Abstract
Endocytosis of Na+,K+-ATPase molecules in response to G protein-coupled receptor stimulation requires activation of class IA phosphoinositide-3 kinase (PI3K-IA) in a protein kinase C-dependent manner. In this paper, we report that PI3K-IA, through its p85α subunit-SH3 domain, binds to a proline-rich region in the Na+,K+-ATPase catalytic α subunit. This interaction is enhanced by protein kinase C-dependent phosphorylation of a serine residue that flanks the proline-rich motif in the Na+,K+-ATPase α subunit and results in increased PI3K-IA activity, an effect necessary for adaptor protein 2 binding and clathrin recruitment. Thus, Ser-phosphorylation of the Na+,K+-ATPase catalytic subunit serves as an anchor signal for regulating the location of PI3K-IA and its activation during Na+,K+-ATPase endocytosis in response to G protein-coupled receptor signals.
The polarized distribution of Na+,K+-ATPase molecules in transporting epithelia is critical for their ability to perform vectorial transport (1–3). In renal epithelial cells, modulation of Na+,K+-ATPase activity plays a key role in sodium reabsorption and potassium secretion (4–6). Decreased Na+,K+-ATPase activity induced by dopamine (DA) is partly responsible for reduced sodium reabsorption during high-salt diet (7–9), and impaired regulation of its activity in the renal tubule has been linked to the development of high blood pressure (10–12).
Inhibition of Na+,K+-ATPase in response to G protein-coupled receptor (GPCR) signals requires the sequential activation and integration of several intracellular signals (13, 14) that lead to activation of protein kinase C (PKC) (15–18). Phosphorylation of the Na+,K+-ATPase catalytic (α) subunit constitutes a triggering signal for removal of both subunits (α/β) from the plasma membrane with the consequent decrease in its activity (19–21). Clathrin-dependent endocytosis of Na+,K+-ATPase subunits requires a dynamic actin-microtubule cytoskeleton (19), as well as activation of class IA subtype-phosphoinositide 3′ kinase activity (PI3K-IA) (22).
Increased PI3K-IA activity results from its binding to phosphorylated tyrosine residues present in integral membrane proteins through Src homology 2 (SH2) domains located within its p85α regulatory subunit (23, 24). However, we demonstrated that in renal epithelial cells, activation of PI3K-IA in response to DA requires activation of PKC (22). Although the activity of PI3K could also be regulated by Ser/Thr phosphorylation (25), the effect of DA was not associated with phosphorylation of the PI3K-IA (our unpublished observation). Because regulation of Na+,K+-ATPase activity and endocytosis are triggered by phosphorylation of Ser18 in the α subunit (20, 21), we have hypothesized that a phosphorylated α subunit may be a requisite for its interaction with the PI3K p85α subunit and thereby increases PI3K-IA activity at the site of Na+,K+-ATPase endocytosis.
Materials and Methods
Experiments were performed in OK cells (an opossum renal epithelial cell line of proximal tubule origin). In this cell line, the mechanisms responsible for regulation of Na+,K+-ATPase activity, as well as DA signaling networks involved that have been described so far, are similar to those of isolated proximal tubule cells from rat kidneys (26, 27).
Site-Directed Mutagenesis and Cell Transfection.
The Pro83→Arg (P83R) mutation was introduced in the Na+,K+-ATPase α subunit. The plasmid pCMV-α (PharMingen) containing the wild-type rodent α1 subunit cDNA was annealed with complementary oligonucleotides containing the mutant bases and then was subjected to 15 cycles of amplification with Pfu polymerase, followed by restriction of the original wild-type template with DpnI. The resulting mutant plasmid was transformed into bacteria for amplification and analysis. Substitution of Pro83 by an Arg residue was accomplished with the oligonucleotide P83R/A (5′-CGCCCTCACGCCCCGTCCAACTACTCCG-3′) and its complement, P83R/B. This substitution created a new site for SacII that was used for screening. Structure of the resulting mutants was evaluated by restriction with appropriate endonucleases and confirmed by dideoxynucleotide sequencing of the altered region. The Ser18→Ala (S18A) mutation has been described previously (21).
Cell transfection and clone selection was performed as described (21, 28), and the levels of S18A and P83R expression were similar to those of wild type.
Peptide Synthesis.
The peptides were synthesized at the Macromolecular Structure Analysis Facility (University of Kentucky, Lexington), except for the PI3K-p85α-SH3 high-affinity binding peptide (Calbiochem). After the synthesis, salts were removed and the peptides were sequenced. Transient cell permeabilization to allow access of the peptide to the cells' interior was performed as described (18, 29, 30).
Determination of Phosphoinositide-3′ Kinase Activity.
After the preincubation protocols with different agonists, cells were homogenized in 400 μl of lysis buffer (140 mM NaCl/10 mM Hepes/10 mM sodium pyrophosphate/10 mM NaF/1 mM CaCl2/1 mM MgCl2/2 mM Na3VO4/10% glycerol/1% Nonidet P-40/10 μg/ml aprotinin/50 μM leupeptin/2 mM PMSF, pH 8.1) and solubilized by continuous stirring for 1 h at 4°C. After centrifugation, the supernatant was collected and 1 mg of protein (in 500 μl) was incubated with an anti-PI3K p85α antibody. After overnight incubation, protein A-Sepharose was added, and the immune complex (p85α antibody coupled to protein A-Sepharose) was washed four times with buffer (100 mM NaCl/1 mM Na3VO4/20 mM Hepes, pH 7.5) and resuspended in 40 μl of buffer (180 mM NaCl/20 mM Hepes, pH 7.5). PI3K-IA activity in the immunoprecipitates (equal amounts of precipitated material as assessed by Western blotting were used in each condition) was assessed directly on the protein A-Sepharose beads as previously described (22, 31).
Determination of Na+,K+-ATPase Activity.
Na+,K+-ATPase activity was determined in OK cells either as the rate of ouabain-sensitive [γ-32P]ATP hydrolysis (19–21) or as the rate of ouabain-sensitive [86Rb+]RbCl transport (28).
Preparation of Intracellular Organelles.
After different protocols, OK cells were gently homogenized to minimize damage of the endosomes by using a motor pestle homogenizer (Kimble-Kontes, Vineland, NJ), and the samples were subjected to a brief (5-min) centrifugation (4°C at 3,000 × g). Isolation of clathrin-coated vesicle was performed as previously described (19, 32). Endosomes were fractionated on a flotation gradient as described (19–21) by using essentially the technique described by Gorvel et al. (33).
Results
Effect of Na+,K+-ATPase α Subunit Phosphorylation on Stimulation of PI3K-IA Activity by DA.
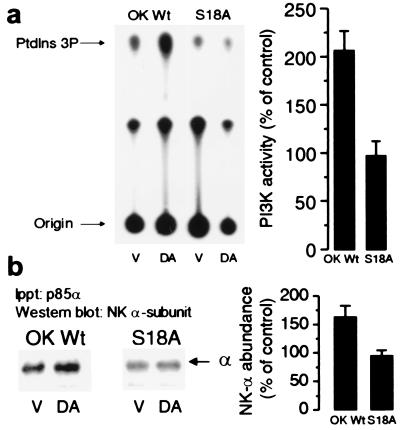
We determined whether DA stimulates PI3K-IA activity in OK cells stably transfected with the rat Na+,K+-ATPase α1 subunit cDNA carrying a mutation in the PKC phosphorylation site (34), Ser18→Ala. In cells expressing S18A mutants, DA did not increase phosphorylation or endocytosis of the Na+,K+-ATPase α subunit, nor did it decrease its activity (21). DA significantly increased PI3K-IA activity in OK cells expressing the wild-type rat Na+,K+-ATPase α1 subunit (OK-Wt), but not in OK cells expressing the S18A mutant (Fig. 1a). These results indicate the possibility that phosphorylation of the Na+,K+-ATPase α1 subunit may induce a conformational change that could favor the interaction with PI3K-IA. Indeed, Western blotting analysis of the p85α-immunoprecipitates (Fig.1b) revealed the presence of the Na+,K+-ATPase α1 subunit; moreover, its abundance was increased in OK-Wt cells treated with DA but not in cells expressing the S18A mutant.
Figure 1.
Effect of Na+,K+-ATPase α subunit phosphorylation on the ability of DA to increase PI3K activity. (a) OK cells expressing the wild-type rat Na+,K+-ATPase α1 subunit or this isoform carrying the S18A mutation were incubated in the presence or absence of 1 μM DA for 2.5 min at 23°C. PI3K activity was determined in immunoprecipitates (equal amounts of precipitated material as assessed by Western blotting were used in each condition) with a polyclonal antibody raised against the p85α subunit whose epitope corresponds to the amino acids 333–430 within the N-terminal SH2 domain (Santa Cruz Biotechnology). (b) The presence of Na+,K+-ATPase α1 subunit in the immunoprecipitates was determined by Western blotting with a monoclonal antibody against the Na+,K+-ATPase α subunit. Experiments in a and b were repeated three and five times, respectively. Basal expression was similar in OK-Wt and S18A mutants as evidenced by Western blotting and Na+,K+-ATPase activity [Rb+ transport (nmol of Rb per mg of protein per min): OK-Wt, 9.8 ± 0.7 vs. S18A, 9.7 ± 1.0; hydrolytic activity in isolated membranes (μmol of Pi per mg of protein per min): OK-Wt, 0.305 ± 0.02 vs. S18A, 0.293 ± 0.01].
Identification of the PI3K-IA Domain That Interacts with the Na+,K+-ATPase α1 Subunit.
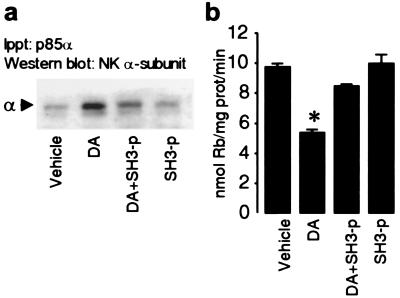
We used a synthetic peptide RRPRPPLKR that binds with high affinity (distribution coefficient Kd = 7.6 μM) (35) to PI3K p85α-SH3 domain to investigate whether this motif might be responsible for the interaction with the Na+,K+-ATPase α subunit. Initially, we determined whether the presence of this peptide (SH3-p) prevented the p85α subunit binding to the Na+,K+-ATPase α1 subunit (Fig. 2a). The Na+,K+-ATPase α1 subunit was present in the p85α-immunoprecipitates in OK cells treated with DA, but this effect was blocked in OK cells previously exposed to the SH3-p; the basal association of the p85α with the Na+,K+-ATPase α1 subunit was unaffected by the peptide alone. The inhibitory effect of DA on Na+,K+-ATPase activity was examined next (Fig. 2b). As predicted, DA decreased Na+,K+-ATPase activity, and the presence of SH3-p prevented this effect, whereas the peptide alone, or the presence of a scrambled peptide (not shown), were without effect.
Figure 2.
Role of p85α-SH3 domain in mediating the interaction of PI3K with the Na+,K+-ATPase α subunits. (a) The presence of Na+,K+-ATPase α1 subunits (see Fig. 1b) in the immunoprecipitates with the p85α antibody was examined in OK cells incubated with 1 μM DA for 2.5 min at 23°C in the presence or absence of 10 μM SH3-binding peptide (Calbiochem). Transient cell permeabilization was performed to allow access of the peptide to the cells' interiors. (b) Rubidium transport (nmol of Rb per mg of protein per min) was used as an index of Na+,K+-ATPase transport activity. Experiments were performed with identical protocols as described in a. Data represent the mean ±SEM of three experiments performed in triplicate. ∗, P < 0.05 (Student's t test).
Identification of the Na+,K+-ATPase α1 Subunit Domain That Interacts with PI3K-IA.
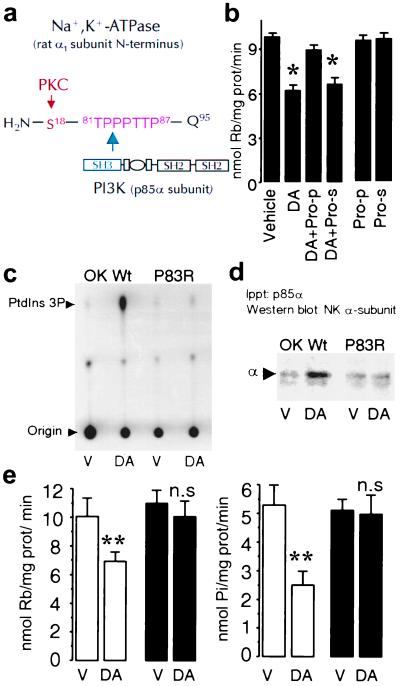
Because SH3 domains recognize and bind proline-rich motifs (36, 37), we have used several strategies to determine whether a conserved polyproline sequence (TPPPTTP87) present in the Na+,K+-ATPase α1 subunit N terminus (Fig. 3a) represents a potential PI3K-IA interaction site required for its activation and Na+,K+-ATPase endocytosis. We examined first whether DA was capable of inhibiting Na+,K+-ATPase activity in the presence of a peptide comprising the polyproline region or a scrambled peptide (Fig. 3b). DA decreased Na+,K+-ATPase activity to the same extent as was reported in Fig. 2, whereas the inhibitory effect was blunted in OK-Wt cells previously exposed to a peptide resembling the proline-rich domain (TPPPTTPE). The scrambled peptide in the presence of DA or either peptide alone did not affect Na+,K+-ATPase activity (Fig. 3b). In view of the above results with peptide competition assays, we next expressed in OK cells the rat Na+,K+-ATPase α1 subunit cDNA carrying the mutation Pro83→Arg (P83R) and measured the effect of DA on PI3K activity. In OK-Wt cells, DA increased PI3K-IA activity (277 ± 85% of control; n = 4; P < 0.05; Fig. 3c), whereas in OK cells expressing the P83R mutation, it failed to induce a significant change in kinase activity (90 ± 13% of control; n = 4; not significant). In addition, failure of DA to stimulate PI3K-IA activity in P83R cells also was associated with an impaired interaction between the Na+,K+-ATPase α1 subunit and PI3K (Fig. 3d), because the Na+,K+-ATPase subunit was not increased in the immunoprecipitated material by using a PI3K-IA p85α antibody in response to DA (shown as percent of control; OK-Wt: 162 ± 22% vs. P83R: 92 ± 11%; both n = 5).
Figure 3.
A proline-rich motif within the Na+,K+-ATPase α1 subunit is necessary for binding and activation of PI3K. (a) Schematic representation of the rat Na+,K+-ATPase α1 subunit N-terminal with the predicted amino acid (Gln95) adjacent to the plasma membrane. A proline-rich motif (TPPPTTP87) is located in the vicinity of the substrate (Ser18) for PKC. (b) Na+,K+-ATPase activity was determined in OK-Wt cells incubated with 1 μM DA in the presence or absence of a peptide (50 μM) comprising the proline motif (Pro-p) or of a scrambled peptide (Pro-s). Each bar represents the mean ±SEM of three experiments performed in triplicate. *, P < 0.02. (c) PI3K activity was determined as described (see legend to Fig. 1) in vehicle (V)- or DA-treated cells. (d) The presence of Na+,K+-ATPase α1 subunits in the immunoprecipitates was determined by Western blotting with a monoclonal antibody against the Na+,K+-ATPase α1 subunit as in Fig. 1. The data are representative of four experiments. (e) Na+,K+-ATPase activity was determined either as Rb+ transport (Left) or ATP hydrolysis (Right). Cells expressing the wild type (open bars) or the P83R mutant (filled bars) were incubated with vehicle (V; Hanks' buffer) or 1 μM DA. Each bar represents the mean ±SEM of five experiments performed in triplicate. **, P < 0.01. n.s., Not significant.
Mutation of Pro83 was performed to minimize possible structural changes that could affect the stability of the Na+,K+-ATPase α1 subunit N terminus and consequently the PKC substrate (Ser18). The ability of DA to increase phosphorylation of the α1 subunit in P83R cells represents a strong indication of N terminus integrity (data not shown). In addition, we previously have determined that simultaneous phosphorylation of Ser11 and Ser18 in the rat Na+,K+-ATPase α1 subunit by phorbol esters resulted in increased Na+,K+-ATPase activity (R.E., A.M.B., T. A. Pressley, M. Rousselot, E. Féraille, and C.H.P., unpublished observations), and this effect was not altered by the P83R mutation. A similar increase in Na+,K+-ATPase activity [Rb+ transport (nmol of Rb per mg of protein per min)] was observed in OK-Wt and P83R cells incubated in the presence of 0.1 μM phorbol 12-myristate 13-acetate (PMA) for 5 min at 23°C (OK-Wt, vehicle: 9.6 ± 0.6% vs. PMA: 16.8 ± 1.7%, P < 0.01; P83R, vehicle: 9.9 ± 0.9% vs. PMA: 14.5 ± 1.1%, P < 0.02; n = 5 in all groups).
Impact of Proline-Rich Motif Disruption on the Functional Properties of Na+,K+-ATPase and Its Regulation by DA.
Na+,K+-ATPase activity was determined in OK-Wt cells, and P83R mutants were preincubated with either vehicle or DA (Fig. 3e). Na+,K+-ATPase activity was determined either as the rate of ouabain-sensitive ATP hydrolysis or ouabain-sensitive rubidium transport, and under both assay conditions, DA exerted a comparable degree of inhibition (percentage inhibition; Na+,K+-ATPase hydrolytic activity: 45 ± 7% vs. rubidium transport: 42 ± 2%; n = 11 in both groups). The permeabilization procedure used for measuring Na+,K+-ATPase hydrolytic activity discriminates between endosomal and plasma membrane Na+,K+-ATPases (21). The basal activity was similar in the two types of cells (Fig. 3e), indicating that translation and membrane location of the molecules was not affected by the mutation. Although in OK-Wt, DA significantly inhibited Na+,K+-ATPase activity, it failed to induce a significant change in cells expressing the P83R mutation.
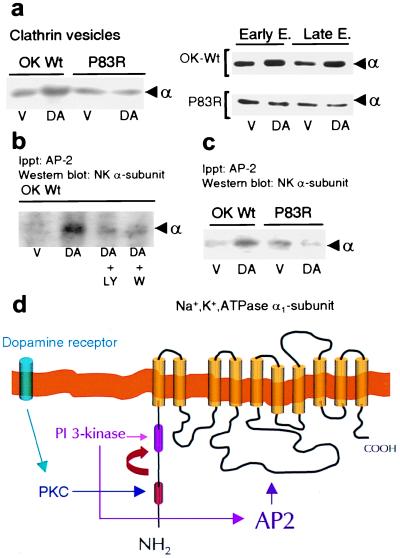
The inhibitory effect of DA on Na+,K+-ATPase is mediated by a decreased number of enzyme units (α/β subunits) within the plasma membrane and their transport into endosomes by clathrin-coated vesicles (19–21). Thus, we anticipated that failure of DA to inhibit Na+,K+-ATPase activity in P83R would be the consequence of impaired endocytosis of the α subunit. Indeed, whereas DA increased the abundance of α subunits in clathrin vesicles in OK-Wt cells (percentage of control, 228 ± 47%; n = 3), it failed to elicit this effect in OK cells expressing the P83R mutant (percentage of control, 90 ± 20; n = 3; Fig. 4a Left). Similarly, DA promoted α subunits' endocytosis into early and late endosomes in OK-Wt cells, but not in OK cells expressing the P83R mutant (Fig. 4a Right). Na+,K+-ATPase molecules originating from the synthetic pathway may explain the presence of α1 subunits in the intracellular organelles under basal conditions in both OK-Wt and P83R mutants.
Figure 4.
PI3K activity is required for recruitment and binding of AP-2 to the Na+,K+-ATPase α1 subunit. (a) Na+,K+-ATPase α1 subunit abundance in clathrin vesicles, early endosomes (EE) and late endosomes (LE) from OK cells expressing the wild type (OK-Wt) or P83R mutant. Cells were incubated at 23°C with vehicle (V; Hanks' buffer) or 1 μM DA for either 2.5 min (clathrin vesicles) or 10 min (EE and LE). Experiments were performed at least three times. (b) OK cells incubated with or without 1 μM DA (2.5 min at 23°C) were previously incubated (20 min at 23°C) in the presence or absence of either 100 nM wortmannin or 25 μM LY294002, two inhibitors of PI3K activity. The experiment shown was reproduced with identical results on three separate occasions. In both a and b, the material immunoprecipitated with an AP2αC antibody (Upstate Biotechnology, Lake Placid, NY) was analyzed by Western blotting using an antibody against the Na+,K+-ATPase α1 subunit. (c) OK wild-type (OK Wt) cells and P83R mutants were incubated with or without 1 μM DA (2.5 min at 23°C). Experiments were repeated three times. (d) Organization of the GPCR signals promoting the internalization of Na+,K+-ATPase α1 subunits (see Discussion for details).
Increased PI3K-IA Promotes the Binding of Adaptor Protein 2 (AP-2) to the Na+,K+-ATPase α Subunit.
DA facilitates the binding of AP-2 to the Na+,K+-ATPase α subunit during its endocytosis in renal epithelial cells (38). In an attempt to determine whether recruitment of AP-2 during Na+,K+-ATPase endocytosis could be promoted by PI3K-IA, we examined the binding of AP-2 to the Na+,K+-ATPase α subunit in the presence or absence of PI3K inhibitors (Fig. 4b). DA increased the abundance of Na+,K+-ATPase α1 subunits in the immunoprecipitated material with an antibody against AP-2, and this effect was blocked in the presence of either wortmannin or LY294002. Moreover, the increased colocalization of AP-2 and Na+,K+-ATPase α subunits in response to DA was absent in cells (P83R mutants) lacking the ability to increase PI3K-IA activity (Fig. 4c), further indicating that PI3K-IA activity is necessary for recruiting AP-2 to the site of Na+,K+-ATPase endocytosis.
Discussion
Inhibition of Na+,K+-ATPase activity by DA is accomplished by phosphorylation of the catalytic α subunit and by activation of PI3K-IA, both in a PKC-dependent manner, resulting in internalization of phosphorylated Na+,K+-ATPase molecules (20–22). Because PKC-dependent activation of PI3K-IA was not associated with phosphorylation of its regulatory or catalytic subunits, we have hypothesized that, similar to tyrosine-phosphorylated substrates, Ser-phosphorylation of the α subunit may serve as an anchor signal for binding and recruitment of PI3K-IA to the site of endocytosis.
To determine whether Ser18 phosphorylation of the Na+,K+-ATPase α subunit plays a relevant role during activation of PI3K-IA by DA, we have examined in cells expressing the Na+,K+-ATPase carrying a mutation in the PKC phosphorylation site (S18A) whether DA activates PI3K activity to the same extent as it does in cells expressing the wild-type rat α subunit. In S18A mutants, DA not only failed to increase PI3K-IA activity but also failed to promote the interaction between the PI3K-IA and Na+,K+-ATPase, because the α subunit did not increase in the immunoprecipitated material with a PI3K-IA p85α antibody. Failure of DA to stimulate PI3K-IA activity in cells expressing the Na+,K+-ATPase α1 subunit lacking the PKC substrate (Ser18 residue) suggests that increased PI3K-IA activity in response to DA is controlled by the state of phosphorylation of the Na+,K+-ATPase α subunit. The constitutive presence of Na+,K+-ATPase α1 subunits in the p85α-immunoprecipitates may represent the basal turnover in Na+,K+-ATPase traffic, and correlates with the presence of Na+,K+-ATPase subunits (α/β) within clathrin vesicles and endosomes under nonstimulating conditions.
Activation of intracellular signaling networks by GPCRs requires a high degree of specificity to ensure proper timing and spatial organization, and to avoid random crosstalk. This effect is partially accomplished by specific protein–protein interactions through domains such as the Src-homology (SH2 and SH3), pleckstrin-homology, phosphotyrosine binding, and PDZ (postsynaptic density protein, PSD-95; Drosophila septate junction protein disc–large, dlg; epithelial tight junction protein zinc-occludens, zo-l) domains, which are present in signaling and/or target molecules (39). The PI3K-IA bears a SH3 domain in its regulatory p85α subunit (40, 41). Although the SH3 domain present in the p85α may represent a site for self-association of the p85 protein (42), it also has been suggested to regulate (by binding to proline-rich motifs present in other proteins) the cellular localization of the PI3K-IA where it may become activated. In this study, by using site-directed mutagenesis to disrupt the proline-rich domain and peptide competition assays (peptides comprising the proline-rich motif and peptides that binds with high affinity to the SH3 domain of PI3K-IA p85 α subunit), we have established that the PI3K-IA interacts with the Na+,K+-ATPase α1 subunit. Furthermore, this interaction, which occurs constitutively, is enhanced by DA and involves the PI3K-IA p85 α subunit SH3 domain and a proline-rich motif within the N terminus of the Na+,K+-ATPase α1 subunit.
Although a selective mutation within the proline-rich motif did not affect the basal activity and expression of the Na+,K+-ATPase, it did impair its regulation, because DA failed to inhibit the Na+,K+-ATPase activity in cells expressing this mutant (P83R). This mutation was chosen to minimize potential changes within the N-terminal structure. Accordingly, DA still was able to phosphorylate the α subunit, and phorbol esters-dependent stimulation of Na+,K+-ATPase activity was not affected by the mutation. Thus, these results further argue in favor of the hypothesis that mutation of Pro83 selectively blocks the inhibitory effect of DA, and that failure of DA to achieve such an effect is unlikely to be the consequence of a possible misfolding, caused by proline disruption, of the Na+,K+-ATPase α1 subunit within the plasma membrane.
Decreased Na+,K+-ATPase activity is a reflection of increased internalization of Na+,K+-ATPase molecules into early and late endosomes by clathrin-coated vesicles (19–22). Therefore, we anticipated that failure of DA to decrease Na+,K+-ATPase activity in cells expressing the P83R mutant would be associated with defective endocytosis. Indeed, whereas in cells expressing the wild-type α subunit DA promoted Na+,K+-ATPase endocytosis, it failed to do so in cells expressing the Na+,K+-ATPase α subunit bearing a disruption in the proline-rich motif (P83R).
Selection of the cargo at the plasma membrane during clathrin-dependent endocytosis (43) requires binding of adaptors (i.e., AP-2) to tyrosine motifs present within plasma membrane proteins (44–46). Moreover, phosphorylation of the Na+,K+-ATPase α1 subunit enhances the binding of AP-2 to the α1 subunit, and this effect was absent in cells expressing the Na+,K+-ATPase lacking the PKC phosphorylation residue Ser-18 (S18A). Although the ability of AP-2 to bind tyrosine-based motifs (YXXØ) in vitro is enhanced by clathrin, such binding is increased severalfold in the presence of phosphatidylinositol 3′ phosphate (47). Our results indicate that activation of PI3K-IA is required to promote the binding of AP-2 to the Na+,K+-ATPase α1 subunit, because DA failed to elicit this effect in the presence of two inhibitors of PI3K-IA (wortmannin or LY294002) and in cells lacking the possibility to increase PI3K-IA activity (P83R). Although the increased PI3K-IA activity may be critical for AP-2 recruitment, it might also be relevant at other stages during endocytosis. Because Rho has been reported to be of importance during constitutive traffic of Na+,K+-ATPase molecules (48), it also may be that active PI3K-IA could modulate Rho activity (49, 50) and thereby affect the actin/microtubule organization during vesicle endocytosis.
In conclusion, we have identified the existence of a proline-rich motif within the Na+,K+-ATPase catalytic α subunit that is essential for its endocytosis in response to a GPCR (DA) signal. Although this study suggests that activation of PI3K-IA activity during Na+,K+-ATPase endocytosis is controlled by the state of Ser-phosphorylation of the Na+,K+-ATPase α subunit, it also defines an alternative pathway by which class IA PI3K could be activated in response to GPCR signals. Unlike the conventional process of activation involving SH2-domain interaction with phosphorylated tyrosine residues, increased PI3K-IA activity in response to DA is controlled by the state of Ser-phosphorylation of the protein (Na+,K+-ATPase α subunit) to be internalized (Fig. 4d). Selective mutation analysis suggests that PI3K-IA activation results from its interaction (through the p85α-SH3 domain) with a proline-rich domain present in the Na+,K+-ATPase α subunit. This proline motif becomes accessible after PKC-dependent phosphorylation of an upstream serine residue (Ser18) within the α subunit N terminus. The results also demonstrate the existence of a region within the Na+,K+-ATPase N terminus that serves as its own scaffold, organizing the receptor signals that ultimately regulate its traffic and activity in epithelial cells.
Acknowledgments
We thank Drs. Michael J. Caplan (Yale University, New Haven, CT), Jack H. Kaplan (Oregon Health Sciences University, Portland), and Bart Vanhaesebroeck (Ludwig Institute for Cancer Research, London, U.K.) for critical reading of the manuscript. We also thank Dr. Michael J. Caplan for the generous gift of Na+,K+-ATPase antibody. This work was supported in part by funds from the Swedish Medical Research Council, the Swedish Heart and Lung Foundation, the Novo Nordisk Foundation, the Karolinska Institutet, and National Institutes of Health Grant DK53460.
Abbreviations
- DA
dopamine
- GPCR
G protein-coupled receptor
- PKC
protein kinase C
- PI3K-IA
phosphoinositide 3′ kinase class IA
- SHn
Src homology n domain
- OK
an opossum renal epithelial cell line of proximal tubule origin
- AP-2
adaptor protein 2
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100128297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100128297
References
- 1.Rodriguez-Boulan E, Nelson W J. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- 2.Fish E M, Molitoris B A. N Engl J Med. 1994;330:1580–1588. doi: 10.1056/NEJM199406023302207. [DOI] [PubMed] [Google Scholar]
- 3.Caplan M J. Am J Physiol. 1997;272:F425–F429. doi: 10.1152/ajprenal.1997.272.4.F425. [DOI] [PubMed] [Google Scholar]
- 4.Jörgensen P L. Physiol Rev. 1980;60:864–917. doi: 10.1152/physrev.1980.60.3.864. [DOI] [PubMed] [Google Scholar]
- 5.Katz A I. Am J Physiol. 1982;242:F207–F219. doi: 10.1152/ajprenal.1982.242.3.F207. [DOI] [PubMed] [Google Scholar]
- 6.Doucet A. Kidney Int. 1988;34:749–760. doi: 10.1038/ki.1988.245. [DOI] [PubMed] [Google Scholar]
- 7.Bertorello A M, Hökfelt T, Goldstein M, Aperia A. Am J Physiol. 1988;254:F795–F801. doi: 10.1152/ajprenal.1988.254.6.F795. [DOI] [PubMed] [Google Scholar]
- 8.Finkel Y, Eklöf A C, Granquist L, Soares-da-Silva P, Bertorello A M. Gastroenterology. 1994;107:675–679. doi: 10.1016/0016-5085(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 9.Vieira-Coelho M A, Teixeira V L, Finkel Y, Soares-da-Silva P, Bertorello A M. Am J Physiol. 1998;275:G1317–G1323. doi: 10.1152/ajpgi.1998.275.6.G1317. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Beach R A, Lokhandwala M F. Hypertension. 1993;21:364–372. doi: 10.1161/01.hyp.21.3.364. [DOI] [PubMed] [Google Scholar]
- 11.Nishi A, Eklof A C, Bertorello A M, Aperia A. Hypertension. 1993;21:767–771. doi: 10.1161/01.hyp.21.6.767. [DOI] [PubMed] [Google Scholar]
- 12.Tripodi G, Valtorta F, Torielli L, Chieregatti E, Salardi S, Trusolino L, Menegon A, Ferrari P, Marchisio P C, Bianchi G. J Clin Invest. 1996;97:2815–2822. doi: 10.1172/JCI118737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertorello A M, Katz A I. Am J Physiol. 1993;265:F743–F755. doi: 10.1152/ajprenal.1993.265.6.F743. [DOI] [PubMed] [Google Scholar]
- 14.Ewart H S, Klip A. Am J Physiol. 1995;269:C295–C311. doi: 10.1152/ajpcell.1995.269.2.C295. [DOI] [PubMed] [Google Scholar]
- 15.Bertorello A M, Aperia A. Am J Physiol. 1989;256:F370–F373. doi: 10.1152/ajprenal.1989.256.2.F370. [DOI] [PubMed] [Google Scholar]
- 16.Bertorello A M, Aperia A, Walaas S I, Nairn A C, Greengard P. Proc Natl Acad Sci USA. 1991;88:11359–11362. doi: 10.1073/pnas.88.24.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ominato M, Satoh T, Katz A I. J Membr Biol. 1996;152:235–243. doi: 10.1007/s002329900101. [DOI] [PubMed] [Google Scholar]
- 18.Efendiev R, Bertorello A M, Pedemonte C H. FEBS Lett. 1999;456:45–48. doi: 10.1016/s0014-5793(99)00925-4. [DOI] [PubMed] [Google Scholar]
- 19.Chibalin A V, Katz A I, Berggren P-O, Bertorello A M. Am J Physiol. 1997;273:C1458–C1465. doi: 10.1152/ajpcell.1997.273.5.C1458. [DOI] [PubMed] [Google Scholar]
- 20.Chibalin A V, Pedemonte C H, Katz A I, Feraille E, Berggren P O, Bertorello A M. J Biol Chem. 1998;273:8814–8819. doi: 10.1074/jbc.273.15.8814. [DOI] [PubMed] [Google Scholar]
- 21.Chibalin A V, Ogimoto G, Pedemonte C H, Pressley T A, Katz A I, Feraille E, Berggren P O, Bertorello A M. J Biol Chem. 1999;274:1920–1927. doi: 10.1074/jbc.274.4.1920. [DOI] [PubMed] [Google Scholar]
- 22.Chibalin A V, Zierath J R, Katz A I, Berggren P O, Bertorello A M. Mol Biol Cell. 1998b;9:1209–1220. doi: 10.1091/mbc.9.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rameh L E, Cantley L C. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 24.Vanhaesebroeck B, Leevers S J, Panayotou G, Waterfield M D. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 25.Reif K, Gout I, Waterfield M D, Cantrell D A. J Biol Chem. 1993;268:10780–10788. [PubMed] [Google Scholar]
- 26.Malström K, Stange G, Murer H. Biochim Biophys Acta. 1987;902:2269–2277. doi: 10.1016/0005-2736(87)90305-1. [DOI] [PubMed] [Google Scholar]
- 27.Nash S R, Godinot N, Caron M G. Mol Pharmacol. 1993;44:918–925. [PubMed] [Google Scholar]
- 28.Pedemonte C H, Pressley T A, Cinelli A R, Lokhandwala M F. Mol Pharmacol. 1997;52:88–97. doi: 10.1124/mol.52.1.88. [DOI] [PubMed] [Google Scholar]
- 29.Erusalimsky J D, Friedberg L, Rozengurt E. J Biol Chem. 1998;263:19188–19194. [PubMed] [Google Scholar]
- 30.Johnson J A, Gray M O, Karliner J S, Chen C-H, Mochly-Rosen D. Circ Res. 1996;79:1086–1099. doi: 10.1161/01.res.79.6.1086. [DOI] [PubMed] [Google Scholar]
- 31.Heydrick S J, Jullien D, Gautier N, Tanti J F, Giorgetti S, Van Obberghen E, Le Marchand-Brustel Y. J Clin Invest. 1993;91:1358–1366. doi: 10.1172/JCI116337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond T G, Verroust P G. Am J Physiol. 1994;266:F554–F562. doi: 10.1152/ajprenal.1994.266.4.F554. [DOI] [PubMed] [Google Scholar]
- 33.Gorvel J-P, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 34.Feschenko M S, Sweadner K J. J Biol Chem. 1994;269:30436–30444. [PubMed] [Google Scholar]
- 35.Chen J K, Lane W S, Brauer A W, Tanaka A, Schreiber S L. J Am Chem Soc. 1993;115:12591–12592. [Google Scholar]
- 36.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 37.Viguera A R, Arrondo J L, Musacchio A, Saraste M, Serrano L. Biochemistry. 1994;33:10925–10933. doi: 10.1021/bi00202a011. [DOI] [PubMed] [Google Scholar]
- 38.Ogimoto G, Yudowski G A, Barker C J, Köhler M, Katz A I, Féraille E, Pedemonte C H, Berggren P-O, Bertorello A M. Proc Natl Acad Sci, USA. 2000;97:3242–3247. doi: 10.1073/pnas.060025597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawson T, Scott J D. Science. 1999;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 41.Pisabarro M T, Serrano L. Biochemistry. 1996;35:10634–10640. doi: 10.1021/bi960203t. [DOI] [PubMed] [Google Scholar]
- 42.Harpur A G, Layton M J, Das P, Bottomley M J, Panayotou G, Driscoll P C, Waterfield M D. J Biol Chem. 1999;274:12323–12332. doi: 10.1074/jbc.274.18.12323. [DOI] [PubMed] [Google Scholar]
- 43.Pearse B M, Robinson M S. Annu Rev Cell Biol. 1990;6:151–171. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- 44.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 45.Kirchhausen T, Bonifacino J S, Riezman H. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 46.Bonifacino J S, Dell'Angelica E C. J Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapoport I, Miyazaki M, Boll W, Duckworth B, Cantley L C, Shoelson S, Kirchhausen T. EMBO J. 1997;16:2240–2250. doi: 10.1093/emboj/16.9.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmalzing G, Richter H P, Hansen A, Schwarz W, Just I, Aktories K. J Cell Biol. 1995;130:1319–1332. doi: 10.1083/jcb.130.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolias K F, Cantley L C, Carpenter C L. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 50.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]