Abstract
Rickettsial pathogens in the genera Anaplasma and Ehrlichia cause acute infection in immunologically naive hosts and are major causes of tick-borne disease in animals and humans. Immunization with purified outer membranes induces protection against acute Anaplasma marginale infection and disease, and a proteomic and genomic approach recently identified 21 proteins within the outer membrane immunogen in addition to the well-characterized major surface proteins MSP1 to MSP5. Among the newly described proteins were the type IV secretion system (TFSS) proteins VirB9, VirB10, and conjugal transfer protein (CTP). In other gram-negative bacteria, TFSS proteins form channels, facilitate secretion of molecules, and are required for intracellular survival. However, TFSS proteins have not been explored as vaccine antigens. In this study we demonstrate that in Anaplasma marginale outer membrane-vaccinated cattle, VirB9, VirB10, and CTP are recognized by serum immunoglobulin G2 (IgG2) and stimulate memory T-lymphocyte proliferation and gamma interferon secretion. VirB9 induced the greatest proliferation in CD4+ T-cell lines, and VirB9-specific CD4+ T-cell clones responded to three A. marginale strains, confirming the VirB9-specific T-cell responses are directed against epitopes in the native protein. The three TFSS proteins are highly conserved with orthologous proteins in Anaplasma phagocytophilum, Ehrlichia chaffeensis, and Ehrlichia canis. Recognition of TFSS antigens by CD4+ T cells and by IgG2 from cattle immunized with the protective outer membrane fraction provides a rationale for including these proteins in development of vaccines against A. marginale and related pathogens.
Anaplasma marginale is a rickettsial hemoparasite of cattle that causes dramatic weight loss, anemia, and often death during acute infection, becoming persistent in animals that recover (2). Among nonliving vaccines, purified outer membranes have provided the best protection against infection and disease, but the protective antigens within the outer membrane have not been well characterized (1, 10, 26, 33-35, 37-39, 48). Antibody responses in outer membrane vaccinees are primarily directed against major surface protein 2 (MSP2) and MSP3, but these proteins continually undergo antigenic variation and do not confer protection (1, 36, 37). We recently identified more than 20 proteins in the outer membrane immunogen by mass spectrometry and genomic mapping, including type IV secretion system (TFSS) proteins VirB9, VirB10, and conjugal transfer protein (CTP) (24).
In gram-negative bacteria, the TFSS mediates transfer of proteins, DNA, or protein-DNA complexes between cells. For example, in the plant pathogen Agrobacterium tumefaciens, VirB7 to VirB10 form the core element of the TFSS and are responsible for transferring substrates across the cell envelope into adjacent bacteria or into the host cell (15). The importance of the TFSS in intracellular survival and virulence has been widely documented for Legionella pneumophila, Helicobacter pylori, Bordetella pertussis, and Brucella suis (12, 14, 20, 23, 31, 42, 50). L. pneumophila contains 26 genes that are designated dot (defect in organelle trafficking) or icm (intracellular multiplication), and a large number of Dot/Icm proteins are homologous to CTPs of other intracellular bacteria. Furthermore, the Dot/Icm proteins are responsible for injecting effector proteins into the host cell phagosome to control its biogenesis (11). The importance of the dot/icm genes in pathogenesis was shown with dot/icm mutant strains of L. pneumophila that exhibited severely inhibited growth in macrophages (44, 50). Similarly, H. pylori uses the TFSS to transport its effector protein CagA into host cells, leading to H. pylori-induced cancer onset and tumor progression (11). The cag pathogenicity island of H. pylori is comprised of a 40-kb stretch of DNA encoding homologues of the TFSS proteins of A. tumefaciens and was shown to be responsible for induction of inflammation and pathogenesis in the gastric lumen of humans (12). TFSS proteins have also been found in rickettsial pathogens, but their functions are less well understood (18, 28, 32, 41).
Because of their surface localization, highly conserved nature, and requirement for intracellular survival, gram-negative bacterial TFSS proteins are logical targets for immunological intervention. However, the immunogenicity of TFSS proteins has been virtually unexplored (18, 28, 32, 41). The present study focused on determining if A. marginale VirB9, VirB10, and CTP, which we previously identified as components of the protective bacterial outer membrane fraction, induced B- and T-lymphocyte responses in outer membrane-immunized cattle.
MATERIALS AND METHODS
Animals used in the study.
Three Holstein steers, designated 04B90, 04B91, and 04B92, were used in this study. Sequencing of the BoLA DQA genes was performed as described previously (40). The nomenclature of bovine class II genes can be found at the following websites: http://www.projects.roslin.ac.uk/bola and http://www.ebi.ac.uk/ipd/mhc/bola. BoLA-DRB3 and DQA haplotypes for the calves in this study are as follows: calf 04B90, DRB3 *1101/*1501, DQA *10011/*2206, DQB *10011/*22021; calf 04B91, DRB3 *1201/*2703, DQA *12011/*2201, DQB *0101/*22031; and calf 04B92, DRB3 *0201/*1201, DQA *0203, DQB *12011/*2201.
At 7 months of age, these animals were immunized four times with outer membranes from the 1.22-g-per-cm3 fraction of a sucrose density gradient as described previously (24). Briefly, each calf received a subcutaneous inoculation at weeks 0, 2, 4, and 8 of 60 μg total protein of A. marginale outer membranes resuspended in 1.3 ml phosphate-buffered saline containing 6 mg saponin. Seroconversion was determined by immunoblotting using pre- and postimmunization sera as described previously (24). Sera used in this study were obtained 2 weeks after the final immunization.
In silico analysis of VirB9, VirB10, and CTP.
The prediction algorithm SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (5) was used to predict signal peptide cleavage sites for VirB9, VirB10, and CTP. Also, TMpred, a transmembrane prediction algorithm (http://www.ch.embnet.org/software/TMPRED_form.html) (21), was used to determine the predicted transmembrane domains in VirB9, VirB10, and CTP. For alignment, presentation, and calculation of percentage identities of predicted amino acids between VirB9, VirB10, or CTP from the A. marginale St. Maries and Florida strains or the A. marginale St. Maries strain and Anaplasma phagocytophilum, Ehrlichia chaffeensis, and Ehrlichia canis, the BoxShade 3.21 server (http://www.ch.embnet.org/software/BOX_form.html) and the AlignX module of the VECTOR NTI software package (Invitrogen, Carlsbad, CA) were used. The accession numbers and sources of the sequences are as follows: A. marginale (St. Maries), YP_154362 (VirB9), YP_154361 (VirB10), and YP_153506 (CTP). The unpublished Florida strain sequences were obtained from the following website: http://genomics.vetmed.wsu.edu/blast/blast.html. For A. phagocytophilum (HZ strain), YP_505897 (VirB9), YP_505896 (VirB10), and YP_504712 (CTP) were used. For E. chaffeensis, YP_506875 (VirB9, Arkansas strain), AAM00413 (VirB10, Arkansas strain), and ZP_00544847 (CTP, Sapulpa strain) were used. For E. canis, AAQ12004 (VirB9, Oklahoma strain), YP_302669 (VirB10, Jake strain), and YP_303416 (CTP, Jake strain) were used.
Amplification, cloning, and sequencing of VirB9, VirB10, and CTP.
The St. Maries A. marginale genomic DNA used to amplify TFSS genes was isolated using the Qiaex II gel extraction kit (QIAGEN Sciences, Valencia, CA) using 3 ml of blood from a persistently infected calf (Table 1). Generally, 10 μmol of forward and reverse primer, containing the 24-nucleotide FLAG tag sequence (Table 1) that encodes the peptide epitope DYKDDDDK, 25 μl of PCR Master Mix (Roche Diagnostics), 2 μl of St. Maries A. marginale genomic DNA, and 22 μl of nuclease-free water were used for PCRs. Amplification consisted of 45 cycles with the melting temperature at 94°C for 30 s, annealing temperature at 60°C for 45 s, and extension temperature at 72°C for 90 s. Amplicons were cloned into the pBAD/TOPO ThioFusion expression vector as specified by the manufacturer (Invitrogen). Plasmid DNA containing correctly oriented inserts was purified from Escherichia coli using the Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI) after overnight growth in LB broth. Inserts were sequenced using the Big Dye kit and an ABI PRISM automated sequencer (PE-Applied Biosystems, Foster City, CA). Sequencing primers used were the Trx forward, pBAD reverse, and forward, reverse, and internal primers from the TFSS genes of interest. Sequences were compiled and analyzed with the VECTOR NTI software package (Invitrogen). Colonies with correct sequences were grown overnight in LB broth and frozen as a glycerol stock (50% glycerol in LB broth).
TABLE 1.
Primer sequences used to amplify TFSS genes
| Gene name | Forward primer | Reverse primera |
|---|---|---|
| VirB9 | 5′-ATGAATTTCTATAAAAACTTGCTTGCGTGCTCGGC-3′ | 5′-TTTGTCGTCGTCGTCTTTATAGTCAAGCACCGTATTCACTACTTCGAC-3′ |
| VirB10 | 5′-TTGAGTTTAGGGATGTCAGACGAAACCAAGG-3′ | 5′-TTTGTCGTCGTCGTCTTTATAGTCCCTACGCACCGCCTCCCTAG-3′ |
| CTP | 5′-ATGAAAAAGGCTTTCATGGTTTGTGCTGTAGCGC-3′ | 5′-TTTGTCGTCGTCGTCTTTATAGTCCCCACGTCCCCTTCTG-3′ |
Reverse primers contain the 30-nucleotide FLAG tag sequence, which is boldfaced.
Expression and purification of VirB9, VirB10, and CTP.
To obtain antigen to be tested in CD4+ T-cell proliferation assays and immunoblot analyses, VirB9, VirB10, CTP, and the C-terminal region (amino acids [aa] 317 to 565) of Babesia bovis rhoptry-associated protein (RAP-1 CT), used as a negative control (30), were expressed as thioredoxin-FLAG fusion proteins according to the manufacturer's specifications (Invitrogen). The FLAG epitope and the thioredoxin protein were positioned at the N and C termini, respectively, of each protein, adding 16 kDa to the molecular weight of each recombinant protein.
For recombinant protein expression, 500 ml of LB broth was inoculated with 5 ml of overnight culture and grown at 37°C. Bacteria were induced with 0.2% arabinose and harvested by centrifugation after 5 h. Bacteria were resuspended in lysis buffer, 6 M guanidine hydrochloride, 20 mM sodium phosphate, pH 7.8, and 500 mM sodium chloride and disrupted by sonication. Recombinant proteins were purified from bacterial lysates with the ProBond purification system (Invitrogen) and dialyzed overnight at 4°C against 10 mM Tris (pH 8.0)-0.1% Triton X-100 using a 12- to 14-kDa molecular mass cutoff D-Tube Dialyzer Maxi (Novagen, Madison, WI). After dialysis, Complete Mini protease inhibitors (Roche) were added to samples per the manufacturer's specifications, and recombinant proteins were further purified by anti-FLAG immunoaffinity chromatography as specified by the manufacturer (Sigma-Aldrich, St. Louis, MO). Purified recombinant proteins of the appropriate molecular masses of 46 kDa (rVirB9 and rCTP) and 65 kDa (rVirB10) were dialyzed against phosphate-buffered saline (pH 7.0) with the 12- to 14-kDa molecular mass cutoff D-Tube Dialyzer Maxi (Novagen). Protein concentrations were determined using the Quick Start Bradford protein assay (Bio-Rad, Hercules, CA). Purity of recombinant proteins was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using an alkaline phosphatase-labeled anti-FLAG monoclonal antibody (MAb) (Sigma-Aldrich). Whole E. coli lysate prior to induction (15 μl) and 5 h postinduction (6 μl) and purified recombinant protein (1 μg) were separated by SDS-PAGE and stained with Coomassie brilliant blue or transferred to a polyvinylidene difluoride (PVDF) membrane. After an overnight incubation in I-Block reagent (Applied Biosystems, Bedford, MA), PVDF membranes were probed with an alkaline phosphatase-labeled anti-FLAG monoclonal antibody (Sigma-Aldrich) diluted 1:5,000 in I-Block reagent and binding was detected with the Western-Star chemiluminescent immunoblot detection system (Applied Biosystems).
IgG2-specific responses and serum titration to recombinant TFSS proteins.
To determine immunoglobulin G2 (IgG2) responses to rVirB9, rVirB10, and rCTP, immunoblotting was performed as described previously (24). Briefly, 5 μg of uninfected erythrocytes and St. Maries A. marginale outer membranes, 15 μl of E. coli extract, and 1 μg of rRAP-1 CT, rVirB9, rVirB10, and rCTP were electrophoresed and transferred to PVDF membranes. Membranes were blocked overnight in I-Block reagent.
Preimmune and immune sera obtained 2 weeks after the fourth antigen immunization from cattle 04B90, 04B91, and 04B92 (24) were adsorbed against E. coli. E. coli was grown overnight in 75 ml of antibiotic-free LB broth, pelleted by centrifugation, resuspended in 20 ml of I-Block reagent (Applied Biosystems) with protease inhibitors, and disrupted by sonication. Sera were subjected to five 30-min rounds of adsorption, pelleting the bacteria after each adsorption at 3,500 × g for 20 min, yielding a final dilution of 1:200. The adsorbed sera were incubated with the membranes for 1 h. Horseradish peroxidase-conjugated sheep anti-bovine IgG2 (Serotec, Raleigh, NC) diluted 1:3,000 was used as the secondary antibody. After washing of immunoblots with I-Block reagent, antigen-antibody binding was detected by chemiluminescence using an ECL Western blotting detection system (Amersham Biosciences, Piscataway, NJ). To determine the presence of rRAP-1 CT, PVDF membranes containing this antigen were then probed with the alkaline phosphatase-labeled anti-FLAG monoclonal antibody diluted at 1:5,000 and developed as described above.
To determine antibody titers against recombinant proteins and MSP2, 30 μg of rCTP, rVirB9, and rVirB10 and 150 μg of outer membranes were electrophoresed and transferred to PVDF membranes. PVDF membranes were cut into individual strips and probed with E. coli-adsorbed immune bovine sera diluted from 1:200 to 1:120,000. Secondary antibody was sheep anti-bovine IgG2 (Serotec) diluted to 1:3,000, and immunoblots were developed as described above. Titers were determined as the highest serum dilutions to react with the specific band in recombinant protein or to the ∼37-kDa MSP2 band in outer membranes on immunoblots.
A. marginale outer membrane-specific T-cell lines.
Peripheral blood mononuclear cells (PBMC) from outer membrane vaccinees were collected 3 years following their final immunizations to establish short-term T-cell lines. PBMC were depleted of CD8+ cells and γδ T cells using MAb 7C2B (anti-CD8 receptor) and MAb CACT-61A (anti-γδ receptor) followed by complement lysis, which consistently depletes 90 to 95% of CD8+ and γδ T cells (7). The resulting PBMC enriched for CD4+ T cells were cultured at 4 × 106 cells per well in 24-well plates (Costar, Cambridge, MA) in a final volume of 1.5 ml of complete RPMI-1640 medium (8) with 3 μg per ml of St. Maries A. marginale outer membrane antigen (24). After 7 days, lymphocytes were washed and subcultured without antigen to a density of 7 × 105 cells per well with 2 × 106 fresh autologous irradiated (3,000 rads) PBMC as a source of antigen-presenting cells (APC) per well for 7 days. These “rested” CD4+ T-cell-enriched lines were then assayed for antigen-specific proliferation or used for establishing T-cell clones.
VirB9-specific T-cell clones.
T-cell clones were established from animals 04B90, 04B91, and 04B92 3 years following their final immunization with outer membranes. In the first cloning experiment with animals 04B90 and 04B91, T-cell lines stimulated and rested as described above were subcultured to 7 × 105 cells per well with 0.5 μg per ml of rVirB9 and 2 × 106 fresh autologous APC per well for 7 days and then cloned. In the second cloning experiment with animal 04B92, T-cell lines were established as described above, followed by restimulation with 0.5 μg per ml of rVirB9 for 7 days, and then subcultured with autologous APC without antigen for 7 days. This two-week round of stimulation with rVirB9 followed by a subculture with autologous APC without antigen was repeated for a total of 6 weeks. Lymphocytes from these T-cell lines were then cloned by limiting dilution in 96-well plates (Costar) as described previously (17). Briefly, cells were diluted to a statistical average of 0.3 or 1 cell per well and stimulated with 0.1 μg (04B91) or 0.5 μg (04B90 and 04B92) of rVirB9 per ml of medium containing 10% T-cell growth factor and 5 × 105 APC. Cells were successively expanded into 48- and 24-well plates and tested for proliferation to rVirB9 and the St. Maries strain of A. marginale outer membranes. The cloning frequencies (percentage of growth-positive wells) were <30%.
Cell surface phenotype analysis.
Differentiation markers on T-cell lines and clones were analyzed by flow cytometry. The MAbs used were specific for bovine CD4 (CACT138), CD8 (CACT80c), and the γδ T-cell receptor (GB21A). These MAbs were purchased from the monoclonal antibody center at Washington State University, Pullman, WA.
Lymphocyte proliferation assays.
To determine proliferative responses to recombinant TFSS proteins, PBMC, T-cell lines, or T-cell clones from outer membrane vaccinees were assayed in triplicate wells with 0.1 to 10 μg per ml of St. Maries A. marginale, outer membranes, recombinant proteins (rVirB9, rVirB10, and rCTP or rRAP-1 CT as a negative control), or uninfected erythrocyte membranes (URBC). After 4 days (T-cell lines and clones) or 6 days (PBMC), cells were radiolabeled for 6 to 18 h with 0.25 μCi of [3H]thymidine (Dupont, New England Nuclear, Boston, MA) and harvested onto glass filters, and radionucleotide incorporation was measured with a Beta-plate 1205 liquid scintillation counter (Wallac, Gaithersburg, MD). Proliferation assays were performed at least three times. Results are presented as the mean counts per minute (cpm) of replicate cultures ± 1 standard deviation (SD) or as the stimulation index, calculated as the mean cpm of cells cultured with antigen/mean cpm of cells cultured with medium alone. Statistical analysis was determined by one-way analysis of variance, where a P value of <0.05 is considered statistically significant.
IFN-γ ELISPOT assays.
PBMC from outer membrane-immunized animals were used for analyzing gamma interferon (IFN-γ) secretion to recombinant proteins using the enzyme-linked immunospot (ELISPOT) assay as described previously (52) with minor modifications. After blocking and washing of nitrocellulose-backed 96-well plates, 1.0 × 106 PBMC per well were added in 100-μl volumes containing complete RPMI 1640 alone or a mixture of 1.0 μg/ml phytohemagglutinin (Sigma-Aldrich) and 0.01 ng/ml human interleukin 12 (IL-12) or with 10 μg per ml of URBC, A. marginale outer membranes, rVirB9, rVirB10, rCTP, or rRAP-1 CT. Cells were incubated at 37°C for 48 h, and plates were then washed, developed, and dried overnight. Spots were visualized using an ELISPOT reader (Cell Technology Inc., Columbia, MD) and AID 2.9 software (AutoImmune Diagnostika, Strasbourg, Germany). For each PBMC sample, the mean number of spots from negative control wells was subtracted from the number of spots in test wells. Results are presented as mean numbers of spot-forming cells per 1 × 106 PBMC. One-way analysis of variance with Bonferonni correction for multiple comparisons was used to determine significance between test antigens, where a P value of <0.05 is considered statistically significant. Recombinant proteins were compared to the negative control antigen rRap1CT, while St. Maries A. marginale outer membranes were compared to URBC.
RESULTS
TFSS protein alignment and prediction of signal peptide cleavage sites and transmembrane domains.
Predicted amino acid sequences from two strains of A. marginale and one strain (each) of A. phagocytophilum, E. chaffeensis, and E. canis were used to align VirB9, VirB10, and CTP. When St. Maries and Florida strains of A. marginale were compared, there was complete amino acid identity in VirB9 and VirB10 and 97.1% identity in CTP (Fig. 1 A to C). When amino acid sequences of VirB9 from A. phagocytophilum, E. chaffeensis, and E. canis were compared to those for the St. Maries strain of A. marginale, there was 55%, 53%, and 54% identity, respectively (Fig. 1A); VirB10 had 60%, 51%, and 51% identity, respectively (Fig. 1B); and CTP had 74%, 66%, and 66% identity, respectively (Fig. 1C). As expected, comparison of these proteins between A. marginale and A. phagocytophilum showed the highest degree of relatedness.
FIG. 1.
Predicted amino acid sequence alignment of VirB9, VirB10, and CTP from the A. marginale Florida and St. Maries strains (A. mar FL and SM), A. phagocytophilum (A. phago), E. chaffeensis (E. chaff), and E. canis. For VirB9 (A), the HZ strain of A. phagocytophilum, Arkansas strain of E. chaffeensis, and Oklahoma strain of E. canis were used. For VirB10 (B), the HZ strain of A. phagocytophilum, Arkansas strain of E. chaffeensis, and Jake strain of E. canis were used. For CTP (C), the HZ strain of A. phagocytophilum, Sapulpa strain of E. chaffeensis, and Jake strain E. canis were used. When the majority of amino acids at a given position are identical between the five organisms, a black background is used, whereas conservative amino acids are indicated by a gray background. Protein sequences were also analyzed by SignalP and TMpred to determine predicted signal peptide cleavage sites (as indicated by an asterisk for CTP and a dashed line for VirB9) or transmembrane regions (as indicated by the underlined amino acids). For all strains analyzed, VirB9 (A) is predicted to include signal peptide cleavage sites within the same region (dashed line), VirB10 (B) is not predicted to contain a signal peptide cleavage site but is predicted to contain two transmembrane domains (underlined amino acids), and CTP (C) is predicted to contain one common signal peptide cleavage site (asterisk).
TFSS proteins were also analyzed with SignalP (5) and TMpred (21) to predict their surface localization. For the strains analyzed, VirB9 (Fig. 1A) was predicted to contain a signal peptide cleavage site within the region of aa 23 to 28, CTP (Fig. 1C) was predicted to contain a common signal peptide cleavage, and SignalP did not predict a signal peptide cleavage site for VirB10 (Fig. 1B). Additionally, VirB10 was predicted to contain two transmembrane domains in the regions of aa 28 to 47 and aa 323 to 357. Topology prediction algorithms calculated VirB9, VirB10, and CTP to be either outer membrane proteins or surface-associated proteins. These predictions are consistent with surface localization of similar TFSS proteins in other bacteria (6, 11, 13, 43).
Expression and purification of recombinant TFSS proteins.
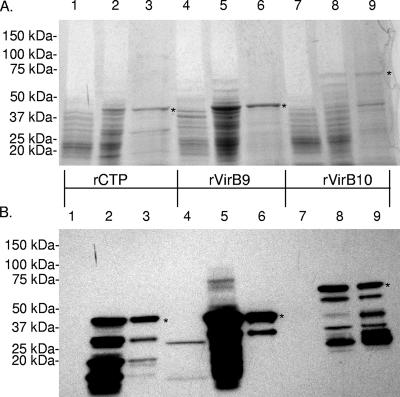
A. marginale (St. Maries strain) VirB9, VirB10, and CTP were expressed as FLAG-tagged proteins and affinity purified for use in immunological assays. SDS-PAGE (Fig. 2A) and immunoblotting analysis (Fig. 2B) of E. coli lysates, prior to and 5 h after induction and of affinity-purified recombinant proteins indicates that the recombinant proteins were relatively free from E. coli contamination. Furthermore, rVirB9 and rCTP fusion proteins of approximately 46 kDa and the rVirB10 fusion protein of approximately 65 kDa were identified in the predicted molecular mass range. However, smaller proteins that reacted with the anti-FLAG MAb were also detected, suggesting that degradation of recombinant proteins occurred during expression, even though protease inhibitors were included.
FIG. 2.
Expression and purification of TFSS proteins. Proteins were separated by SDS-PAGE and stained with Coomassie blue (A) or transferred to PVDF membranes and probed with an alkaline phosphatase-labeled anti-FLAG MAb and detected by chemiluminescence (B). Lysates (15 μl) of uninduced E. coli containing rCTP (lane 1), rVirB9 (lane 4), or VirB10 (lane 7), lysates (6 μl) of induced E. coli containing rCTP (lane 2), rVirB9 (lane 5), or rVirB10 (lane 8), and 5 μg (Coomassie blue-stained gel) or 1 μg (immunoblots) of purified rCTP (lane 3), rVirB9 (lane 6), or rVirB10 (lane 9) were tested. rCTP and rVirB9 fusion proteins are approximately 46 kDa, while rVirB10 is approximately 65 kDa, indicated by asterisks.
IgG2 from outer membrane vaccinees recognizes recombinant proteins.
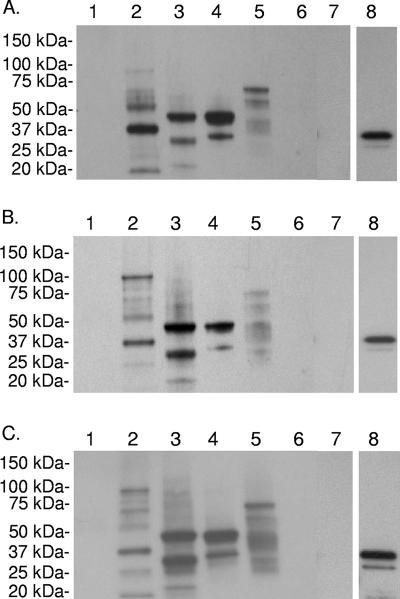
To evaluate serologic responses to recombinant proteins, immunoblotting was performed with E. coli-adsorbed immune sera from outer membrane-immunized cattle and the blots were developed with IgG2-specific antibody. IgG2 was measured because this isotype is the best opsonizing antibody in cattle (27) and is associated with protective immunity in outer membrane vaccinees (10). IgG2 from animals 04B90 (Fig. 3A), 04B91 (Fig. 3B), and 04B92 (Fig. 3C) reacted with all three recombinant proteins but not with the negative control, rRAP-1 CT. To confirm the presence of the rRAP-1 CT fusion protein, immunoblots containing this antigen were reprobed with anti-FLAG MAb (Fig. 3A, B, and C), revealing a protein of the expected molecular mass. Preimmune sera did not react with any of the antigens tested (data not shown).
FIG. 3.
Serological reactivity to recombinant proteins. Five micrograms of URBC (lane 1) or St. Maries A. marginale outer membranes (lane 2), 1 μg of rCTP (lane 3), rVirB9 (lane 4), rVirB10 (lane 5), or rRAP-1 CT (lane 6), or 15 μl of Top 10 E. coli (lane 7) was separated by SDS-PAGE and transferred to a PVDF membrane. Membranes were probed with E. coli-adsorbed immune serum (1:200 dilution) from bovine 04B90 (A), 04B91 (B), or 04B92 (C). Binding was detected by horseradish peroxidase-conjugated anti-bovine IgG2 secondary antibody and chemiluminescence. To confirm the presence of rRAP-1 CT, PVDF membranes were then probed with an alkaline phosphatase-labeled anti-FLAG monoclonal antibody and binding was detected by chemiluminescence (lane 8).
Bovine serum IgG2 titers against recombinant TFSS proteins or MSP2 were also determined by immunoblotting (Table 2). IgG2 titers to TFSS proteins ranged from 1,000 to 5,000, whereas titers against MSP2 were 30,000 or 60,000, consistent with the immunodominant nature of this protein.
TABLE 2.
Titers of IgG2 to recombinant proteins in cattle immunized with outer membranes
| Animal | Titer (reciprocal of dilution)a to:
|
|||
|---|---|---|---|---|
| rCTP | rVirB9 | rVirB10 | MSP2 | |
| 04B90 | 5,000 | 5,000 | 1,000 | 30,000 |
| 04B91 | 1,000 | 1,000 | 1,000 | 60,000 |
| 04B92 | 1,000 | 1,000 | 2,000 | 30,000 |
Sera were diluted 1:200 to 1:120,000 and tested for reactivity to recombinant protein or native MSP2 on immunoblots. The titer is defined as the reciprocal of the highest serum dilution giving a positive signal.
Lymphocyte proliferation and IFN-γ secretion against recombinant TFSS.
Proliferation assays were performed with PBMC from cattle 04B90, 04B91, and 04B92 prior to immunization and 4 weeks after the final immunization. Preimmunization PBMC did not respond to either the A. marginale St. Maries strain or uninfected erythrocytes, whereas PBMC from all three immunized animals proliferated strongly to the A. marginale St. Maries strain but insignificantly to URBC (data not shown).
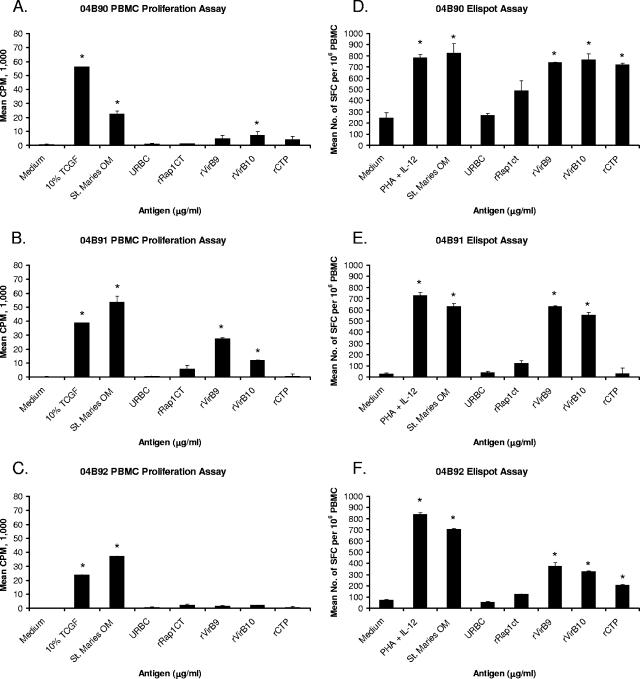
To measure memory T-cell responses to TFSS proteins, PBMC obtained 3 years or more after the last immunization were assayed for proliferation and IFN-γ-secreting cells (Fig. 4). For animal 04B90, only rVirB10 induced significant proliferation, whereas all three recombinant TFSS proteins induced IFN-γ secretion (Fig. 4A and D). For animal 04B91, significant proliferation and IFN-γ secretion were obtained with VirB9 and VirB10, whereas the responses to rCTP were insignificant (Fig. 4B and E). For animal 04B92, there were no detectable proliferative responses to any TFSS protein, whereas rVirB9 and rVirB10 induced significant IFN-γ responses (Fig. 4C and F). These results are representative of proliferation, and IFN-γ ELISPOT assays were performed at least three times for each animal.
FIG. 4.
PBMC from outer membrane vaccinees proliferate and secrete IFN-γ in response to recombinant TFSS proteins. PBMC from bovine 04B90 (A and D), 04B91 (B and E), or 04B92 (C and F) were tested for proliferation (A to C) or IFN-γ secretion (D to F). For proliferation, PBMC were stimulated with T-cell growth factor (TCGF) (positive control) or 0.1 to 10 μg per ml of St. Maries outer membranes (OM), URBC, or recombinant proteins. Results are presented as the mean cpm plus one SD, and responses significantly greater than those to medium or negative-control antigens are indicated by an asterisk. For IFN-γ ELISPOT assays, PBMC were stimulated with 1.0 μg per ml phytohemagglutinin plus 0.01 ng per ml IL-12 (positive control) or 10 μg per ml indicated antigen. Data are presented as mean numbers of spot-forming cells (SFC) per 106 PBMC plus one SD. Statistical significance of a response compared that to medium or negative-control antigen is indicated by an asterisk where the P value is ≤0.05.
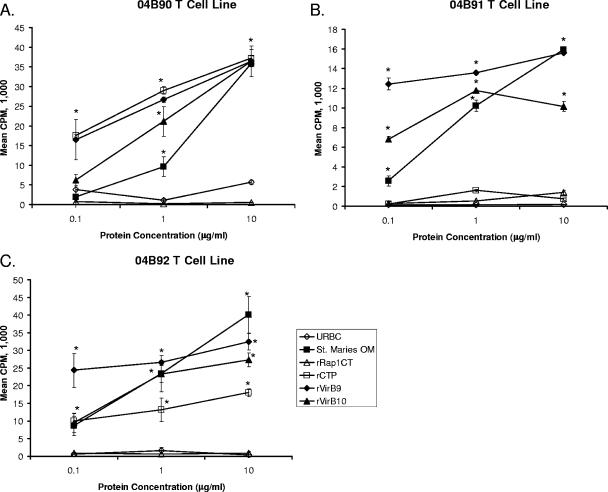
To attempt to enhance the frequency of antigen-specific T cells within PBMC, T-cell lines were established by incubating CD8+ T-cell-depleted and γδ T-cell-depleted PBMC with A. marginale outer membranes for 1 week followed by a 1-week resting period. Such short-term T-cell lines from animals 04B90 (Fig. 5A) and 04B92 (Fig. 5C) had significant proliferative responses to all three recombinant TFSS proteins at antigen concentrations ranging from 0.1 to 10 μg/ml. In contrast, T-cell lines from animal 04B91 still responded only to VirB9 and VirB10 (Fig. 5B). These assays were performed at least three times with similar results.
FIG. 5.
CD4+ T-cell proliferative responses to rCTP, rVirB9, and rVirB10. Two-week T-cell lines enriched for CD4+ T cells from animal 04B90 (A), 04B91 (B), or 04B92 (C) were cultured for 4 days with 0.1, 1, or 10 μg per ml of the indicated antigen, radiolabeled, and harvested. Results are presented as the mean cpm ± 1 SD for duplicate cultures. Statistical significance of a response compared to that for medium or negative-control antigen is indicated by an asterisk where the P value is ≤0.05.
VirB9-specific CD4+ T-cell clones.
To verify that the response to antigen was mediated by CD4+ T cells, T-cell clones specific for VirB9 were obtained. rVirB9 was selected, since this antigen was consistently recognized by memory T cells and induced strong proliferative responses from all three cattle. For each animal, the cloning frequency was <30%, indicating that the clones were derived from a single precursor cell (17). All clones expressed the CD4+ CD8− γδ-TCR− surface phenotype (data not shown). Importantly, clones from each animal responded to the St. Maries strain of A. marginale in addition to rVirB9 (Table 3) but did not respond to negative control antigens, including URBC, rRAP-1 CT, or native MSP2, indicating the response to rVirB9 by PBMC and isolated CD4+ T cells is specific for the native protein antigen. In all cases, the clones had higher proliferative responses to rVirB9 antigen than to A. marginale outer membranes, which was not unexpected, since VirB9 is not an abundant protein in the outer membrane fraction (24). However, the difference in response was most marked for clone 91.4C12. Neither epitope specificity nor major histocompatibility complex (MHC) class II restriction elements have been determined for the T-cell clones, so it is possible that certain epitopes are processed and presented more effectively than others from outer membranes, resulting in comparatively better responses to this antigen.
TABLE 3.
Proliferative responses of A. marginale VirB9-specific CD4+ T-cell clones
| Expt no. and animal | T-cell clone | Proliferation (SI)c in response to:
|
||||||
|---|---|---|---|---|---|---|---|---|
| URBC | St. M | FL | S. Idaho | rVirB9 | MSP2 | rRAP-1 CT | ||
| Expt. 1a | ||||||||
| 04B90 | 90.2F1 | 1.0 | 203.7 | 382.2 | 0.7 | NDd | ||
| 04B91 | 91.3D11 | 0.8 | 3.4 | 12.9 | 1.0 | 0.3 | ||
| 91.1H8 | 0.7 | 5.4 | 53.3 | ND | 0.7 | |||
| 91.2G10 | 0.4 | 2.6 | 26.6 | ND | 1.1 | |||
| 91.4C12 | 1.3 | 3.4 | 162.9 | ND | 0.5 | |||
| 04B92 | 92.3A6 | 0.5 | 9.2 | 74.0 | ND | 0.5 | ||
| 92.3B3 | 0.4 | 7.3 | 97.6 | ND | 0.4 | |||
| 92.4C5 | 1.0 | 3.8 | 22.8 | ND | 0.8 | |||
| Expt. 2b | ||||||||
| 04B92 | 92.3E2 | 0.9 | 37.8 | 144.7 | 105.4 | 182.4 | 1.3 | |
| 92.4E4 | 0.7 | 14.0 | 44.8 | 20.4 | 107.6 | 0.5 | ||
In experiment 1, T-cell clones were stimulated with antigen and APC in a 4-day proliferation assay for responses to URBC, St. Maries strain A. marginale outer membranes, rVirB9, MSP2, or rRap1CT. Clone 90.2F1 was stimulated with 1.0 μg per ml VirB9 and 10 μg per ml of the other antigens, clone 91.3D11 was stimulated with 1 μg per ml antigen, and the remaining clones were stimulated with 5 μg per ml antigen.
In experiment 2, T-cell clones 92.3E2 and 92.4E4 were stimulated as above and tested for proliferation responses to the URBC, St. Maries, Florida, and S. Idaho strains of A. marginale at 10 μg per ml and rVirB9 and rRAP-1 CT at 5 μg per ml.
Data are presented as the stimulation index (SI), which is the mean cpm of duplicate cultures of cells plus antigen/mean cpm of duplicate cultures of cells plus medium. Statistically significant responses are indicated in boldfaced type. URBC, St. M, FL, and S. Idaho, URBC, St. Maries, Florida, and S. Idaho strains of A. marginale.
ND, not determined.
DISCUSSION
TFSS proteins are important for survival and virulence of gram-negative bacteria (12, 14, 20, 23, 31, 42, 50), but these proteins have been virtually unexplored as vaccine candidates. We recently showed that VirB9, VirB10, and CTP are components of the protective A. marginale outer membrane fraction (24). Additionally, because TFSS proteins are highly conserved due to functional constraints, they may be more appropriate candidates for vaccines than the antigenically variant MSP2 and MSP3. In this study we show that in outer membrane-immunized cattle, these three A. marginale TFSS proteins elicit significant CD4+ T-lymphocyte proliferation, IFN-γ secretion, and IgG2 production, immune responses associated with protective immunity (10, 48).
VirB9, VirB10, and CTP were originally identified by mass spectrometry of proteins eluted from two-dimensional gel spots that reacted with immune bovine sera (24). However, we could not definitively identify these as the immunoreactive proteins, because the spot containing VirB9 also contained two additional proteins (Opag2 and OMP7), and it is possible that the spots containing VirB10 and CTP also contained additional proteins not identified by mass spectrometry. In the current study we definitively demonstrate the antigenicity of VirB9, VirB10, and CTP in purified outer membranes, since T cells and antibody from outer membrane vaccinees recognize recombinant forms of these proteins. Interestingly, serum IgG2 titers to these recombinant TFSS proteins were 5- to 60-fold lower than those to MSP2, indicating the relative immunodominance of MSP2 compared with TFSS proteins in these animals. Because the well-characterized outer membrane proteins, including serologically immunodominant and antigenically variant MSP2 and MSP3 of A. marginale, either have been ineffective or have provided only partial protection against challenge compared with outer membranes (1, 26, 33-35, 37-39, 48), subdominant conserved antigens are of interest for vaccine development.
Even though IgG2 responses to the three recombinant TFSS proteins were detected in all animals, differential T-lymphocyte responses were observed. For animal 04B90, the use of short-term T-cell lines increased the ability to detect responses to rCTP and rVirB9 at all antigen concentrations tested. Also, while PBMC from animal 04B92 had undetectable proliferative responses to recombinant TFSS proteins, T-cell lines did respond significantly to all three TFSS proteins. In contrast, responses to rCTP were never detected by either PBMC or short-term T-cell lines from animal 04B91. The discrepancy in CTP-specific IgG2 production but lack of CTP-specific T-cell recognition could be explained by a frequency of CTP-specific T cells that is below detection limits. Alternatively, CTP could be covalently associated with another protein containing T-cell epitopes that could serve as a carrier, as observed for A. marginale MSP1a and MSP1b (9, 25, 49). In support of this possibility, extensive covalent associations between TFSS proteins have been documented for A. tumefaciens (4, 22, 51).
Another interesting finding is the lack of strict correlation between proliferation and IFN-γ secretion by PBMC from animal 04B92, where proliferative responses to recombinant TFSS proteins were not observed, but rVirB9 and rVirB10 did induce IFN-γ secretion. Our data using T-cell lines which were presumably enriched for outer membrane protein-specific T cells (29), including TFSS-specific T cells, support the likelihood that the frequency of TFSS-specific lymphocytes in PBMC was below detection limits for a proliferation assay. This lack of strict correlation between lymphocyte proliferation and IFN-γ secretion is similar to results that have been reported with PBMC from humans exposed to malaria (19). Furthermore, differential IFN-γ and IL-2 gene regulation in memory and effector T cells has been documented with other models (3, 46, 47).
Importantly, the TFSS proteins are highly conserved among geographically distant strains of A. marginale. The strong responses of two VirB9-specific CD4+ T-cell clones against the St. Maries, Florida, and South Idaho strains result from conserved T-cell epitopes, as expected because of the complete conservation of VirB9 in the St. Maries and Florida strains of A. marginale. Furthermore, VirB10 and CTP from the St. Maries and Florida strains of A. marginale have 100% and 97.1% amino acid identity, respectively, so that CD4+ T-cell epitopes will be conserved in these proteins as well. While the cattle used in this study were selected for their expression of common MHC class II DRB3, DQA, and DQB alleles, which are present in more than 50% of Holstein-Friesian cattle (16, 45), it will also be important to determine if TFSS protein epitopes are recognized by additional MHC class II haplotypes within the Holstein population to ensure recognition of these proteins in the population if these antigens are to be incorporated into a vaccine.
Predicted amino acid sequence alignment of A. marginale VirB9, VirB10, and CTP and orthologous proteins in related rickettsial pathogens A. phagocytophilum, E. canis, and E. chaffeensis revealed the presence of highly conserved regions and an overall high amino acid identity between A. marginale VirB9, VirB10, and CTP and the orthologous proteins. Transcriptional analyses of TFSS genes in E. chaffeensis and A. phagocytophilum have shown that VirB8, VirB9, VirB10, VirB11, and VirD4 are transcribed polycistronically (32). In E. canis, VirB9 is expressed throughout various stages of infection and is antigenic in E. canis-infected dogs (18). Additionally, elevated levels of VirB9 were expressed during replication of A. phagocytophilum within neutrophils, whereas basal levels of VirB9 were expressed upon neutrophil lysis (28), demonstrating differential levels of expression of TFSS proteins during the mammalian infection cycle. It will be useful to determine if immune sera from infected human and canine patients recognize TFSS proteins, which might indicate their relevance as vaccine candidates for these organisms. Antibodies specific for TFSS proteins might prevent bacterial attachment to host cells or transport of macromolecules and in this way provide protective immunity by neutralizing the organism.
There are several examples demonstrating the importance of the TFSS in virulence and intracellular survival for several gram-negative bacteria (12, 14, 20, 23, 31, 42, 50). We have shown that three TFSS proteins, present in the bacterial outer membrane, are recognized by immune effectors from A. marginale outer membrane-immunized cattle. In addition, these proteins are highly conserved between geographically distinct strains of A. marginale and immunogenic for cattle that have common MHC class II haplotypes. These findings, together with the regions of sequence identity in orthologous proteins of related rickettsial pathogens, indicate the potential use of TFSS proteins in vaccine development for A. marginale and related pathogens of animals and humans.
Acknowledgments
We thank Kim Kegerreis, Shelly Whidbee, Beverly Hunter, Emma Karel, and Ralph Horn for excellent technical assistance and Kevin Lahmers for help with flow cytometry.
This research was supported by an American Society for Microbiology Robert D. Watkins Fellowship, by National Institutes of Health NIAID grant R01-AI053692, and by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2004-35600-14175. Job Lopez was partially supported by a fellowship from the Seattle Chapter of the ARCS Foundation.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Abbott, J. R., G. H. Palmer, K. A. Kegerreis, P. F. Hetrick, C. J. Howard, J. C. Hope, and W. C. Brown. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 174:6702-6715. [DOI] [PubMed] [Google Scholar]
- 2.Alderink, F. J., and J. Dietrich. 1981. Anaplasmosis in Texas: epidemiologic and economic data from a questionnaire survey, p. 27-44. In Proceedings of the Seventh National Anaplasmosis Conference. Mississippi State University Press, Starkville, MS.
- 3.Aune, T. M., L. A. Penix, M. R. Rincon, and R. A. Flavell. 1997. Differential transcription directed by discrete gamma interferon promoter elements in naive and memory (effector) CD4 T cells and CD8 T cells. Mol. Cell. Biol. 17:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaupre, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Bourzac, K. M., and K. Guillemin. 2005. Helicobacter pylori-host cell interactions mediated by type IV secretion. Cell Microbiol. 7:911-919. [DOI] [PubMed] [Google Scholar]
- 7.Brown, W. C., W. C. Davis, D. A. Dobbelaere, and A. C. Rice-Ficht. 1994. CD4+ T-cell clones obtained from cattle chronically infected with Fasciola hepatica and specific for adult worm antigen express both unrestricted and Th2 cytokine profiles. Infect. Immun. 62:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, W. C., K. S. Logan, G. G. Wagner, and C. L. Tetzlaff. 1991. Cell-mediated immune responses to Babesia bovis merozoite antigens in cattle following infection with tick-derived or cultured parasites. Infect. Immun. 59:2418-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, W. C., G. H. Palmer, H. A. Lewin, and T. C. McGuire. 2001. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect. Immun. 69:6853-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, W. C., V. Shkap, D. Zhu, T. C. McGuire, W. Tuo, T. F. McElwain, and G. H. Palmer. 1998. CD4+ T lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., and R. Rappuoli. 1993. Pertussis toxin export requires accessory genes located downstream from the pertussis toxin operon. Mol. Microbiol. 8:429-434. [DOI] [PubMed] [Google Scholar]
- 15.Das, A., and Y. H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, C. J., L. Andersson, I. Joosten, P. Mariani, L. C. Gasbarre, and E. J. Hensen. 1992. Characterization of bovine MHC class II polymorphism using three typing methods: serology, RFLP and IEF. Eur. J. Immunogenet. 19:253-262. [DOI] [PubMed] [Google Scholar]
- 17.de St. Groth, F. 1982. The evaluation of limiting dilution assays. J. Immunol. Methods 49:R11-R23. [DOI] [PubMed] [Google Scholar]
- 18.Felek, S., H. Huang, and Y. Rikihisa. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71:6063-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan, K. L., E. A. Lee, M. B. Gravenor, W. H. Reece, B. C. Urban, T. Doherty, K. A. Bojang, M. Pinder, A. V. Hill, and M. Plebanski. 2001. Unique T cell effector functions elicited by Plasmodium falciparum epitopes in malaria-exposed Africans tested by three T cell assays. J. Immunol. 167:4729-4737. [DOI] [PubMed] [Google Scholar]
- 20.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 68:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, K., and W. Stoffel. 1993. A database of membrane spanning proteins segments. Biol. Chem. 374:166. [Google Scholar]
- 22.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lammertyn, E., and J. Anne. 2004. Protein secretion in Legionella pneumophila and its relation to virulence. FEMS Microbiol. Lett. 238:273-279. [DOI] [PubMed] [Google Scholar]
- 24.Lopez, J. E., W. F. Siems, G. H. Palmer, K. A. Brayton, T. C. McGuire, J. Norimine, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macmillan, H., K. A. Brayton, G. H. Palmer, T. C. McGuire, G. Munske, W. F. Siems, and W. C. Brown. 2006. Analysis of the Anaplasma marginale major surface protein 1 complex protein composition by tandem mass spectrometry. J. Bacteriol. 188:4983-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire, T. C., W. C. Davis, A. L. Brassfield, T. F. McElwain, and G. H. Palmer. 1991. Identification of Anaplasma marginale long-term carrier cattle by detection of serum antibody to isolated MSP-3. J. Clin. Microbiol. 29:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGuire, T. C., A. J. Musoke, and T. Kurtti. 1979. Functional properties of bovine IgG1 and IgG2: interaction with complement, macrophages, neutrophils and skin. Immunology 38:249-256. [PMC free article] [PubMed] [Google Scholar]
- 28.Niu, H., Y. Rikihisa, M. Yamaguchi, and N. Ohashi. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell Microbiol. 8:523-534. [DOI] [PubMed] [Google Scholar]
- 29.Norimine, J., S. Han, and W. C. Brown. 2006. Quantitation of Anaplasma marginale major surface protein (MSP)1a and MSP2 epitope-specific CD4+ T lymphocytes using bovine DRB3*1101 and DRB3*1201 tetramers. Immunogenetics 58:726-739. [DOI] [PubMed] [Google Scholar]
- 30.Norimine, J., J. Mosqueda, C. Suarez, G. H. Palmer, T. F. McElwain, G. Mbassa, and W. C. Brown. 2003. Stimulation of T-helper cell gamma interferon and immunoglobulin G responses specific for Babesia bovis rhoptry-associated protein 1 (RAP-1) or a RAP-1 protein lacking the carboxy-terminal repeat region is insufficient to provide protective immunity against virulent B. bovis challenge. Infect. Immun. 71:5021-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70:2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, G. H., A. F. Barbet, G. H. Cantor, and T. C. McGuire. 1989. Immunization of cattle with the MSP-1 surface protein complex induces protection against a structurally variant Anaplasma marginale isolate. Infect. Immun. 57:3666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299-1302. [DOI] [PubMed] [Google Scholar]
- 35.Palmer, G. H., A. F. Barbet, K. L. Kuttler, and T. C. McGuire. 1986. Detection of an Anaplasma marginale common surface protein present in all stages of infection. J. Clin. Microbiol. 23:1078-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer, G. H., J. E. Futse, D. P. Knowles, Jr., and K. A. Brayton. 2006. Insights into mechanisms of bacterial antigenic variation derived from the complete genome sequence of Anaplasma marginale. Ann. N. Y. Acad. Sci. 1078:15-25. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, G. H., and T. F. McElwain. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 57:233-253. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, G. H., D. Munodzana, N. Tebele, T. Ushe, and T. F. McElwain. 1994. Heterologous strain challenge of cattle immunized with Anaplasma marginale outer membranes. Vet. Immunol. Immunopathol. 42:265-273. [DOI] [PubMed] [Google Scholar]
- 39.Palmer, G. H., S. M. Oberle, A. F. Barbet, W. L. Goff, W. C. Davis, and T. C. McGuire. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 56:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y. H., Y. S. Joo, J. Y. Park, J. S. Moon, S. H. Kim, N. H. Kwon, J. S. Ahn, W. C. Davis, and C. J. Davies. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 5:29-39. [PubMed] [Google Scholar]
- 41.Renesto, P., H. Ogata, S. Audic, J. M. Claverie, and D. Raoult. 2005. Some lessons from Rickettsia genomics. FEMS Microbiol. Rev. 29:99-117. [DOI] [PubMed] [Google Scholar]
- 42.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohde, M., J. Puls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 44.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharif, S., B. A. Mallard, B. N. Wilkie, J. M. Sargeant, H. M. Scott, J. C. Dekkers, and K. E. Leslie. 1999. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) with production traits in Canadian dairy cattle. Anim. Genet. 30:157-160. [DOI] [PubMed] [Google Scholar]
- 46.Srikiatkhachorn, A., and T. J. Braciale. 1997. Virus-specific memory and effector T lymphocytes exhibit different cytokine responses to antigens during experimental murine respiratory syncytial virus infection. J. Virol. 71:678-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szabo, S. J., B. M. Sullivan, S. L. Peng, and L. H. Glimcher. 2003. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 21:713-758. [DOI] [PubMed] [Google Scholar]
- 48.Tebele, N., T. C. McGuire, and G. H. Palmer. 1991. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect. Immun. 59:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidotto, M. C., T. C. McGuire, T. F. McElwain, G. H. Palmer, and D. P. Knowles, Jr. 1994. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect. Immun. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 51.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., G. H. Palmer, J. R. Abbott, C. J. Howard, J. C. Hope, and W. C. Brown. 2003. CpG ODN 2006 and IL-12 are comparable for priming Th1 lymphocyte and IgG responses in cattle immunized with a rickettsial outer membrane protein in alum. Vaccine 21:3307-3318. [DOI] [PubMed] [Google Scholar]