Abstract
Individual or multiple resistance to clindamycin, tetracycline, erythromycin, levofloxacin, or mupirocin was detected in a large proportion of methicillin-resistant Staphylococcus aureus pulsed-field type USA300 isolates collected at an ambulatory health center in Boston. The clindamycin, tetracycline, and mupirocin resistance genes identified in these isolates are commonly associated with plasmids.
Methicillin-resistant Staphylococcus aureus (MRSA) has long been recognized as an important cause of nosocomial morbidity and mortality and, more recently, as a cause of disease arising in the community among previously healthy persons without traditional risk factors for infection (referred to as community-associated MRSA). One particular strain of MRSA, designated USA300-0114 on the basis of pulsed-field gel electrophoresis (PFGE) typing, has emerged as a predominant and widely disseminated strain linked to transmission in community settings nationwide (12). Most USA300 isolates are resistant only to β-lactam and macrolide antimicrobial agents; however, isolates resistant to tetracycline, clindamycin, fluoroquinolones, and mupirocin have been reported (3, 4, 12, 13). This report describes (i) a high prevalence of clindamycin and tetracycline resistance among USA300 isolates collected at a single urban ambulatory health center in Boston and (ii) the identification of multiple resistance to erythromycin, clindamycin, levofloxacin, tetracycline, and mupirocin in an MRSA strain closely related to USA300-0114 (USA300-0247) and in one isolate of USA300-0114.
All of the MRSA isolates collected at the health center during the 19-month period between May 2004 and November 2005 were forwarded to the Massachusetts Department of Public Health State Laboratory Institute for PFGE typing. More than 50% of the patients attending this health center are men who report that they have sex with men, and many health center patients are human immunodeficiency virus infected (7, 9). PFGE was performed with SmaI enzyme digestion as previously described (8). Gel analyses were performed with BioNumerics software, version 4.0 (Applied Maths, Kortrijk, Belgium). Antimicrobial susceptibility testing (AST) was performed with the Dade MicroScan WalkAway instrument (Dade MicroScan, Inc., West Sacramento, CA). Isolates resistant to erythromycin and susceptible to clindamycin were subjected to disk diffusion testing for detection of inducible clindamycin resistance (D-zone test) (2). AST was also performed on a second set of USA300 isolates comprising all of the isolates collected during a similar time period from outpatients tested at a Boston area community health network serving adult and pediatric patients at three hospitals and 20 primary-care practices. A subset of isolates resistant to erythromycin, clindamycin, levofloxacin, and tetracycline was forwarded to the Centers for Disease Control and Prevention, Atlanta, GA, for broth microdilution susceptibility and PCR testing. Susceptibility to minocycline, doxycycline, and mupirocin was determined by the reference broth microdilution method described by the Clinical and Laboratory Standards Institute, with cation-adjusted Mueller-Hinton broth (Becton Dickinson Microbiology Systems, Cockeysville, MD) (2). Quality control strains included S. aureus ATCC 29213, Enterococcus faecalis ATCC 25922, and S. aureus ATCC 43300. PCR testing was performed to identify genes conferring tetracycline resistance (tetK, tetM), inducible or constitutive clindamycin resistance (ermA, ermC), and mupirocin resistance (mupA) (10, 11, 12).
Between May 2004 and November 2005, culture specimens yielding MRSA were obtained from 123 health center patients. Only the first MRSA isolate collected from each patient was included in this analysis. Among 115 isolates with a known source, 103 (90%) were collected from skin and soft-tissue sites, 11 (10%) were from the nares or the nasopharynx, and 1 (1%) was from urine. Among 123 total isolates, 102 (83%) had PFGE patterns corresponding to either MRSA strain type USA300-0114 (73 isolates, 59% of the total) or USA300-0247 (29 isolates, 24% of the total) (Fig. 1). AST data are summarized in Table 1. All 12 multiresistant isolates (11 USA300-0247 and 1 USA300-0114) tested at the Centers for Disease Control and Prevention contained the tetK and ermC genes; none contained the tetM or ermA gene. All were susceptible to minocycline and doxycycline, all contained the mupA resistance gene, and the mupirocin MICs were ≥128 μg/ml. Among 26 USA300-0114 and 4 USA300-0247 isolates collected from the nearby health network, only 2 were clindamycin resistant and none were tetracycline resistant.
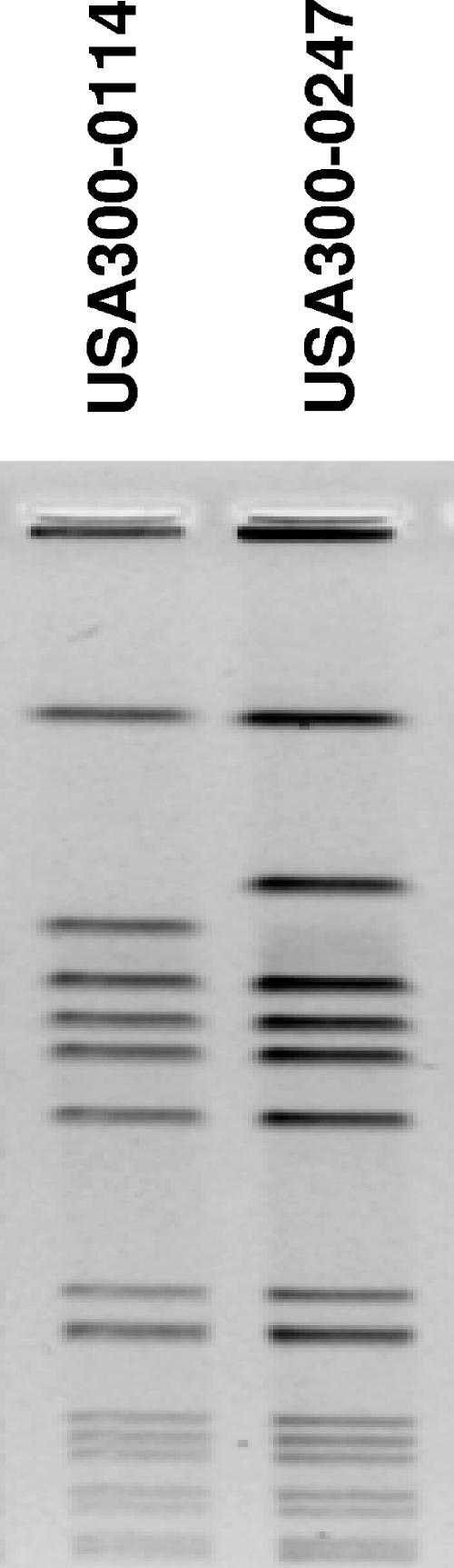
FIG. 1.
Comparison of PFGE SmaI macrorestriction patterns of the two major USA300 strain types found at the health center.
TABLE 1.
Prevalence of resistance to antimicrobial agents among predominant USA300 MRSA strain types at a Boston ambulatory health center in 2004 and 2005
| Test result | No. of resistant isolates/no. tested (% resistant)
|
|
|---|---|---|
| USA300-0114 | USA300-0247 | |
| Resistance to: | ||
| Erythromycin | 72/73 (99) | 26/29 (90) |
| Levofloxacin | 58/73 (79) | 29/29 (100) |
| Clindamycin | 36/73 (49) | 22/29 (76) |
| Tetracycline | 10/73 (14) | 21/29 (72) |
| Gentamicin | 0/73 (0) | 0/29 (0) |
| Linezolid | 0/73 (0) | 0/29 (0) |
| Rifampin | 0/73 (0) | 0/29 (0) |
| Trimethoprim-sulfamethoxazole | 0/73 (0) | 0/29 (0) |
| Vancomycin | 0/73 (0) | 0/29 (0) |
| Erythromycin, levofloxacin, clindamycin, and tetracycline | 2/73 (3) | 16/29 (55) |
| Inducible clindamycin resistance in D-zone test | 0/33 (0) | 0/6 (0) |
| Mupirocin MIC of ≥128 μg/mla | 1/1 | 11/11 |
Mupirocin MICs were determined for a subset of isolates found to be resistant to erythromycin, levofloxacin, clindamycin, and tetracycline.
In a recent report of MRSA skin infections among adult emergency department patients in 11 U.S. cities, 97% of the MRSA isolates were USA300, 5% were resistant to clindamycin, 8% were resistant to tetracycline (9a), and 1% were resistant to both clindamycin and tetracycline (Gregory E. Fosheim, unpublished data). The prevalence of resistance to these agents among USA300 isolates in our investigation, particularly USA300-0247 isolates, was considerably higher and was also higher than the prevalence of resistance among isolates collected at the nearby health network. Furthermore, we identified multiple resistance to erythromycin, clindamycin, levofloxacin, and tetracycline in 55% of the USA300-0247 isolates collected, and elevated mupirocin MICs (≥128 μg/ml) were detected in all 12 isolates for which mupirocin MICs were determined. USA300 isolates with multiple resistance to the same agents have previously been reported among patients of a San Francisco community health network (4). Multiple resistance has also been identified in non-USA300 MRSA strains isolated from children in community settings in Taiwan (1). In a study involving human immunodeficiency virus-infected men with skin infections in Los Angeles who report that they have sex with men, 3.2% and 35.5% of the MRSA isolates were resistant to clindamycin and tetracycline, respectively, and multiple resistance was not reported (6). Examination of clinical and demographic features of our case patients, including antibiotic exposure and underlying disease conditions, may prove useful in identifying risk factors and elucidating mechanisms associated with resistance acquisition in this community. On a molecular level, an important mechanism of resistance acquisition in USA300 MRSA appears to be the transfer of plasmids from bacterial reservoirs; in the isolates described in this report, resistance to clindamycin, tetracycline, and mupirocin was mediated by resistance genes that are typically located on plasmids (4, 11, 12).
Among USA300 isolates resistant to tetracycline, we consistently found in vitro susceptibility to minocycline, doxycycline, and trimethoprim-sulfamethoxazole. These findings suggest several inexpensive oral treatment options for skin infections with multidrug-resistant USA300 isolates. However, the clinical efficacy of these drugs in the treatment of MRSA infections has not been extensively documented and incision and drainage should still be considered the primary therapy when skin abscesses are present (5). The high prevalence of elevated mupirocin MICs described in this report raises concerns about the in vivo efficacy of mupirocin for eradication of MRSA nasal colonization in certain populations. The appearance of multidrug-resistant MRSA in the community setting emphasizes the importance of routine collection of specimens for culture and AST, not only for individual patient management but also for development of community-wide treatment and prevention strategies.
Acknowledgments
We thank our colleagues Karen Anderson, Barbara Bolstorff, Dan Cohen, Al DeMaria, Bela Matyas, and John Szumowski for their contributions.
The findings and conclusions in this report are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Boyle-Vavra, S., B. Ereshefsky, C.-C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—seventh edition, M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Diep, B. A., H. A. Carleton, R. F. Chang, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 193:1495-1503. [DOI] [PubMed] [Google Scholar]
- 4.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 5.Gorwitz, R. J., D. B. Jernigan, J. H. Powers, J. A. Jernigan, and Participants in the CDC-Convened Experts' Meeting on Management of MRSA in the Community. 2006. Strategies for clinical management of MRSA in the community: summary of an experts' meeting convened by the Centers for Disease Control and Prevention. [Online.] http://www.cdc.gov/ncidod/dhqp/ar_mrsa_ca.html.
- 6.Lee, N. E., M. M. Taylor, E. Bancroft, P. J. Ruane, M. Morgan, L. McCoy, and P. A. Simon. 2005. Risk factors for community-associated methicillin-resistant Staphylococcus aureus skin infections among HIV-positive men who have sex with men. Clin. Infect. Dis. 40:1529-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer, K., J. Appelbaum, T. Rogers, W. Lo, J. Bradford, and S. Boswell. 2001. The evolution of the Fenway Community Health model. Am. J. Public Health 91:892-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mimiaga, M. J., H. Goldhammer, C. Belanoff, A. M. Tetu, and K. H. Mayer. 2007. Men who have sex with men: perceptions about sexual risk, HIV and sexually transmitted disease testing, and provider communication. Sex Transm. Dis. 34(2):113-119. [DOI] [PubMed] [Google Scholar]
- 9a.Moran, G. J., A. Krishnadasan, R. G. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan for the EMERGEncy ID Net Study Group. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666-674. [DOI] [PubMed] [Google Scholar]
- 10.Olsvik, B., F. C. Tenover, I. Olsen, and J. K. Rasheed. 1996. Three subtypes of the tet(M) gene identified in bacterial isolates from periodontal pockets. Oral Microbiol. Immunol. 11:299-303. [DOI] [PubMed] [Google Scholar]
- 11.Ramsey, M. A., S. F. Bradley, C. A. Kauffman, and T. M. Morton. 1996. Identification of chromosomal location of mupA gene encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob. Agents Chemother. 40:2820-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wener, K. M., H. S. Gold, M. Wong, L. Venkataraman, K. H. Mayer, D. E. Cohen, and S. B. Wright. 2006. High prevalence of methicillin-resistant Staphylococcus aureus (MRSA) colonization in an urban outpatient population, p. 118. Abstr. 44th Ann. Meet. Infect. Dis. Soc. Am. Infectious Disease Society of America, Alexandria, VA.