Abstract
Genomic analysis of Mycoplasma pneumoniae revealed the existence of a large number of putative lipoprotein genes compared with the numbers in other bacteria. However, the pathogenic roles of M. pneumoniae lipoproteins are still obscure. In this study, we synthesized a lipopeptide (designated M. pneumoniae paralogous lipoprotein 1 [MPPL-1]) in which an S-dipalmitoylglyceryl cysteine was coupled to a peptide with a consensus sequence of a putative paralogous lipoprotein group characteristic of M. pneumoniae. The cytokine-inducing activity of MPPL-1 in human monocytic cells was much weaker (∼700-fold weaker) than that of the known mycoplasmal S-dipalmitoylated lipopeptide FSL-1 or MALP-2. MPPL-1 required Toll-like receptor (TLR2) to activate NF-κB-dependent gene transcription in HEK293 cells, although a 1,000-fold-larger amount of MPPL-1 was needed to exert activity similar to that of FSL-1 in the cells. TLR2-mediated recognition of MPPL-1 was synergistically upregulated by TLR6 but not by TLR1 or TLR10, although the activity was still weak. In addition, MPPL-1 did not antagonize FSL-1 recognition in human monocytic cells and TLR2/TLR6-expressing HEK293 cells. Thus, these results suggest that there is preferential selective recognition of diacylated lipopeptides due to the magnitude of an affinity with TLR2 and TLR6 and the roles of increased paralogous lipoprotein genes of M. pneumoniae in evasion of TLR2 recognition.
Membrane-bound lipoproteins are thought to play important roles in the survival of bacteria through four main functions: a structural function, a transport function, an adhesion function, and an enzymatic function (7). Many lipoproteins have been identified in various species of bacteria and have been shown to comprise a framework structure containing a lipidated N-terminal cysteine residue coupled to distinct polypeptides. The maturation of bacterial lipoproteins generally comprises three steps; the first step involves diacylglyceryl modification of a cysteine residue by diacylglycerol transferase, the second step involves cleavage of the leader peptide by signal peptidase II, and the final step involves N acylation of the N-terminal diacylglyceryl cysteinyl residue, with which lipoproteins are synthesized as triacylated lipoproteins (7). It has also been shown that lipoproteins derived from Rhodopseudomonas viridis and several mycoplasmal species do not undergo modification in the final step and are synthesized as diacylated lipoproteins (7).
In contrast to their crucial functions in the survival of bacteria, bacterial lipoproteins act as pathogenic substances to stimulate the immune systems of humans and animals through the recognition receptors that monitor exogenous pathogens (3). Toll-like receptors (TLRs) are central pattern recognition receptors of the innate immune system that recognize a wide range of invading microorganisms through conserved chemical structures in their cells (34). TLR2 is essential for mediation of immune responses to the most diverse set of molecular structures of microbes, including peptidoglycans, lipoteichoic acids, porins, lipoarabinomannans, and lipoproteins/lipopeptides (21, 34). TLR2 forms heteromers with either TLR1 or TLR6, probably to discriminate the structures of molecular patterns, especially the N-terminal lipidated cysteinyl portions of bacterial lipoproteins as active sites (4, 29). TLR1 and TLR6 have been reported to be involved in simple discrimination of the difference between triacylated and diacylated lipoproteins/lipopeptides (36, 37). However, recent arduous work by several study groups has shown that such diverse potentials of TLR1 and TLR6 are largely dependent on more subtle structures of lipoproteins/lipopeptides, such as the length of an N-terminal fatty acid chain, the chirality of the central carbon of the diacylglycerol, and the charge of the C-terminal amino acids (5, 6, 28). It has been suggested that in addition to TLR1 and TLR6, TLR10, which is not encoded in the murine genome, is related to TLR2 recognition because of its sequence similarity and the possibility that it forms a heteromer with TLR2 (8, 12).
Mycoplasmas are microbes in regressive evolution and differ from other microbes in many respects. For example, they completely lack a cell wall, and their bilipid membrane is therefore the only structure that regulates interactions with the external environment (31). Some mycoplasmas cause severe respiratory, arthritic, and urogenital diseases in humans and animals. Mycoplasma pneumoniae is a human pathogen that causes “atypical pneumonia,” particularly in older children and young adults (38). The genome size of M. pneumoniae is ∼820 kb, and the genomic sequence has been completely analyzed (13, 14). Interestingly, a large number of putative lipoprotein-encoding genes have been identified in the genome (46 of 689 genes; 6.68%) compared with the numbers of such genes in the genomes of other microbes, such as Escherichia coli K-12 (22 of 4,243 genes; 0.52%) and Bacillus subtilis (26 of 4,105 genes; 0.63%) (7). Even in the closely related sister species Mycoplasma genitalium, only 21 putative lipoproteins (encoded by 477 genes; 4.4%) could be found. Despite the existence of such genetic data, little is known about the roles of lipoproteins in M. pneumoniae pathogenicity, although there has been much interest in the pathogenic roles of membrane lipoproteins of other mycoplasmal species during infection because of their diverse functions, including adherence to host cells, antigenic variation, and TLR2- and TLR6-mediated immunostimulation (30).
In this study, we attempted to synthesize a lipopeptide having an S-(2,3-bispalmitoyloxypropyl)-cysteine residue coupled to an N-terminal consensus peptide of M. pneumoniae-specific lipoproteins encoded by paralogous genes. Interestingly, the level of immunostimulatory activity of this lipopeptide was much lower than that of the known mycoplasmal lipopeptide MALP-2 or FSL-1 despite the structural uniformity. We also investigated the recognition of this lipopeptide by TLRs.
MATERIALS AND METHODS
Preparation of synthetic lipopeptides.
The synthetic lipopeptides FSL-1 and MALP-2 were prepared as described previously (17). S-(2,3-bisacyloxypropyl)-cysteinyl TGIQADLRNLIK, designated M. pneumoniae paralogous lipoprotein 1 (MPPL-1), was synthesized using a method similar to the method used for synthesis of FSL-1 and MALP-2. Briefly, the side chain-protected sequence TGIQADLRNLIK was constructed with an automated peptide synthesizer (model 433; Applied Biosystems). (9-Fluorenylmethoxy carbonyl)-S-(2,3-bispalmitoyloxypropyl)-cysteine (Novabiochem) was manually coupled to the peptide resin by using a 1-hydroxy-7-azabenzotriazole-1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide/CH2Cl2-dimethylformamide solvent system. The 9-fluorenylmethoxy carbonyl and resin were removed from the lipopeptide by using trifluoroacetic acid. The lipopeptide was extracted into 90% acetic acid, lyophilized, and purified by preparative high-pressure liquid chromatography with a reversed-phase C18 column (30 by 250 mm). The level of purity of the lipopeptide was confirmed by analytical high-pressure liquid chromatography with a reversed-phase C18 column (4.6 by 150 mm) to be 96%. All of the lipopeptides were used without separation of the S-form and R-form stereoisomers. The lipopeptides were dissolved in phosphate-buffered saline containing 10 mM n-octyl-β-glucopyranoside at a concentration 0.5 mM and stored at −80°C until they were used.
Cell culture.
Dulbecco modified Eagle medium, RPMI 1640 medium, penicillin G, streptomycin, and trypsin-EDTA were obtained from Sigma. Human monocytic cell line THP-1 was cultured in RPMI 1640 medium as described previously (19). Human embryonic kidney HEK293 cells were grown in Dulbecco modified Eagle medium as described previously (18).
Determination of IL-6 and IL-8 by enzyme-linked immunosorbent assays (ELISA).
A total of 1 × 105 THP-1 cells were stimulated for 12 h with various concentrations of mycoplasmal lipopeptides, and the amounts of interleukin-6 (IL-6) and IL-8 released into the media were determined by using human IL-6 Cytoset and human IL-8 Cytoset (Invitrogen), respectively, according to the instructions of the manufacturer. The results described below are representative of three separate experiments, and the data are expressed as means and standard deviations.
DNA cloning.
Plasmids encoding human TLR1, TLR2, and TLR6 have been described previously (18). Human TLR10 cDNA was obtained by reverse transcription-PCR of RNA isolated from human umbilical vein endothelial cells and then cloned into a pEF6 vector (Invitrogen). The DNA sequences were confirmed by the dideoxy chain termination method by using an ABI Prism 3100 genetic analyzer.
Luciferase reporter gene assay.
HEK293 cells were plated at a concentration of 0.5 × 105 cells per well in 24-well plates before transfection. The cells were transiently transfected with an NF-κB-driven firefly luciferase reporter plasmid (pNF-κB-Luc; Stratagene) and a construct directing expression of Renilla luciferase under the control of a constitutively active thymidine kinase promoter (pRL-TK; Promega) together with TLR-encoding plasmids. After 24 h of incubation, the cells were stimulated for 6 h with MPPL-1 or FSL-1 in media containing 1% fetal bovine serum. Then the cells were lysed, and the luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega) according to instructions of the manufacturer. The results below, expressed as the means and standard deviations of values for triplicate wells, are representative of three separate experiments. The experiment using HEK293 cells stably expressing TLR2 has been described previously (20).
Statistics.
All values were evaluated by statistical analysis using Student-Newman-Keul's test. Differences were considered to be statistically significant at a P value of <0.05.
RESULTS
Preparation of MPPL-1.
Himmelreich et al. reported that 46 protein genes were identified as genes encoding putative lipoproteins in the M. pneumoniae M129 (=ATCC 29342) genome based on the following characteristic lipoprotein-specific features: (i) the presence of one or more basic amino acids among the first five to seven amino acids of the N terminus, (ii) the presence of a hydrophobic signal peptide, and (iii) the presence of a cysteine residue immediately downstream of the signal peptide (13). However, we found that 48 proteins had these lipoprotein signatures. The N-terminal lipoprotein moieties of all putative lipoproteins are shown in Table S1 in the supplemental material. The amino acid sequences of these lipoproteins are included in the data at a website (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=genome), and the protein designations were based on the MPN numbering scheme described by Himmelreich et al. (13). Importantly, many of these putative lipoproteins have recently been confirmed to be functionally expressed in the microorganism (11, 33, 39). In addition to 48 putative lipoproteins, there are several proteins with high levels of similarity to the lipoproteins without the lipoprotein signature at the N terminus (13), but we did not include these proteins in the list.
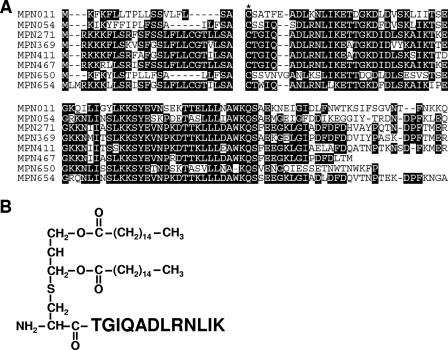
Comparison of 30 amino acids of N-terminal lipoprotein moieties revealed that the M. pneumoniae lipoproteins include members of seven subgroups, which are probably groups of paralogous lipoproteins (see Table S1 in the supplemental material). We focused on group 1 composed of MPN011, MPN054, MPN271, MPN369, MPN411, MPN467, MPN650, and MPN654 because the N-terminal sequences of these putative lipoproteins could not be identified by a BLAST search in other known organisms, even the sister species M. genitalium, suggesting that the lipoprotein genes were propagated uniquely in the evolution of this microorganism. The sequence of MPN505 is also very similar to the sequences of these lipoproteins, but MPN505 lacks the lipoprotein signature. Importantly, the study of Hallamaa et al. showed that there was expression of mRNAs for all group 1 lipoproteins and the detectable proteins MPN271, MPN411, and MPN650 (11). Comparison of N-terminal sequences of these lipoproteins revealed that the levels of similarity of MPN271, MPN369, MPN411, MPN467, and MPN654 are particularly high (Fig. 1A). These putative lipoproteins have a characteristic feature; namely, the C-terminal amino acid residue flanking the cysteine residue immediately downstream of the signal peptide is threonine, although the corresponding amino acid of common bacterial lipoproteins is glycine, alanine, or serine.
FIG. 1.
Synthesis of MPPL-1. (A) Alignment of putative paralogous lipoproteins. The N-terminal sequences of MPN011, MPN054, MPN271, MPN369, MPN411, MPN467, MPN650, and MPN654 were compared. The cysteine residue immediately downstream of the signal peptide is indicated by an asterisk. (B) Structure of MPPL-1.
To analyze the pathological roles of this paralogous lipoprotein group, we attempted to synthesize a lipopeptide having an N-terminal sequence common to these lipoproteins. The results of previous work suggested that synthetic lipopeptides with original peptide sequences with more than 10 amino acids could mimic the immunostimulatory activity of natural lipoproteins (24, 27, 32). Therefore, we determined that the partial consensus sequence of MPN271, MPN369, MPN411, MPN467, and MPN654 is TGIQADLRNLIK, which should couple to an S-dipalmitoylglyceryl cysteine. The structure was chemically synthesized using a method similar to the method used for synthesis of known mycoplasmal lipopeptides described previously (17, 32), and the protein was designated MPPL-1 (Fig. 1B). All of our preparations of mycoplasmal lipopeptides were synthesized as mixtures of the S-form and R-form stereoisomers.
Immunostimulatory activity of MPPL-1.
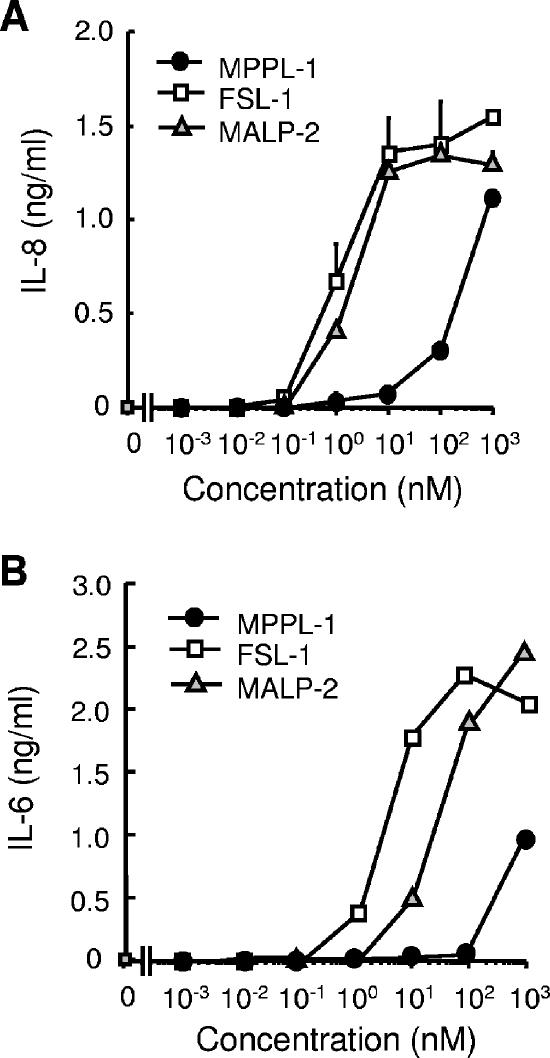
To investigate the immunostimulatory activity of MPPL-1, we examined the induction of cytokine production in human monocytic THP-1 cells, comparing the activity of MPPL-1 with the activities of two synthetic mycoplasmal lipopeptides. MALP-2 (S-dipalmitoylglyceryl CGNNDESNISFKEK) derived from Mycoplasma fermentans was first identified and characterized by Mühlradt's group as a compound that can activate macrophages even at picomolar concentrations (24). FSL-1 (S-dipalmitoylglyceryl CGDPKHPKSF) derived from Mycoplasma salivarium has recently been characterized by our group as a potent immunostimulatory compound, whose activity has been proposed to be stronger than that of MALP-2 (16, 17, 27). MPPL-1 could induce production of IL-8 in a dose-dependent manner at a concentration of ≥10 nM, whereas FSL-1 and MALP-2 could induce the production of IL-8 at picomolar concentrations (Fig. 2A). To induce a level of IL-8 production similar to the level induced by 1 nM FSL-1, a 300-fold-higher concentration of MPPL-1 was required (Fig. 2A). Moreover, similar weak activity of MPPL-1 was also observed when IL-6 production in THP-1 cells was examined (Fig. 2B). In this case, the concentration of FSL-1 needed to induce a level of IL-6 production similar to that induced by 1 μM MPPL-1 was 700-fold lower (Fig. 2B). Thus, the immunostimulatory activity of MPPL-1 is much weaker than the activities of structurally similar lipopeptides.
FIG. 2.
Cytokine-inducing activity of MPPL-1. A total of 1 × 105 THP-1 cells were stimulated for 12 h with the concentrations of MPPL-1, FSL-1, and MALP-2 indicated. Then the amounts of IL-8 (A) and IL-6 (B) released into the media were determined by ELISA. The results are representative of three separate experiments, and the data are means and standard deviations.
TLR recognition of MPPL-1.
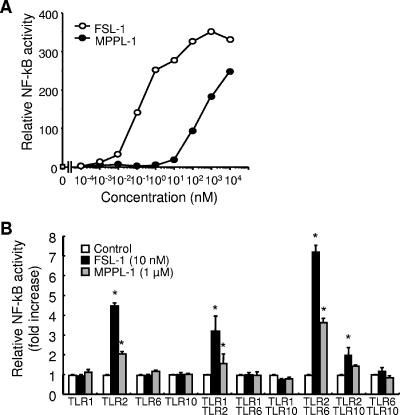
It has been shown that FSL-1 and MALP-2 stimulate human cells via recognition by TLR2 and TLR6 (26, 27, 35). We first examined whether MPPL-1 was recognized by TLR2 using HEK293 cells intrinsically lacking expression of TLR2 and responsiveness to TLR2 ligands (1). MPPL-1 could not stimulate parental HEK293 cells at concentrations ranging from 100 fM to ∼10 μM (data not shown) but could stimulate the cells stably transfected with TLR2, leading to induction of NF-κB activation, in a dose-dependent manner (Fig. 3A). Therefore, MPPL-1 recognition was completely dependent on TLR2 in the same way that FSL-1 and MALP-2 recognition was. However, an approximately 1,000-fold-higher concentration of MPPL-1 was required for activity similar to that of FSL-1 in TLR2-expressing HEK293 cells (Fig. 3A).
FIG. 3.
TLR usage of MPPL-1. (A) HEK293 cells stably transfected with TLR2 were prepared and transiently transfected with an NF-κB-driven firefly luciferase reporter plasmid. The cells were stimulated for 6 h with the concentrations of MPPL-1 and FSL-1 indicated. Then the cells were lysed, and the luciferase activity was measured. The results, expressed as the means of values for triplicate wells, are representative of three separate experiments. (B) HEK293 cells were transiently transfected with an NF-κB-driven firefly luciferase reporter plasmid together with the TLR-encoding plasmids indicated. The cells were stimulated for 6 h with 1 μM MPPL-1 or 10 nM FSL-1. Then the cells were lysed, and the luciferase activity was measured. The results, expressed as means and standard deviations of values for triplicate wells, are representative of three separate experiments. An asterisk indicates that the P value was <0.05 for a comparison with the control group.
We further investigated the requirement for TLR1, TLR6, and TLR10 for recognition of MPPL-1, since TLR2 has been shown to form not only a homomer but also heteromers with these TLRs (29). MPPL-1 could not activate HEK293 cells transfected with TLR1, TLR6, or TLR10 alone (Fig. 3B). Similarly, MPPL-1 could not activate cells transfected with a combination of TLR1 and TLR6, TLR1 and TLR10, or TLR6 and TLR10 (Fig. 3B). Compared with the MPPL-1 activity in the cells transfected with TLR2 alone, cotransfection of TLR6 with TLR2 synergically augmented the activity of MPPL-1 in a way similar to way observed with FSL-1, whereas cotransfection of TLR1 or TLR10 with TLR2 did not (Fig. 3B). Thus, MPPL-1 is preferentially recognized by TLR2/TLR6 in human cells in a manner similar to the recognition of FSL-1 and MALP-2.
Possibility of an antagonistic effect of MPPL-1 on TLR2 recognition.
TLR4 recognition of E. coli lipopolysaccharide can be antagonized by structurally similar compounds that have weak TLR4-stimulating activities (9, 10, 23, 25). However, it is still not clear whether TLR2 recognition of lipopeptides can be antagonized by structurally similar compounds. The results described above raise the possibility that MPPL-1 has an antagonistic effect on FSL-1 recognition by TLR2/TLR6, because MPPL-1 exhibits a much lower level of activity than FSL-1 exhibits through recognition by TLR2/TLR6. We therefore examined the IL-6-producing activity of FSL-1 in the presence and absence of a higher concentration of MPPL-1. IL-6 production induced by 1 or 10 nM FSL-1 was not altered by the presence of 1 μM MPPL-1 (Fig. 4A). Moreover, the presence of MPPL-1 was found to slightly increase the activity of FSL-1 as determined by analysis of NF-κB activation in HEK293 cells (Fig. 4B), and this analysis was more sensitive than an IL-6 ELISA with THP-1 cells. In addition, the MPPL-1 effect on FSL-1 recognition was not altered in the presence or absence of TLR1, TLR6, or TLR10 cotransfection (Fig. 4B). Similar results were obtained in experiments using MALP-2 (data not shown).
FIG. 4.
Antagonistic effect of MPPL-1. (A) A total of 1 × 105 THP-1 cells were stimulated for 12 h with 1 or 10 nM FSL-1 in the presence or absence of 1 μM MPPL-1. Then the amounts of IL-6 released into the media were determined by ELISA. The results, expressed as means and standard deviations, are representative of three separate experiments. (B) HEK293 cells were transiently transfected with an NF-κB-driven firefly luciferase reporter plasmid together with the TLR-encoding plasmids indicated. The cells were stimulated for 6 h with 1 nM FSL-1 in the presence or absence of 100 nM MPPL-1. Then the cells were lysed, and the luciferase activity was measured. The results, expressed as means and standard deviations of values for triplicate wells, are representative of three separate experiments.
DISCUSSION
We have been interested in the immunostimulatory activity of mycoplasmal diacylated lipoproteins/lipopeptides and the pathological roles of these proteins in mycoplasmal infections. So far, lipopeptides FSL-1 and MALP-2 have been identified as potent immunostimulatory compounds (22, 24). In this study, we synthesized lipopeptide MPPL-1 having a structure common in mycoplasmal lipopeptides, an S-dipalmitoylglyceryl cysteine residue coupled to a distinct peptide, which was determined on the basis of paralogous lipoproteins characteristic of M. pneumoniae. The cytokine-inducing activity of MPPL-1 in human cells was very weak compared with that of FSL-1 or MALP-2. At a higher concentration, MPPL-1 could weakly stimulate cells via TLR2/TLR6 recognition. However, MPPL-1 could not antagonize FSL-1 recognition by TLR2. These findings raised several important possibilities for biological activities of mycopalsmal lipopeptides, as discussed below.
Recent studies have revealed that the immunostimulatory activity of bacterial lipoproteins is completely dependent on the recognition and signal transduction by TLR2 that functions together with several associated molecules. TLR6 has been considered to be an essential participant in the discrimination of mycoplasmal diacylated lipoproteins/lipopeptides by TLR2, because MALP-2 recognition was impaired in macrophages from TLR6-deficient mice (36) and was reduced by a blocking antibody to TLR6 in human cells (26). However, Buwitt-Beckmann et al. found that C-terminal addition of SKKKK to the peptide moiety of MALP-2 converted the MALP-2 recognition by TLR2/TLR6 into recognition by a TLR6-independent mechanism (6). In addition, we previously reported that substitution of the C-terminal amino acid of FSL-1 (F to R) greatly impaired the immunostimulatory activity (27). Therefore, discrimination of diacylated lipopeptides by TLR2 and TLR6 has been suggested to be dependent on the amino acid sequence or structure of the peptide portion, although recognition of the lipolyated cysteine residue may be dependent on other molecules, such as CD36 (15). Furthermore, a recent report suggested that TLR1 participates in the recognition of a dipalmitoylated lipoprotein derived from M. pneumoniae (MPN602) (33). In this study, MPPL-1 was shown to be recognized by TLR2 and TLR6 but not by TLR1 or TLR10, as observed for MALP-2 and FSL-1. We could not discern a role for TLR10 in the recognition of mycoplasmal lipopeptides, although it is possible that TLR10 participates in accurate discrimination of bacterial lipoproteins/lipopeptides in human cells.
It is possible that studies of TLR antagonists may lead to the development of efficient therapeutic regulators of microbial infection or excess inflammation. In this study, however, MPPL-1 could not antagonize TLR2 recognition of FSL-1 (Fig. 4). The weak TLR2-stimulating activity of MPPL-1 raises the possibility that the peptide moiety of MPPL-1 has a low affinity for TLR6 but does not have an affinity for either TLR1 or TLR10. This possibility may be supported by our results showing that a small amount of FSL-1, which may have a stronger affinity than MPPL-1 has, could be preferentially recognized by TLR2 and TLR6 more than a larger amount of MPPL-1 could be recognized (Fig. 4). Moreover, our results may provide strong evidence for different ligand recognition mechanisms of TLR2 and TLR4, because TLR4 recognition of lipopolysaccharide is known to be antagonized by structurally similar compounds that have weak TLR4-stimulating activities (9, 10, 23, 25). Further study is needed to determine the detailed recognition machinery of mycoplasmal lipoproteins/lipopeptides.
The magnitude of the immunostimulatory activity of bacterial lipoproteins has been thought to be one of the crucial factors for pathogenicity of bacteria (3) which may be involved in the severity of host immune responses after bacterial infection. However, the presence of immunostimulatory compounds on the surface of bacterial cells leads to efficient clearance of bacteria through activation of immune cells, resulting in great reductions in efficient propagation and colonization on the host cell surface. To avoid activation of immune responses, several pathogenic bacteria have been shown to modify their surface molecules so they do not stimulate the TLR recognition system. For example, α- and ɛ-Proteobacteria, including Campylobacter jejuni, Helicobacter pylori, and Bartonella bacilliformis, modify the N-terminal D1 domain of flagellin, leading to evasion of TLR5 recognition (2). Therefore, structural modification of pathogen-activated molecular patterns may be important for bacterial pathogenicity. However, it has not been determined whether M. pneumoniae has the ability to evade immune systems. So far, mycoplasmal lipoproteins/lipopeptides have been identified to determine strong activators of immune cells in crude mixtures of lipoproteins obtained using methods such as Triton X-114 phase separation (24, 32, 33). In a recent study performed by Shimizu et al. (33), lipoprotein MPN602, which may have the strongest activities in M. pneumoniae lipoprotein mixtures, was identified by using a method to separate the fraction that strongly stimulates 293T cells transfected with TLR2 to activate NF-κB (33). MPN602 does not belong to a paralogous lipoprotein family, as shown in Table S1 in the supplemental material. Interestingly, it was also found that only a few lipoproteins possessed strong immunostimulatory activities and that the majority of lipoproteins had weak or no immunostimulatory activity (24, 32, 33). Consistent with this possibility, only a few lipoproteins with potent immunostimulatory activity have been identified so far, although there are many lipoproteins in mycoplasmal species. These observations suggest that the majority of lipoproteins of M. pneumoniae, including paralogous lipoprotein family members, have weak immunostimulatory activities. Moreover, our results suggest that propagation of genes encoding lipoproteins with weak immunostimulatory activity may be an important factor for the pathogenicity of M. pneumoniae through which the microorganism may evade TLR2 recognition. Further detailed investigations of the functions and immunostimulatory activities of lipoproteins found in M. pneumoniae are needed to address this possibility.
The bacterial lipoprotein structure has been found to be a lipidated (commonly palmitoylated) triacylated or diacylated S-glyceryl cysteine residue coupled to distinct polypeptides. However, the coupled peptide sequence has been shown to have a great effect on the immunostimulatory activity of the whole molecule. Therefore, synthesis and characterization of lipopeptides based on the known lipoprotein sequences of the N terminus may be an effective method for determining unknown biological activities of bacterial lipoproteins. Moreover, exhaustive screening of synthetic lipopeptides can lead to the identification of novel bacterial pathogenicities and to the development of biologically beneficial compounds or immune regulators. Also, it is possible that a cognate ligand for TLR10 will be identified by screening of these lipopeptides.
Supplementary Material
Acknowledgments
This work was supported by grants-in-aid for young scientists (B):16791102 and (B):18791363 to T.I. provided by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Editor: D. L. Burns
Footnotes
Published ahead of print on 26 February 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 2.Andersen-Nissen, E., K. D. Smith, K. L. Strobe, S. L. Barrett, B. T. Cookson, S. M. Logan, and A. Aderem. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102:9247-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 4.Bulut, Y., E. Faure, L. Thomas, O. Equils, and M. Arditi. 2001. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167:987-994. [DOI] [PubMed] [Google Scholar]
- 5.Buwitt-Beckmann, U., H. Heine, K. H. Wiesmuller, G. Jung, R. Brock, S. Akira, and A. J. Ulmer. 2006. TLR1- and TLR6-independent recognition of bacterial lipopeptides. J. Biol. Chem. 281:9049-9057. [DOI] [PubMed] [Google Scholar]
- 6.Buwitt-Beckmann, U., H. Heine, K. H. Wiesmuller, G. Jung, R. Brock, S. Akira, and A. J. Ulmer. 2005. Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur. J. Immunol. 35:282-289. [DOI] [PubMed] [Google Scholar]
- 7.Chambaud, I., H. Wroblewski, and A. Blanchard. 1999. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 7:493-499. [DOI] [PubMed] [Google Scholar]
- 8.Chuang, T., and R. J. Ulevitch. 2001. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim. Biophys. Acta 1518:157-161. [DOI] [PubMed] [Google Scholar]
- 9.Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 175:4490-4498. [DOI] [PubMed] [Google Scholar]
- 10.Coats, S. R., R. A. Reife, B. W. Bainbridge, T. T. Pham, and R. P. Darveau. 2003. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at Toll-like receptor 4 in human endothelial cells. Infect. Immun. 71:6799-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallamaa, K. M., G. F. Browning, and S. L. Tang. 2006. Lipoprotein multigene families in Mycoplasma pneumoniae. J. Bacteriol. 188:5393-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasan, U., C. Chaffois, C. Gaillard, V. Saulnier, E. Merck, S. Tancredi, C. Guiet, F. Briere, J. Vlach, S. Lebecque, G. Trinchieri, and E. E. Bates. 2005. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J. Immunol. 174:2942-2950. [DOI] [PubMed] [Google Scholar]
- 13.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoebe, K., P. Georgel, S. Rutschmann, X. Du, S. Mudd, K. Crozat, S. Sovath, L. Shamel, T. Hartung, U. Zahringer, and B. Beutler. 2005. CD36 is a sensor of diacylglycerides. Nature 433:523-527. [DOI] [PubMed] [Google Scholar]
- 16.Hubschle, T., J. Mutze, P. F. Muhlradt, S. Korte, R. Gerstberger, and J. Roth. 2006. Pyrexia, anorexia, adipsia, and depressed motor activity in rats during systemic inflammation induced by the Toll-like receptors-2 and −6 agonists MALP-2 and FSL-1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290:R180-187. [DOI] [PubMed] [Google Scholar]
- 17.Into, T., M. Fujita, T. Okusawa, A. Hasebe, M. Morita, and K. Shibata. 2002. Synergic effects of mycoplasmal lipopeptides and extracellular ATP on activation of macrophages. Infect. Immun. 70:3586-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Into, T., K. Kiura, M. Yasuda, H. Kataoka, N. Inoue, A. Hasebe, K. Takeda, S. Akira, and K. Shibata. 2004. Stimulation of human Toll-like receptor (TLR) 2 and TLR6 with membrane lipoproteins of Mycoplasma fermentans induces apoptotic cell death after NF-κB activation. Cell. Microbiol. 6:187-199. [DOI] [PubMed] [Google Scholar]
- 19.Into, T., Y. Nodasaka, A. Hasebe, T. Okuzawa, J. Nakamura, N. Ohata, and K. Shibata. 2002. Mycoplasmal lipoproteins induce Toll-like receptor 2- and caspases-mediated cell death in lymphocytes and monocytes. Microbiol. Immunol. 46:265-276. [DOI] [PubMed] [Google Scholar]
- 20.Into, T., and K. Shibata. 2005. Apoptosis signal-regulating kinase 1-mediated sustained p38 mitogen-activated protein kinase activation regulates mycoplasmal lipoprotein- and staphylococcal peptidoglycan-triggered Toll-like receptor 2 signalling pathways. Cell. Microbiol. 7:1305-1317. [DOI] [PubMed] [Google Scholar]
- 21.Kirschning, C. J., and R. R. Schumann. 2002. TLR2: cellular sensor for microbial and endogenous molecular patterns. Curr. Top. Microbiol. Immunol. 270:121-144. [DOI] [PubMed] [Google Scholar]
- 22.Kiura, K., H. Kataoka, T. Nakata, T. Into, M. Yasuda, S. Akira, N. Inoue, and K. Shibata. 2006. The synthetic analogue of mycoplasmal lipoprotein FSL-1 induces dendritic cell maturation through Toll-like receptor 2. FEMS Immunol. Med. Microbiol. 46:78-84. [DOI] [PubMed] [Google Scholar]
- 23.Lepper, P. M., M. Triantafilou, C. Schumann, E. M. Schneider, and K. Triantafilou. 2005. Lipopolysaccharides from Helicobacter pylori can act as antagonists for Toll-like receptor 4. Cell. Microbiol. 7:519-528. [DOI] [PubMed] [Google Scholar]
- 24.Mühlradt, P. F., M. Kiess, H. Meyer, R. Sussmuth, and G. Jung. 1997. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullarkey, M., J. R. Rose, J. Bristol, T. Kawata, A. Kimura, S. Kobayashi, M. Przetak, J. Chow, F. Gusovsky, W. J. Christ, and D. P. Rossignol. 2003. Inhibition of endotoxin response by e5564, a novel Toll-like receptor 4-directed endotoxin antagonist. J. Pharmacol. Exp. Ther. 304:1093-1102. [DOI] [PubMed] [Google Scholar]
- 26.Nakao, Y., K. Funami, S. Kikkawa, M. Taniguchi, M. Nishiguchi, Y. Fukumori, T. Seya, and M. Matsumoto. 2005. Surface-expressed TLR6 participates in the recognition of diacylated lipopeptide and peptidoglycan in human cells. J. Immunol. 174:1566-1573. [DOI] [PubMed] [Google Scholar]
- 27.Okusawa, T., M. Fujita, J. Nakamura, T. Into, M. Yasuda, A. Yoshimura, Y. Hara, A. Hasebe, D. T. Golenbock, M. Morita, Y. Kuroki, T. Ogawa, and K. Shibata. 2004. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by Toll-like receptors 2 and 6. Infect. Immun. 72:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omueti, K. O., J. M. Beyer, C. M. Johnson, E. A. Lyle, and R. I. Tapping. 2005. Domain exchange between human Toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 280:36616-36625. [DOI] [PubMed] [Google Scholar]
- 29.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawadi, G. 2000. Mycoplasma fermentans interaction with monocytes/macrophages: molecular basis. Microbes Infect. 2:955-964. [DOI] [PubMed] [Google Scholar]
- 31.Rottem, S. 2003. Interaction of mycoplasmas with host cells. Physiol. Rev. 83:417-432. [DOI] [PubMed] [Google Scholar]
- 32.Shibata, K., A. Hasebe, T. Into, M. Yamada, and T. Watanabe. 2000. The N-terminal lipopeptide of a 44-kDa membrane-bound lipoprotein of Mycoplasma salivarium is responsible for the expression of intercellular adhesion molecule-1 on the cell surface of normal human gingival fibroblasts. J. Immunol. 165:6538-6544. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu, T., Y. Kida, and K. Kuwano. 2005. A dipalmitoylated lipoprotein from Mycoplasma pneumoniae activates NF-κB through TLR1, TLR2, and TLR6. J. Immunol. 175:4641-4646. [DOI] [PubMed] [Google Scholar]
- 34.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, O., A. Kaufmann, K. Grote, T. Kawai, K. Hoshino, M. Morr, P. F. Muhlradt, and S. Akira. 2000. Preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a Toll-like receptor 2- and MyD88-dependent signaling pathway. J. Immunol. 164:554-557. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi, O., T. Kawai, P. F. Muhlradt, M. Morr, J. D. Radolf, A. Zychlinsky, K. Takeda, and S. Akira. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13:933-940. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi, O., S. Sato, T. Horiuchi, K. Hoshino, K. Takeda, Z. Dong, R. L. Modlin, and S. Akira. 2002. Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10-14. [DOI] [PubMed] [Google Scholar]
- 38.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner, J., 3rd, R. Herrmann, and G. F. Browning. 2000. Transcription in Mycoplasma pneumoniae. Nucleic Acids Res. 28:4488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.