCTG). Third, the A. fumigatus riboB marker was amplified as a 1,943-bp fragment with primers 5ribo (CTGGCTCGTTTGATCACATGG) and 6ribo (GCGCTGCAGAACCGTTACATG) using plasmid pAfriboPstE1Skt(ssp1)-37 as a template. The three PCR fragments were purified, mixed, and subjected to fusion PCR with nested primers 7dcfwA (GTCTCACGGCAACAATATCACC) and 8dcfwA (CATTTGCCATTCGTTGACTTGC). The final 4,640-bp cfwA-A. fumigatus riboB (AfriboB)-cfwA cassette was purified with a QIAquick gel purification kit (QIAGEN, Hilden, Germany) and used to transform A. nidulans strain 11035 (43) or A770. Transformed protoplasts were plated onto minimal medium supplemented with 10 mM lysine and 50% siderophore-containing conditioned medium.
Strains, media, and growth conditions.
The A. nidulans strains used in this work are listed in Table 1. All strains were grown in glucose minimal nitrate medium (25) plus supplements. To prepare siderophore-containing medium, 1 × 109 conidiospores from strain FGSC26 were used to inoculate Pontecorvos's liquid minimal medium with 20 mM glutamine as the sole nitrogen source and without iron (13, 46). After 48 h, mycelium was removed by filtration, and the medium was mixed 1:1 with fresh minimal medium (25) plus supplements and autoclaved. The presence of siderophores was confirmed by the chrome azurol S liquid assay (45). Pure siderophore triacetylfusarine C (ECM Microcollections, Tübingen, Germany) was used in some experiments. Massive solid cultures of strains CLS2 (cfwA2) and CLS12 (cfwA+) were carried out using polyurethane foam (PUF) cubes of ∼0.5 cm per side, which were prepared and sterilized as reported previously (11). A total of 25 to 50 ml of supplemented liquid minimal medium with 5% glucose was inoculated with 106 spores per ml and used to saturate 1 to 2 g of PUF in 250- to 500-ml flasks. These cultures were incubated for 10 days at 32 to 34°C. Flasks containing the same medium and support but without spores were monitored as contamination controls. Samples from these cultures were examined under a microscope every 24 h to monitor growth, development, and lack of microbial contamination.
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotype | Source and/or reference |
|---|---|---|
| FGSC26 | biA1 veA1 | FGSC |
| CJA15.3 | cfwA2 biA1 metG1 brlA::lacZ::argB veA1 | This work, UV mutagenesis of TJA22 |
| TJA22 | biA1 metG1 brlA(p)::lacZ::argB veA1 | 52 |
| CRO1 | cfwA2 pabaA1 yA2 veA1 | This work, progeny from CJA15.3 × RMS011 |
| WX24 | biA1 npgA1 sB3 chaA1 trpC801 veA1 | K. Jahng |
| AJC7.1 | biA1 brlA1 veA1 | J. Clutterbuck |
| NK002 | pabaA1 yA2 biA1 wA1 trpC801 veA1 | M. Mayorga |
| TJH3.40 | biA1 wA3 metG1 ΔstcE::argB veA1 | 7 |
| CLK43 | pabaA1 yA2 veA1 | 28 |
| 11035 | pyrG89 pyroA4 ΔnkuA::argB riboB2 veA1a | M. Hynes; 43 |
| PW-1 | biA1 argB2 metG1 veA1 | P. Weglenski |
| A770 | pyrG89 pabaB22 riboB2 | FGSC |
| RBN119 | biA1 yA2 ΔfluG::trp, argB2 metG1 veA1 | 64 |
| CGS49 | pabaA1 yA2 ΔfluG::trpC veA1 | G. Soid, progeny from RBN119 × CLK43 |
| TGS6 | pabaA1 yA2 ΔargB::trpCΔB trpC801 ΔtmpA::argB veA1 | 52 |
| CLS2 | cfwA2 pabaA1 wA3 ΔstcE::argB veA1 | This work, progeny from CRO1 × TJH3.40 |
| CLS5 | cfwA2 biA1 ΔstcE::argB veA1 | This work, progeny from CRO1 × TJH3.40 |
| CLS8 | cfwA2 biA1 metG1 stcE::argB veA1 | This work, progeny from CRO1 × TJH3.40 |
| CLS12 | pabaA1 wA3 stcE::argB veA1 | This work, progeny from CRO1 × TJH3.40 |
| CLSN12 | npgA1 pabaA1 trpC801 veA1 | This work, progeny from WX24 × RMS011 |
| CJR5 | cfwA2 pabaA1 trpC801 veA1 | This work, progeny from CLS8 × CLSN12 |
| TJRΔcfwA1 | ΔcfwA::AfriboB pyrG89 pyroA4 ΔnkuA::argB riboB2 veA1a | This work, 11035 transformed with PCR construct cfwA-AfriboB-cfwA |
| TJRΔcfwA2 | ΔcfwA::AfriboB pyrG89 pyroA4 ΔnkuA::argB riboB2 veA1a | This work, 11035 transformed with PCR construct cfwA-AfriboB-cfwA |
| CJRΔtmpAcfwA2-4 | cfwA2 pabaA1 yA2 ΔtmpA::argB veA1 | This work, progeny from CLS5 × TGS6 |
| CJRΔfluGcfwA2-4 | cfwA2 pabaA1 yA2 ΔfluG::trpC veA1 | This work, progeny from CRO1 × RBN119 |
Partial genotype; it may contain argB2.
Extraction of secondary metabolites.
After the indicated incubation times, the cultures (PUF plus mycelia plus fermentation broth) were collected in a single container and incubated with ethyl acetate for 1 month. After this, the liquid phase was separated from the fungal biomass and PUF by vacuum filtration using Whatman paper (Whatman Inc., NJ). The aqueous and solvent phases were separated by solubility difference, and the solvent was evaporated under reduced pressure. The aqueous phase was reextracted with ethyl acetate three times, and extracts were pooled. The extracts were analyzed by thin-layer chromatography (TLC) on silica gel plates (Merck 60 GF254, 0.25-mm thickness) using different mixtures of n-hexane-ethyl acetate.
Isolation and identification of secondary metabolites.
The composite extracts of cultures from each strain were subjected to gravitatory column chromatography using silica gel (Kieselgel 60, particle sizes of 0.063 to 0.200 mm, 0.040 to 0.063 mm, and 0.2 to 0.5 mm; Merck) as the stationary phase and eluted with n-hexane and n-hexane-ethyl acetate in a polarity gradient. The isolated compounds were purified by column chromatography and recrystallized with organic dissolvents. From the CLS12 extract, elution with hexane-ethyl acetate (95:5, 8:2, 7:3, and 5:5) yielded compounds 1, 2, 3 (18, 26, and 35 mg), 4 (45 mg), 5, and 6 (52 and 35 mg). From the CLS2 extract, elution with hexane-ethyl acetate (8:2 and 5:5) yielded compounds 3 (52 mg) and 6 (6 mg). Nuclear magnetic resonance (NMR) spectroscopic data and comparison with authentic samples (58, 59) identified compounds 3, 4, and 6 as being ergosterol, ergosterol peroxide, and cerevisterol, respectively.
Melting-point analysis was carried out with a Fisher-Johns apparatus, and data were not corrected. 1H and 13C NMR spectroscopic experiments were recorded using a Bruker DMX-500 spectrometer with deuterated chloroform and tetramethylsylane as an internal reference. The mass spectra were determined with a Jeol-102 spectrometer; the X-ray spectra for shamixanthone and dehydroaustinol were obtained with a Bruker Smart Apex charge-coupled-device diffractometer.
Isolation of shamixanthone (compound 1) from CLS12.
Eighteen milligrams of a yellow compound was obtained from the fractions eluted with hexane-ethyl acetate (95:5). TLC plates (hexane-ethyl acetate [9:1]; silica gel [Merck]; Rf of 0.35) were revealed with UV light at 254 nm and iodine steam. 1H NMR (500 MHz, CDCl3) δ ppm: 7.44 (1H, d, J = 4.0 Hz, H-6), 7.29 (1H, s, H-4); 6.74 (1H, d, J = 4.0 Hz, H-7); 5.41 (1H, t, J = 3.4 and 1.13 Hz, H-20); 5.31 (1H, t, J = 7.38 Hz, H-16), 5.05 (1H, d, J = 3.4 Hz, H-O-Ar), 4.80 (1H, t, J = 1.4 and 7 Hz, H-13a); 4.59 (1H, d, J = 0.7 Hz, H-13b); 4.42 (1H, ddd, J = 0.8, 3.3 and 1.3 Hz, H-10a); 4.35 (1H, dd, J = 10.8 and 3.0 Hz, H-10b); 3.50 (2H, t, J = 6.6 and 3.3 Hz, H-15); 2.73 (1H, d, J = 3.3 Hz, H-11); 2.35 (3H, s, H-21); 1.84 (3H, s, H-14); 1.79 (3H, s, H-19); 1.75 (3H, s, H-18);. 13C NMR (500 MHz, CDCl3) δ ppm: 184.5 (C-9); 159.7 (C-8); 153.5 (C-4a); 152.3 (C-5a); 149.5 (C-2); 145.0 (C-12); 142.6 (C-3); 134.0 (C-17); 136.6 (C-6); 133.5 (C-1); 121.7 (C-17); 119.3 (C-4); 118.8 (C-8a); 117.0 (C-1a); 112.3 (C-13); 109.7 (C-7); 108.1 (C-5); 64.6 (C-10); 63.3 (C-20); 45.0 (C-11); 27.5 (C-15); 25.8 (C-18); 22.6 (C-14); 17.8 (C-19); 17.4 (C-21).
Isolation of emericellin (compound 2) from CLS12.
Twenty-six milligrams of a yellow compound was obtained from fractions eluted with hexane-ethyl acetate (95:5). TLC plates (hexane-ethyl acetate [9:1]; silica gel [Merck]; Rf of 0.29) were revealed with UV light at 254 nm and by iodine steam. 1H NMR (500 MHz, CDCl3) δ ppm: 7.47 (1H, s, H-O-Ar); 7.46 (1H, d, J = 8.5 Hz, H-6); 7.34 (1H, s, H-4); 6.75 (1H, d, J = 8.5 Hz, H-7); 5.63 (1H, m J = 7.5, 4.5, 3.0 and 1.5 Hz, H-11); 5.31 (1H, m, J = 7.5, 4.5, 3.0 and 1.5 Hz, H-16); 5.10 (2H, d, J = 8.0 Hz, H-20); 4.48 (1H, t, J = 8.0 Hz, H-O-R); 4.46 (2-H, d, J = 7.5 Hz, H-10); 3.51 (2H, d, J = 7.5 Hz, H-15); 2.48 (3H, s, H-21); 1.82 (3-H, s, H-14); 1.80 (3H, s, H-13); 1.78 (3H, s, H-19); 1.74 (3H, s, H-18). 13C NMR (500 MHz, CDCl3) δ ppm: 185.0 (C-9); 159.8 (C-8); 153.8(C-4a); 152.8 (C-2); 152.5 (C-5a); 142.5 (C-3); 138.9 (C-12); 136.8 (C-6); 134.1 (C-1); 133.1 (C-17); 121.5 (C-16); 119.5 (C-11); 119.3 (C-4); 117.8 (C-1a); 109.8 (C-7); 108.7 (C-5); 72.09 (C-10); 56.9 (C-20); 27.3 (C-15); 25.7 (C-14); 25.6 (C-19); 17.9 (C-21); 17.7 (C-13); 17.6 (C-18).
Isolation of dehydroaustinol (compound 5) from CLS12.
Fifty-two milligrams of a colorless crystalline compound was obtained from the fractions eluted with hexane-ethyl acetate (7:3). TLC plates (hexane-ethyl acetate [7:3]; silica gel [Merck]; Rf of 0.23) were revealed with iodine steam. 1H NMR (500 MHz, CDCl3) δ ppm: 6.90 (1H, d, J = 2 Hz, H-1); 6.33 (1H, s, H-13a); 5.91 (1H, d, J = 10.2 Hz, H-2); 5.84 (1H, s, H-1′a); 5.76 (1H, s, H-13b); 5.67 (1H, s, H-1′b); 5.27 (1H, q, J = 6.8 Hz, H-5′); 4.37 (1H, d, J = 4.0 Hz, H-11); 2.30 (1H, d, J = 4.0Hz, −OH); 2.10 (1H, td, J = 4.6, 13 and 27.7 Hz, H-7a); 1.78 (1H, td, J = 4.6, 13 and 27.7 Hz, H-6b); 1.72 (1H, c, H-6a); 1.71 (3H, s, H-9′); 1.65 (3H, d, J = 6.8 Hz, H-10′); 1.53 (3H, s, H-14); 1.51 (3H, s, H-15); 1.34 (1H, dt, J = 3 and 13 Hz, H-7b); 1.27 (3H, s, H-12). 13C NMR (500 MHz, CDCl3) δ ppm: 169.0 (C-4′); 167.2 (C-8′); 163.5 (C-3); 150.9 (C-1); 140.9 (C-10); 137.4 (C-2′); 124.3 (C-13); 116.2 (C-2); 114.5 (C-1′); 90.1 (C-9); 86.4 (C-4); 84.9 (C-6′); 83.5 (C-3′); 76.1 (C-5′); 75.0 (C-11); 64.0 (C-7′); 50.1 (C-8); 44.3 (C-5); 26.9 (C-6); 26.5 (C-7); 25.7 (C-14); 23.7 (C-15); 19.8 (C-9′); 16.7 (C-12); 13.5 (C-10′).
Bioassays.
Emericellin, shamixanthone, and dehydroaustinol purified from strain CLS12 were tested for their ability to remediate the developmental defects of mutant strains cfwA2, ΔfluG, and ΔtmpA. These compounds, as well as total crude extracts, were dissolved in ethyl acetate, and a volume equivalent to 4 mg was dried on a sterile filter paper disc. Discs with compounds were placed ∼1 cm from the border of colonies pregrown for 48 h. Colonies were observed after 24, 48, and 72 h of incubation with the different compounds.
RESULTS
cfwA1, cfwA2, and npgA1 are gene alleles encoding an essential PPTase.
UV light mutagenesis experiments aimed at isolating developmental mutants allowed us to obtain a temperature-sensitive mutant that was unable to grow at 42°C and that showed developmental defects at lower temperatures. Between 32 and 37°C, this mutant showed a delay in conidiophore development characterized by the proliferation of aerial hyphae and a cotton-like or “fluffy” morphology to finally develop conidiophores with wild-type morphology but lacking the conidiospore pigment. In contrast, a nearly wild-type phenotype was observed at 25°C. Notably, contiguous wild-type or other developmental mutant colonies were able to suppress both the conidiation and pigmentation defects at 37°C, suggesting “cross feeding” of conidiation-inducing and pigment precursor compounds (Fig. 1). Genetic mapping and diploid complementation tests between our mutant and a cfwA1 mutant unable to grow on its own, isolated by John Clutterbuck (University of Glasgow), confirmed cfwA (cross-feedable white) as a gene located on chromosome I closely linked to pyroB. A third white conidiospore mutant allowed the identification and cloning of npgA (null pigmentation) as a gene encoding a putative PPTase needed for conidiospore pigmentation (23, 32). Our diploid complementation tests between cfwA2 and npgA1 mutants indicated that cfwA and npgA corresponded to the same gene. To confirm this, we amplified and sequenced the cfwA/npgA gene from wild-type and cfwA2 strains. Analysis of the cfwA2 sequence showed a single nucleotide change in which the wild-type leucine (L) 217 codon CTC was replaced by the arginine (R) codon CGC. This has been confirmed independently, but it was incorrectly stated that cfwA2 contained a second point mutation (31). These results suggested that the L217R replacement resulted in temperature-sensitive PPTase enzyme activity and that CfwA/NpgA was essential for growth. To test this further, we generated a complete-lack-of-function allele by deleting the entire cfwA open reading frame. A cfwA replacement construct, with A. fumigatus riboB as a genetic marker, was generated by double-joint PCR (65) and used to transform an A. nidulans ΔkuA strain in which virtually all DNA integration events occur by homologous recombination (43). Transformed protoplasts were plated onto minimal medium supplemented with lysine and conditioned medium containing siderophores, as we anticipated that CfwA activity could be required for lysine (12, 62) and siderophore (13) biosynthesis. Two Ribo+ transformants were obtained, which grew as visible heterokaryons with green and white sectors. Although white sectors conidiated very poorly and only after several days, pure white colonies were isolated from these two transformants by single-spore colony isolation. Deletion of the cfwA gene in transformants TJRΔcfwA1 and TJRΔcfwA2 was confirmed by diagnostic PCR (Fig. 2A and B) and Southern blot analysis (not shown). The same gene disruption strategy using kuA+ strain A770 yielded additional mutants with similar white, cotton-like appearances that were not further analyzed. As shown in Fig. 2C, deletion of cfwA resulted in an absolute requirement for both lysine and siderophores such as triacetylfusarinine C. Oberegger et al. (44) reported previously that lysine and triacetylfusarinine C supplementation restored the growth of a cfwA2 mutant at the partially restrictive temperature of 37°C. Our results with total-lack-of-function ΔcfwA mutants indicated that CfwA is not involved in additional essential functions (i.e., biosynthesis of essential fatty acids) and that the presence of lysine and siderophores did not remediate the severe sporulation defects observed in these strains. Indeed, ΔcfwA mutants showed a cotton-like “fluffy” morphology and were unable to differentiate any conidiophore structure before 3 to 4 days at 37°C, with few conidiophores bearing white conidiospores being formed between 4 and 6 days. Quantification of conidiospore number per square centimeter shows that a ΔcfwA mutant produced only 0.06% of the spores formed by cfwA+ strain 11035 after 5 days. In summary, these results indicate that the PPTase CfwA/NpgA is not only essential for lysine and siderophore biosynthesis but also needed for asexual development.
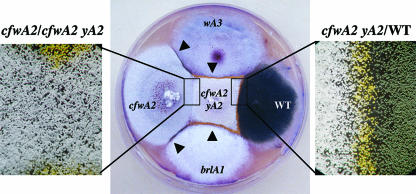
FIG. 1.
Sporulation and pigmentation defects of cfwA2 mutants are remediated by contiguous wild-type and developmental mutant colonies. Strains NK002 (wA3), CJA15.3 (cfwA2), CRO1 (cfwA2 yA2), FGSC26 (wild type [WT]), and AJC7.1 (brlA1) were point inoculated next to each other and grown for 5 days at 37°C. Bands of yellow or green conidiophores formed between cfwA2 and wild-type, wA3, or brlA1 strains are indicated by arrowheads (center panel). Enlarged areas in the left and right panels show the color of conidiophores formed at the region where colonies of indicated strains merge. A band of yellow conidiophores is formed at the interface between green spore wild-type and cfwA2 yA2 strains (right), whereas no pigmented conidiophores are formed between two strains carrying a cfwA2 allele (left). The yA2 allele confers a yellow spore phenotype. See Table 1 for full genotypes.
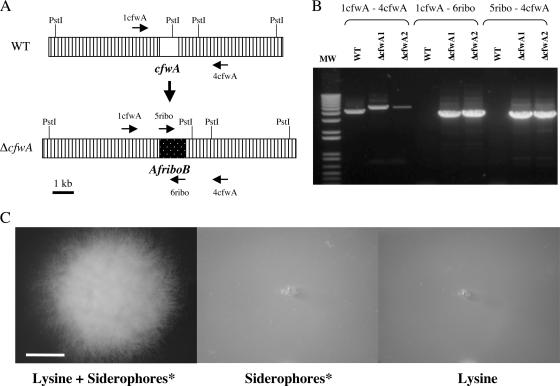
FIG. 2.
cfwA encodes an essential PPTase required for asexual development and the biosynthesis of lysine and siderophores. (A) A cfwA gene deletion construct based on AfriboB was generated by double-joint PCR and used to transform strain 11035. (B) The correct integration event was confirmed by PCR in two independent strains using genomic DNA and the indicated primers (1cfwA-4cfwA, 1cfwA-6ribo, and 5ribo-4cfwA). (C) Strain TJRΔcfwA1 was point inoculated into plates containing 10 mM lysine and/or siderophores and photographed after 48 h of incubation at 37°C. *, siderophores were provided as siderophore-containing conditioned medium; the same result was obtained with 10 μM of the siderophore triacetylfusarine C (see Materials and Methods). The white scale bar in C corresponds to 0.5 cm. WT, wild type. MW, molecular weight.
CfwA/NpgA is a member of a single-domain large-type PPTase family.
A BLAST search analysis of the A. nidulans genome (18) showed that in addition to CfwA/NpgA, this fungus contains four other putative PPTases. Three PPTases are integral domains of the fatty acid synthase α-subunits FasA, StcJ (5), and AN3380.2, whereas the fourth enzyme (AN7043.2) is a small-type PPTase (193 amino acids) similar to Ppt2p from yeast, which is involved in the activation of a mitochondrial fatty acid synthase (53). With sizes between 272 and 359 amino acids, CfwA/NpgA and its orthologues constitute the largest single-domain members of the PPTase superfamily (see Fig. S1 in the supplemental material).
A phylogenetic analysis based on fungal CfwA/NpgA orthologues showed that these PPTases cluster into four major groups, one from basidiomycetes and three subgroups from ascomycetes (Fig. 3). As the clustering of the different species is coherent with phylogenies based on complete fungal genomes, it reflects the similarity between analyzed PPTases. However, the relationship between the four groups is not consistent with established phylogenies (16), perhaps due to the low overall conservation among PPTases. This analysis also indicated that all fungi with a complete genome sequence available appear to have a single cfwA/npgA orthologue. Furthermore, the leucine replaced by an arginine in A. nidulans CfwA2 is part of a region that is conserved in all large-type PPTases and is present in 14 out of 18 fungal PPTases analyzed, except for the proteins from the ascomycetes Schizosaccharomyces pombe (cysteine) and Kluveromyces lactis (methionine) and the basidiomycetes Ustilago maydis (tyrosine) and Cryptococcus neoformans (valine). This leucine, invariant in all filamentous ascomycetes analyzed (see Fig. S1 in the supplemental material), is next to a positively charged amino acid (lysine or arginine) that also appears to be invariant in all PPTases but whose function is unknown (47).
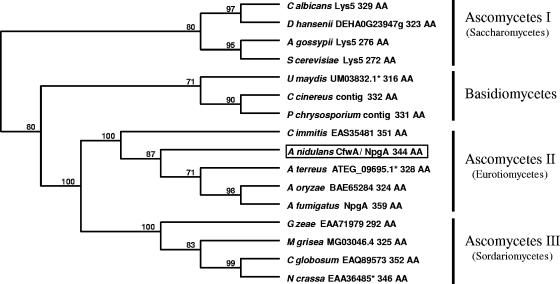
FIG. 3.
Many fungi contain a single orthologous PPTase. Indicated PPTases were aligned with ClustalW. The phylogenetic tree was generated with the distance-based unweighted-pair group method using average linkages using MacVector 7.2. Numbers at the nodes indicate percentages of 2,000 bootstrap replicates in which the node was recovered. Sequences are identified by species name, followed by protein name or GenBank accession number and protein size. Coprinus cinereus (C cinereus) and Phanerochaete chrysosporium (P chrysosporium) proteins were manually deduced from contigs AACS01000118 (Broad Institute of MIT and Harvard [http://www.broad.mit.edu]) and AADS01000639 (DOE Joint Genome Institute [http://genome.jgi-psf.org/whiterot1/whiterot1.home.html]), respectively. Asterisks indicate proteins that seem to have been incorrectly annotated and were manually deduced as follows: only the last 316 and 328 amino acids (AA) of Ustilago maydis (U maydis) UM03832.1 and Aspergillus terreus (A terreus) ATEG_09695.1 were considered to be CfwA/NpgA homologues, respectively. Neurospora crassa (N crassa) EAAA36485 is missing the first exon and is incorrectly annotated as being a protein of only 284 amino acids. Abbreviations for other organism names are as follows: C albicans, Candida albicans; D hansenii, Debaryomyces hansenii; A gossypii, Ashbya gossypii; S cerevisiae, Saccharomyces cerevisiae; C immitis, Coccidioides immitis; A oryzae, Aspergillus oryzae; A fumigatus, Aspergillus fumigatus; G zeae, Gibberella zeae; M grisea, Magnaporthe grisea; C globosum, Chaetomium globosum.
Secondary metabolites produced during development in solid-phase fermentation.
The failure of cfwA2 mutants to produce conidiospore pigments and the mycotoxin sterigmatocystin (21) suggested that CfwA/NpgA PPTase was needed to activate polyketide synthases WA (17, 40) and StcA (6), which are required to produce these conidiation-associated secondary metabolites. Results in Fig. 1 show that mutants lacking WA activity (wA3) or unable to develop conidiophores (brlA1) and to express wA (40) were still capable of suppressing both the conidiation and pigmentation defects of cfwA2 partial-loss-of-function mutants. These results suggested that the PPTase CfwA/NpgA was required for the production of other metabolites, whose production was independent of WA and BrlA activities.
We grew large-scale cultures from cfwA+ (CLS12) and cfwA2 (CLS2) strains to try to identify small-molecular-weight compounds related to the cross-feeding phenomenon observed in cfwA2 mutants. Since the production of conidiospore pigments and ST varies with temperature in cfwA2 mutants and we wanted to avoid the purification of such well-characterized compounds, isogenic strains CLS2 and CLS12 also carried wA3 and ΔstcE null mutations to completely block conidiospore pigment and ST production, respectively (7, 17, 40). Solid-phase fermentation on an inert support (11), rather than growth in liquid medium, was chosen because secondary metabolism and development are generally associated with growth in solid or highly aerated medium. Solid-phase fermentation also facilitated the chemical extraction procedures.
Small pieces of PUF were soaked with liquid minimal glucose medium inoculated with conidiospores from the indicated strains and incubated at 32 to 34°C for 10 days. Microscopic observation of CLS12 samples taken every 24 h showed growing hyphae after 1 day. First, conidiophores were observed by day 2, with increasing numbers between days 3 and 4. Pink pigments, Hülle cells, and immature sexual fruit bodies (cleistothecia) were observed by day 7. Day 10 samples contained mature hyphae, conidiophores, conidia, Hülle cells, cleistothecia, and a reddish pigment generally associated with the cleistothecial cell wall. A similar pattern of development was observed for cfwA2 strain CLS2, except that the development of conidiophores and cleistothecia was delayed. Although CLS2 10-day samples also contained hyphae, conidiophores, conidia, Hülle cells, and cleistothecia, the number of cleistothecia and overall culture pigmentation were clearly reduced compared with CLS12 samples.
The ethyl acetate extracts from CLS12 and CLS2 strains were subjected to repeated column chromatography on silica gel, allowing the isolation of six compounds (compounds 1 to 6) from strain CLS12 and two compounds (compounds 3 and 6) from strain CLS2 (see Table S1 in the supplemental material). Based on their physical and spectral properties, compounds 1, 2, and 5 from strain CLS12 (Fig. 4) were identified as being the polyketides shamixanthone (8), emericellin (26, 27), and dehydroaustinol (20, 39). Spectral data and comparisons with authentic samples showed that compounds 3, 4, and 6 were ergosterol, ergosterol peroxide (59), and cerevisterol (58). Shamixanthone is a moderately cytotoxic molecule first isolated from Aspergillus variecolor (8) but not reported previously in A. nidulans.
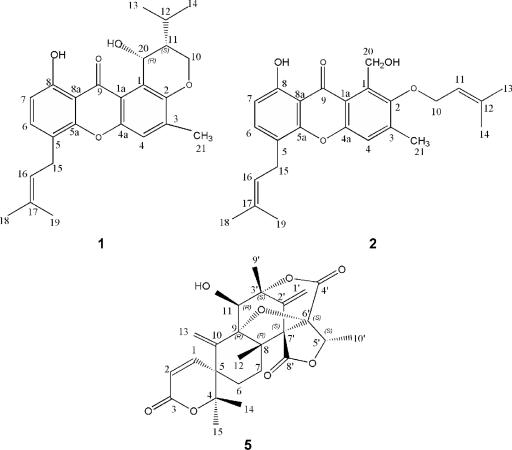
FIG. 4.
Chemical structures of emericellin (compound 1), shamixanthone (compound 2), and dehydroaustinol (compound 5) purified from A. nidulans developmental cultures. The indicated compounds were purified from ethyl acetate extracts obtained from strain CLS12 incubated for 10 days under conditions of solid substrate fermentation. Chemical structures were determined from spectral data and comparisons with published structures. Spectral analysis consisted of one-dimensional and two-dimensional NMR measurements (1H, 13C, DEPT, COSY, HSQC, and HMBC) (Bruker DMX500) and X-ray analysis (see Table S2 in the supplemental material) (see Materials and Methods for details).
The structure of compound 5 (Fig. 4) was furnished by one-dimensional and two-dimensional NMR measurements (1H, 13C, DEPT, COSY, HSQC, and HMBC) (Bruker DMX500) and X-ray analysis (Bruker Smart Apex X-Ray Difractometer). The 1H and 13C NMR spectra established the presence of 28 protons and 25 carbons. The gas chromatography-mass spectrometry spectrum indicated a molecular ion peak at m/z 456 [M]+, suggesting the molecular formula C25H28O8. The 1H NMR spectrum (see Table S2 in the supplemental material) showed six vinylic signals at δ 6.90 (10.2 Hz), 6.33, 5.91 (10.2 Hz), 5.84, 5.76, and 5.67, corresponding to H-1, H-13a, H-2, H-1′a, H-13b, and H-1′b, respectively. Signals corresponded to two methine protons, geminals to one lactonic ring and a hydroxyl group at δ 5.27 (multiple signal) and 4.37 (double signal, 4.0 Hz). The presence of two methylenes on compounds 6 and 7 was evident from four multiple signals at δ 2.1, 1.34, 1.78, and 1.72, corresponding to protons H-7a, H-7b, H-6b, and H-6a, respectively. One signal at δ 2.30 (exchangeable with D2O) corresponded to one hydrogen on oxygen, with this proton coupled to H-11, according to the 1H COSY experiment. Finally, four single signals at δ 1.71, 1.53, 1.51, and 1.27 and one double signal at 1.65 (6.8 Hz) corresponded to methyl protons H-9′, H-14, H-15, H-12, and H-10′, respectively. The presence of three lactonic groups (δ 169.0, 167.2, and 163.5), six unsaturated carbons (δ 150.9, 140.9, 137.4, 124.3, 116.2, and 114.5), and one ether group (δ 90.1 and 84.9) was confirmed by 13C NMR data. Therefore, compound 5 showed 1H and 13C NMR data (see Table S2 in the supplemental material) virtually identical to those reported previously for dehydroaustin (20), with minor differences around the hydroxile on carbon 11 (Fig. 4). We confirmed our results by C-H long-range couplings (HMBC) and X-ray crystallography. Dehydroaustinol has not been reported previously for A. nidulans.
Emericellin, shamixanthone, dehydroaustinol, and ergosterol peroxide are not detected in extracts from the cfwA2 mutant.
As indicated, only compounds 3 and 6 were obtained from cfwA2 mutant strain CLS2 crude extracts, which were identified as being ergosterol and cerevisterol, respectively. Although the yield of the total extract of strain CLS2 was ∼30% lower than that obtained from strain CLS12, the amount of ergosterol was ∼48% higher than that in CLS12 extracts (see Table S1 in the supplemental material). We carried out TLC analysis of total ethyl acetate crude extracts from strains CLS2 and CLS12 using compounds purified from strain CLS12 as standards. The results confirmed that emericellin, shamixanthone, dehydroaustinol, and ergosterol peroxide are either absent or below the detection limit in cfwA2 mutant extracts. In contrast, TLC analysis of the apolar extracts from both strains showed similar profiles, which, along with the presence of ergosterol and cerevisterol, suggests that the biosynthesis of small-molecular-weight lipids and sterols is not affected in the cfwA2 mutant. Furthermore, we used gas chromatography-mass spectrometry to analyze fatty acid contents and found similar amounts of palmitic, stearic, oleic, and linoleic acids in both strains (not shown), a result consistent with previous studies from our laboratory (D. Schnabell et al., unpublished data). Based on these data, we conclude that the PPTase CfwA/NpgA is required for the production of emericellin, shamixanthone, dehydroaustinol, and ergosterol peroxide and dispensable for ergosterol, cerevisterol, and fatty acid biosynthesis.
Emericellin, shamixanthone, and dehydroaustinol do not suppress the sporulation or pigmentation defects of cfwA2 and other developmental mutants.
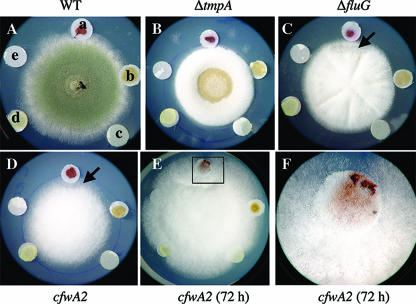
As shown in Fig. 1, contiguous wild-type colonies are able to suppress the conidiation and pigmentation defects of cfwA2 mutants. As cfwA2 mutants failed to produce or accumulate emericellin, shamixanthone, and dehydroaustinol, we asked whether these compounds could suppress cfwA2 mutant defects. In addition, we tested these compounds on developmental mutants ΔfluG (38) and ΔtmpA (52), as these mutants appear to be affected in the production of different unknown sporulation signals, and their conidiation defects are remedied by contiguous wild-type colonies. We found that none of the purified compounds was able to rescue the pigmentation or conidiation defects of ΔtmpA, ΔfluG, or cfwA2 mutants (Fig. 5B to D), even after extended incubation (not shown). However, the CLS12 (cfwA+) but not the CLS2 (cfwA2) crude extract was able to induce a slight reduction in the production of aerial hyphae in ΔfluG (Fig. 5C) and cfwA2 (Fig. 5D) mutants as well as the development of cfwA2 white conidiophores and conidia after 72 h (Fig. 5E and F). These results indicate that a cfwA-dependent activity capable of inducing conidiation, but not conidiospore pigmentation, is present in CLS12 crude extracts but was not purified by our experimental procedures.
FIG. 5.
Shamixanthone, emericellin, and dehydroaustinol do not remediate the sporulation defects of ΔtmpA, ΔfluG, and cfwA2 mutants. Strains FGSC26 (wild type [WT]), TGS6 (ΔtmpA), rBN119 (ΔfluG), and CRO1 (cfwA2) were point inoculated and grown for 48 h at 37°C. After this time, sterile paper discs impregnated with 4 mg of each tested compound were placed ∼1 cm from the edge of the colonies, and pictures were taken after 48 or 72 h. a, CLS12 (cfwA+) crude extract; b, CLS2 (cfwA2) crude extract; c, shamixanthone; d, emericellin; e, dehydroaustinol. Arrows in C and D indicate areas showing a reduction in proliferation of aerial hyphae; the rectangle in E indicates the region enlarged in F, showing large numbers of white conidiophores around the paper disc.
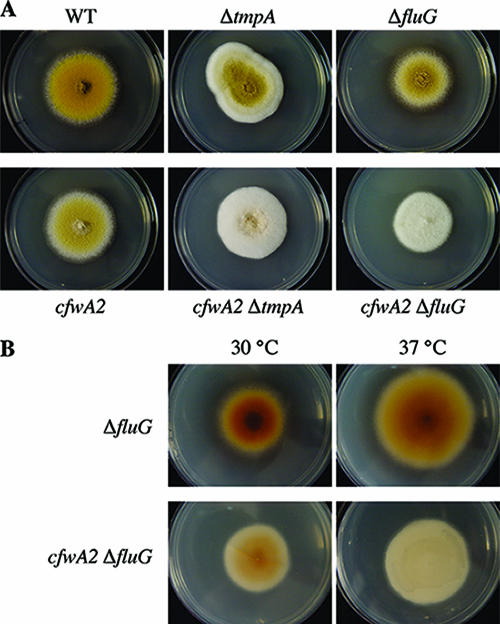
To further explore the role of the PPTase CfwA in conidiation, we generated cfwA2 ΔtmpA (CJRΔtmpAcfwA2-4) and cfwA2 ΔfluG (CJRΔfluGcfwA2-4) double mutants. The presence of the cfwA2 mutation enhanced the conidiation defects of both ΔtmpA and ΔfluG single mutants at 30°C (Fig. 6A) and 37°C (not shown). On the other hand, it has been shown (37, 38) that fluG inactivation results in the production of a characteristic yellow-orange pigment (Fig. 6B, top panels), which might be a precursor of the FluG sporulation signal. In cfwA2 ΔfluG mutants, such pigment was clearly reduced at 30°C and virtually absent at 37°C (Fig. 6B), indicating that its production depends on CfwA activity. As tmpA and fluG genes have been shown to regulate A. nidulans conidiation through independent pathways (52), our results suggest that both pathways involve the participation of CfwA-dependent NRPSs and/or PKSs.
FIG. 6.
cfwA2 mutation enhances the sporulation defects of ΔtmpA and ΔfluG mutants and prevents ΔfluG mycelial pigment accumulation. (A) Isogenic strains CLK43 (wild type [WT]), TGS6 (ΔtmpA), CGS49 (ΔfluG), CRO1 (cfwA2), CJRDtmpAcfwA2-4 (cfwA2 ΔtmpA), and CJRDfluGcfwA2-4 (cfwA2 ΔfluG) were point inoculated and grown for 5 days at 30°C. (B) Strains CGS49 (ΔfluG) and CJRDfluGcfwA2-4 (cfwA2 ΔfluG) were grown as in A at 30 or 37°C, and pictures were taken from the back of the plates. The yellow color of colonies in A corresponds to conidiospores, except in strain CGS49, where it corresponds to both conidiospores and Hülle cells, which are associated with sexual development.
DISCUSSION
The roles of the PPTase CfwA/NpgA in primary metabolism.
After identifying cfwA2 and npgA1 as being allelic mutations, sequence analysis of the corresponding gene, cloned previously by Kim and coworkers (32), indicated to us that cfwA/npgA encoded a PPTase. The cfwA2 thermosensitive phenotype, resulting from an L217R replacement in the protein, indicated that CfwA/NpgA is required for essential functions. By genetic complementation of an S. cerevisiae lys5 mutant, Mootz et al. (42) previously demonstrated that CfwA/NpgA is able to restore lysine biosynthesis and therefore to activate the AAR Lys2 (42). Indeed, lysine supplementation, in the presence of the siderophore triacetylfusarinine C, restored the growth of cfwA2 (44) and ΔcfwA mutant strains. In contrast to some other fungi, NRPS-mediated siderophore biosynthesis is essential in A. nidulans (13). As lysine biosynthesis in fungi occurs through the AAR pathway (62) and a single cfwA/npgA orthologue is found in fungi with a genome sequence available (Fig. 3), cfwA/npgA orthologues should be required for lysine biosynthesis in most if not all fungi. Indeed, a deletion of the cfwA/npgA orthologue in Neurospora crassa results in lysine auxotrophy (J. Ramos-Balderas and J. Aguirre, unpublished data).
PPTase CfwA/NpgA plays an essential role in secondary metabolism.
The inability of cfwA2 mutants to produce conidial pigments and the mycotoxin ST (21) first demonstrated the involvement of CfwA/NpgA in secondary metabolism. The fact that lysine and siderophore supplementation restored the growth of ΔcfwA mutants but not the conidiation and pigmentation defects clearly distinguishes the roles of CfwA/NpgA in primary and secondary metabolism. Here, we have shown that A. nidulans produces emericellin, shamixanthone, dehydroaustinol, and ergosterol peroxide during development in solid-phase medium and that CfwA/NpgA is needed for the production of these compounds. Based on chemical structure and xanthone biosynthesis in A. variecolor (3, 4), the xanthone emericellin can be synthesized from an anthrone precursor through a pathway involving at least one PKS. Shamixhantone may be formed by cyclization of one of the emericellin isoprenylated chains.
The biosynthesis of dehydroaustinol would involve a different mixed polyketide-terpenoid route, as has been demonstrated for the compound austin in Aspergillus ustus, which is derived from the polyketide dimethylorsellinate (49, 51). Therefore, CfwA would be needed for the activation of at least two as-yet-unidentified PKSs involved in the biosynthesis of emericellin (and shamixhantone) and dehydroaustinol.
Ergosterol peroxide was not detected in the cfwA2 extract despite the finding that higher amounts of ergosterol were isolated from this strain (see Table S1 in the supplemental material). Ergosterol peroxide is produced from the reaction of ergosterol with singlet oxygen. The formation of singlet oxygen requires a photosensitizer molecule, visible light, and oxygen (10, 59). This suggests that CfwA activity is needed to produce a natural photosensitizer capable of producing singlet oxygen in A. nidulans. Indeed, some plant-pathogenic fungi use a PKS pathway to produce the photosensitizer cercosporin and form singlet oxygen, which is required for the successful pathogenesis of plants (10, 56). Here, it is interesting that reactive oxygen species have been linked to different developmental processes in A. nidulans and other fungi (2, 36).
In addition, cfwA2 mutants are compromised in penicillin (31) and siderophore (44) biosynthesis, indicating that the NRPSs AcvS and SidC are substrates of the PPTase CfwA/NpgA. The phenotype of cfwA2 and ΔcfwA mutants indicates that no other PPTase can replace CfwA functions. The A. nidulans genome predicts 27 PKSs and 14 NRPSs (18, 30). Our results suggest that the PPTase CfwA/NpgA is needed for the activation of all these enzymes and that a similar situation occurs in the human pathogen Aspergillus fumigatus and many other economically important fungi such as Aspergillus oryzae, Giberella zea, etc. (Fig. 3).
The roles of CfwA/NpgA in asexual sporulation.
Two different conidiation signaling pathways, fluG (38) and tmpA (52), have been identified in A. nidulans. As shown in this study, lysine, triacetylfusarinine C, emericellin, shamixanthone, or dehydroaustiol was not able to suppress the conidiation defects of cfwA, ΔfluG, and ΔtmpA mutants, indicating that other cfwA-dependent compounds are involved in the regulation of conidiation. The crude extract form of cfwA+ CLS12 grown for 10 days contained a conidiation-inducing activity (Fig. 5E and F). The fact that 48 to 72 h was required to observe abundant conidiophores suggests that the responsible molecule(s) was present in low amounts or had to be transformed into the actual sporulation signals.
As shown here, a cfwA2 mutation enhances the conidiation defects of both ΔtmpA and ΔfluG single mutants, and the production of mycelial pigmentation in ΔfluG mutants is cfwA dependent (Fig. 6). These results predict that fluG and tmpA conidiation pathways involve the participation of PKSs and/or NRPSs that are yet to be identified. On the other hand, the fact that ΔcfwA mutants can produce some conidiophores and conidiospores indicates the presence of a cfwA-independent conidiation pathway(s). Although different secondary metabolites have been involved in several aspects of fungal development (61), little is known about their biosynthetic pathways or mechanisms of action. In particular, there are few examples of PKS- or NRPS-derived products involved in the regulation of cell differentiation in eukaryotic cells. In Aspergillus parasiticus, disruption of the PKS-encoding gene fluP (also called pksL2) results in a reduction of radial growth, a cotton-like morphology, and decreased conidiospore and conidiophore development (67). Nevertheless, the A. nidulans genome does not predict a PKS that is clearly homologous to FluP, which belongs to the Penicillium patulum 6-methylsalicylic acid synthase type (33). In the amebozoid Dictyostelium discoideum, a chlorinated polyketide called DIF-1 and other related molecules regulate the differentiation of a specific cell type during sporulation (48).
CfwA/NpgA-orthologous PPTases as tools to study secondary metabolism and as possible drug targets.
Our results suggest that a conditional-null PPTase mutant approach similar to the one reported here could be used to evaluate the roles of secondary metabolism in the biology of different fungi. On the other hand, the requirement of PPTase activity for lysine biosynthesis is specific to fungi, and different PKSs and NRPSs are involved in the production of mycotoxins (54) and virulence factors in plant (56) and animal (35) pathogens. This suggests that CfwA/NpgA PPTase can be considered a potential novel antifungal drug target, as its inhibition would not only block the production of melanin and other pathogen virulence factors but also interfere with essential amino acid biosynthesis. As fungal CfwA homologues differ significantly from their human, animal, and plant counterparts, it might be possible to design fungus-specific inhibitors.
Supplementary Material
Acknowledgments
We are grateful to Rogelio Ortíz, Maura Cárdenas, and Denhí Schnabell for initial characterization of cfwA2 mutants. We acknowledge Dong-Min Han (Wonkwang University) and Kwang-Yeop Jahng (Chonbuck National University) from the Republic of Korea for providing us with npgA mutants and for useful discussions and John Clutterbuck (University of Glasgow) for kindly providing a cfwA1 strain. We also thank Olivia Sánchez for cloning cfwA alleles and technical assistance, Luis Segura for CLS strains, and Atilano Gutierrez Carrillo (Universidad Autónoma Metropolitana-Iztapalapa) for NMR spectral experiments. The IFC-UNAM molecular biology unit is acknowledged for DNA synthesis and sequencing.
This work was supported by grant 2002-C01-1713 from SAGARPA-CONACYT (México) and a partnership grant from Volkswagen Stiftung (Germany).
Footnotes
Published ahead of print on 2 February 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguirre, J., M. Rios-Momberg, D. Hewitt, and W. Hansberg. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111-118. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, S. A., E. Bardshiri, C. R. McIntyre, and T. J. Simpson. 1992. Biosynthetic studies on tajixanthone and shamixanthone, polyketide hemiterpenoid metabolites of Aspergillus variecolor. Aust. J. Chem. 45:249-274. [Google Scholar]
- 4.Bardshiri, E., C. R. McIntyre, T. J. Simpson, R. N. Moore, L. A. Trimble, and J. C. Vederas. 1984. Biosynthesis of tajixanthone and shamixanthone by Aspergillus variecolor: incorporation of oxygen-18 gas. J. Chem. Soc. Chem. Commun. 21:1404-1406. [Google Scholar]
- 5.Brown, D. W., T. H. Adams, and N. P. Keller. 1996. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 93:14873-14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butchko, R. A., T. H. Adams, and N. P. Keller. 1999. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics 153:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chexal, K. K., J. S. Holker, and T. J. Simpson. 1975. The biosynthesis of fungal metabolites. Part VI. Structures and biosynthesis of some minor metabolites from variant strains of Aspergillus variecolor. J. Chem. Soc. 1975:549-554. [PubMed] [Google Scholar]
- 9.Clutterbuck, A. J. 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daub, M. E., S. Herrero, and K. R. Chung. 2005. Photoactivated perylenequinone toxins in fungal pathogenesis of plants. FEMS Microbiol. Lett. 252:197-206. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Godinez, G., J. Soriano-Santos, C. Augur, and G. Viniegra-Gonzalez. 2001. Exopectinases produced by Aspergillus niger in solid-state and submerged fermentation: a comparative study. J. Ind. Microbiol. Biotechnol. 26:271-275. [DOI] [PubMed] [Google Scholar]
- 12.Ehmann, D. E., A. M. Gehring, and C. T. Walsh. 1999. Lysine biosynthesis in Saccharomyces cerevisiae: mechanism of alpha-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry 38:6171-6177. [DOI] [PubMed] [Google Scholar]
- 13.Eisendle, M., H. Oberegger, I. Zadra, and H. Haas. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 49:359-375. [DOI] [PubMed] [Google Scholar]
- 14.Fichtlscherer, F., C. Wellein, M. Mittag, and E. Schweizer. 2000. A novel function of yeast fatty acid synthase. Subunit alpha is capable of self-pantetheinylation. Eur. J. Biochem. 267:2666-2671. [DOI] [PubMed] [Google Scholar]
- 15.Finking, R., and M. A. Marahiel. 2004. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58:453-488. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick, D. A., M. E. Logue, J. E. Stajich, and G. Butler. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii, I., A. Watanabe, U. Sankawa, and Y. Ebizuka. 2001. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem. Biol. 8:189-197. [DOI] [PubMed] [Google Scholar]
- 18.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 19.Geiger, O., and I. M. Lopez-Lara. 2002. Rhizobial acyl carrier proteins and their roles in the formation of bacterial cell-surface components that are required for the development of nitrogen-fixing root nodules on legume hosts. FEMS Microbiol. Lett. 208:153-162. [DOI] [PubMed] [Google Scholar]
- 20.Geris dos Santos, R. M., and E. Rodrigues-Filho. 2003. Structures of meroterpenes produced by Penicillium sp, an endophytic fungus found associated with Melia azedarach. J. Braz. Chem. Soc. 14:722-727. [Google Scholar]
- 21.Guzman-de-Pena, D., J. Aguirre, and J. Ruiz-Herrera. 1998. Correlation between the regulation of sterigmatocystin biosynthesis and asexual and sexual sporulation in Emericella nidulans. Antonie Leeuwenhoek 73:199-205. [DOI] [PubMed] [Google Scholar]
- 22.Haarmann, T., C. Machado, Y. Lubbe, T. Correia, C. L. Schardl, D. G. Panaccione, and P. Tudzynski. 2005. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66:1312-1320. [DOI] [PubMed] [Google Scholar]
- 23.Han, K. H., J. H. Kim, W. S. Kim, and D. M. Han. 2005. The snpA, a temperature-sensitive suppressor of npgA1, encodes the eukaryotic translation release factor, eRF1, in Aspergillus nidulans. FEMS Microbiol. Lett. 251:155-160. [DOI] [PubMed] [Google Scholar]
- 24.Hicks, J. K., J. H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill, T. W., and E. Käfer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium stock solution. Fungal Genet. Newsl. 48:20-21. [Google Scholar]
- 26.Ishida, M., T. Hamasaki, Y. Hatsuda, K. Fukuyama, T. Tsukihara, and Y. Katsube. 1975. Emericellin, a new metabolite from Aspergillus nidulans. Agric. Biol. Chem. 39:291-292. [Google Scholar]
- 27.Ishida, M., T. Hamasaki, Y. Hatsuda, K. Fukuyama, T. Tsukihara, and Y. Katsube. 1975. The structure of two new metabolites, emerin and emericellin from Aspergillus nidulans. Agric. Biol. Chem. 39:2181-2184. [Google Scholar]
- 28.Kawasaki, L., O. Sanchez, K. Shiozaki, and J. Aguirre. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153-1163. [DOI] [PubMed] [Google Scholar]
- 29.Keating, T. A., and C. T. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3:598-606. [DOI] [PubMed] [Google Scholar]
- 30.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 31.Keszenman-Pereyra, D., S. Lawrence, M. E. Twfieg, J. Price, and G. Turner. 2003. The npgA/cfwA gene encodes a putative 4′-phosphopantetheinyl transferase which is essential for penicillin biosynthesis in Aspergillus nidulans. Curr. Genet. 43:186-190. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J. M., D. M. Han, K. S. Chae, and K. Y. Jahng. 2001. Isolation and characterization of the npgA gene involved in pigment formation in Aspergillus nidulans. Fungal Genet. Newsl. 48(Suppl.):52. [Google Scholar]
- 33.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 35.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol 38:143-158. [DOI] [PubMed] [Google Scholar]
- 36.Lara-Ortiz, T., H. Riveros-Rosas, and J. Aguirre. 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50:1241-1255. [DOI] [PubMed] [Google Scholar]
- 37.Lee, B. N., and T. H. Adams. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8:641-651. [DOI] [PubMed] [Google Scholar]
- 38.Lee, B. N., and T. H. Adams. 1996. fluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlAβ activation. EMBO J. 15:299-309. [PMC free article] [PubMed] [Google Scholar]
- 39.Maebayashi, Y., E. Okuyama, M. Yamazaki, and Y. Katsube. 1982. Structure of ED-1 isolated from Emericella dentata. Chem. Pharm. Bull. 30:1911-1912. [Google Scholar]
- 40.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 41.Mofid, M. R., R. Finking, and M. A. Marahiel. 2002. Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4′-phosphopantetheinyl transferases AcpS and Sfp. J. Biol. Chem. 277:17023-17031. [DOI] [PubMed] [Google Scholar]
- 42.Mootz, H. D., K. Schorgendorfer, and M. A. Marahiel. 2002. Functional characterization of 4′-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS Microbiol. Lett. 213:51-57. [DOI] [PubMed] [Google Scholar]
- 43.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberegger, H., M. Eisendle, M. Schrettl, S. Graessle, and H. Haas. 2003. 4′-phosphopantetheinyl transferase-encoding npgA is essential for siderophore biosynthesis in Aspergillus nidulans. Curr. Genet. 44:211-215. [DOI] [PubMed] [Google Scholar]
- 45.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 46.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-283. [DOI] [PubMed] [Google Scholar]
- 47.Reuter, K., M. R. Mofid, M. A. Marahiel, and R. Ficner. 1999. Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 18:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito, T., G. W. Taylor, J. C. Yang, D. Neuhaus, D. Stetsenko, A. Kato, and R. R. Kay. 2006. Identification of new differentiation inducing factors from Dictyostelium discoideum. Biochim. Biophys. Acta 1760:754-761. [DOI] [PubMed] [Google Scholar]
- 49.Scott, F. E., T. J. Simpson, L. A. Trimble, and J. C. Vederas. 1986. Biosynthesis of meroterpenoid austin by Aspergillus ustus: synthesis and incorporation of 13C, 18O-labelled ethyl 3,5 dimethylorsellinate. J. Chem. Soc. Chem. Commun. 1986:214-215. [Google Scholar]
- 50.Seo, J. A., Y. Guan, and J. H. Yu. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson, T. J., S. A. Ahmed, C. R. McIntyre, F. E. Scott, and I. H. Sadler. 1997. Biosynthesis of polyketide-terpenoid (meroterpenoid) metabolites andibenin B and andilesin A in Aspergillus variecolor. Tetrahedron 53:4013-4034. [Google Scholar]
- 52.Soid-Raggi, G., O. Sanchez, and J. Aguirre. 2006. TmpA, a member of a novel family of putative membrane flavoproteins, regulates asexual development in Aspergillus nidulans. Mol. Microbiol. 59:854-869. [DOI] [PubMed] [Google Scholar]
- 53.Stuible, H. P., S. Meier, C. Wagner, E. Hannappel, and E. Schweizer. 1998. A novel phosphopantetheine:protein transferase activating yeast mitochondrial acyl carrier protein. J. Biol. Chem. 273:22334-22339. [DOI] [PubMed] [Google Scholar]
- 54.Sweeney, M. J., and A. D. Dobson. 1999. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol. Lett. 175:149-163. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, A., B. A. Tapper, A. Popay, E. J. Parker, and B. Scott. 2005. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57:1036-1050. [DOI] [PubMed] [Google Scholar]
- 56.Thines, E., J. Aguirre, A. J. Foster, and H. B. Deising. 2006. Genetics of phytopathology: secondary metabolites as virulence determinants of fungal plant pathogens. Prog. Bot. 67:134-161. [Google Scholar]
- 57.Timberlake, W. E., and A. J. Clutterbuck. 1994. Genetic regulation of conidiation, p. 383-427. In S. D. Martinelli and J. R. Kinghorn (ed.), Aspergillus: 50 years on, vol. 29. Elsevier, Amsterdam, The Netherlands. [PubMed] [Google Scholar]
- 58.Trigos, A., C. Amezcua, S. Reyna, and G. Carrion. 1997. Cerevisterol from cultures of Verticillum lecanii. Micol. Neotrop. Apl. 10:57-62. [Google Scholar]
- 59.Trigos, A., and A. Ortega-Regules. 2002. Selective destruction of microscopic fungi through photo-oxidation of ergosterol. Mycologia 94:563-568. [DOI] [PubMed] [Google Scholar]
- 60.Tudzynski, B. 2005. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl. Microbiol. Biotechnol. 66:597-611. [DOI] [PubMed] [Google Scholar]
- 61.Ugalde, U. 2006. Autoregulatory signals in mycelial fungi, p. 203-213. In U. Kues and R. Fischer (ed.), The mycota, 2nd ed., vol. I. Growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 62.Velasco, A. M., J. I. Leguina, and A. Lazcano. 2002. Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 55:445-459. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, C. T., A. M. Gehring, P. H. Weinreb, L. E. Quadri, and R. S. Flugel. 1997. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1:309-315. [DOI] [PubMed] [Google Scholar]
- 64.Wieser, J., and T. H. Adams. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 9:491-502. [DOI] [PubMed] [Google Scholar]
- 65.Yu, J. H., Z. Hamari, K. H. Han, J. A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 66.Yu, J. H., and N. Keller. 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43:437-458. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, R., R. Rasooly, and J. E. Linz. 2000. Isolation and analysis of fluP, a gene associated with hyphal growth and sporulation in Aspergillus parasiticus. Mol. Gen. Genet. 264:514-520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.