Abstract
A survey of 152 methicillin-resistant Staphylococcus aureus (MRSA) strains from medical centers in Las Cruces, NM, and El Paso, TX, revealed the presence of spa types 2 and 24 (clone USA100) and spa type 1 (clone USA300-0114). Las Cruces MRSA displayed relatively high vancomycin MICs, and one hetero-vancomycin-intermediate S. aureus strain was identified.
Staphylococcus aureus is the leading cause of nosocomial infections (2), and methicillin-resistant S. aureus (MRSA) is a common cause of community skin and soft-tissue infections (10). The USA100 MRSA clone represents up to 44% of MRSA isolates in the United States (8), and a unique community-associated clone, USA300-0114, is widely disseminated (6, 8, 10, 17). The use of vancomycin for the treatment of serious MRSA infections is threatened by the appearance of vancomycin-intermediate S. aureus (VISA) and hetero-VISA (hVISA) (1, 3, 5, 18). hVISA can present initial vancomycin MICs of ≤2 μg/ml, yet upon exposure to vancomycin, VISA subpopulations appear (MICs of ≥4 μg/ml) (15, 19). VISA and vancomycin-resistant S. aureus (MIC of ≥32 μg/ml) are often USA100 derivatives (4, 8, 15).
A survey of two medical centers in El Paso, TX, revealed that 71% of the MRSA isolates were related USA100 clones (12). Northwest from El Paso (∼50 miles) lies Las Cruces, NM (population ∼80,000), a popular U.S. retirement destination. In order to understand MRSA epidemiology in a Southern New Mexico border city with a large Hispanic (∼52%) and growing retirement population, we have analyzed MRSA isolates collected from Las Cruces medical centers.
(This work was presented in part at poster sessions of the 105th [Atlanta, GA, 2005] and 106th [Orlando, FLA, 2006] General Meetings of the American Society for Microbiology).
Seventy-four MRSA strains from the Memorial Medical Center and 67 MRSA strains from the Mountain View Regional Medical Center (isolated 15 September 2003 to 20 August 2004; “MM” and “MV” strains, respectively), both located in Las Cruces, ∼3.5 miles apart, as well as 11 El Paso MRSA isolates (“LP” strains [12]) were analyzed (Fig. 1; Table 1). Kirby-Bauer antibiograms and fusidic acid and inducible clindamycin resistance were determined by using disc diffusion (11, 12). Vancomycin MICs were determined by using AB BIODISK E-test strips (Remel, Lenexa, KS) and agar dilution (11), and vancomycin resistance population analyses were preformed as described previously (13). VISA mutants of hVISA strain MM66 were isolated on brain heart infusion agar (Becton-Dickinson and Co., Sparks, MD) containing 3 μg/ml vancomycin (Sigma Chemical Co., St Louis, MO) by inoculating these plates with 100 μl of brain heart infusion broth culture (18 h, 200 rpm, 37°C). Pulsed-field gel electrophoresis (PFGE) of SmaI-digested DNA (14) was carried out by using the CHEF DR III system (Bio-Rad Laboratories, Inc., Hercules, CA), and SmaI restriction fragment length polymorphism similarities were determined (12). Chromosomal DNA for spa typing was isolated using the Qiamp DNA Mini kit (QIAGEN, Inc., Valencia, CA), and spaA polymorphic X regions were amplified (9) and sequenced. spa polymorphic X sequences were analyzed using eGenomics, and spa type numbers were assigned (7) (Table 1).
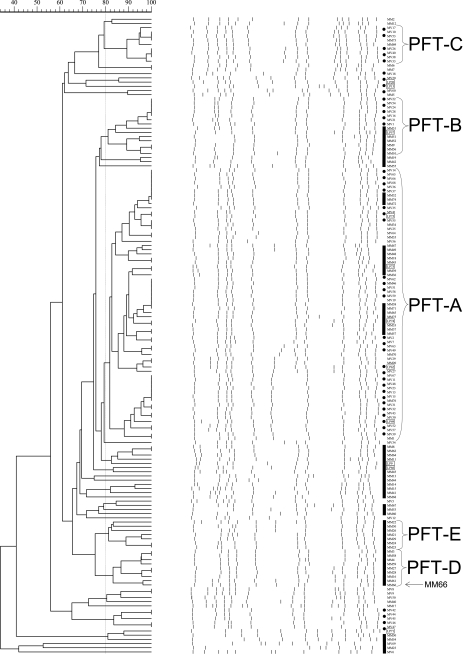
FIG. 1.
PFGE patterns of SmaI-restricted chromosomal DNA and dendrogram of percent relatedness. Bars indicate “MM” strains, and dotted lines indicate “MV” strains (if within a strain group, LP strains are included under the respective lines). “LP” strains are boxed, and MM66 is indicated with an arrow.
TABLE 1.
Characteristics of investigated strains
| Strain | Source | Antibiotypea | E-test MIC (μg/ml) of vanb | Pulsotype | spa type; repeat |
|---|---|---|---|---|---|
| MM2 | Buttock | CD*EOT | 2.0 | 1; YHGFMBQBLO | |
| MV53 | Head | EO | 2.0 | C | 1; YHGFMBQBLO |
| MV38 | Body fluid | EO | 1.5 | C | 1; YHGFMBQBLO |
| MV20 | Urine | CDEO | 2.0 | 24; TJMEMDMGMK | |
| LP20 | Bronchial sample | CDEGIOTm | 2.0 | 24; TJMEMDMGMK | |
| MM5 | Thigh | O | 2.0 | 175; UJFKPE | |
| MV1 | Wound | CDEO | 1.5 | B | 2; TJMBMDMGMK |
| LP57 | Wound | CD*EO | 1.5 | B | 2; TJMBMDMGMK |
| MM56 | Pleural sample | CD*EO | 3.0 | B | NDc |
| MM19 | Sputum | CDEO | 2.0 | 2; TJMBMDMGMK | |
| MV66 | Blood | CDEO | 2.0 | A | 2; TJMBMDMGMK |
| MM32 | Thigh | CEO | 1.5 | A | 2; TJMBMDMGMK |
| MM74 | Sputum | CD*EO | 2.0 | A | 2; TJMBMDMGMK |
| MV35 | Blood | CDEO | 2.0 | A | 24; TJMEMDMGMK |
| LP75 | Nasal | CD*EO | 2.0 | A | 2; TJMBMDMGMK |
| MM49 | Leg | CD*EO | 3.0 | A | ND |
| MM48 | Bronchial sample | CD*EO | 3.0 | A | ND |
| MM43 | Abscess | CD*EO | 2.0 | A | 24; TJMEMDMGMK |
| LP73 | Sputum | CDEO | 2.0 | A | 2; TJMBMDMGMK |
| MM38 | Leg | CDEO | 3.0 | A | ND |
| LP74 | Sputum | CD*EOT | 1.5 | A | 2; TJMBMDMGMK |
| MV49 | Blood | CD*EO | 2.0 | A | 2; TJMBMDMGMK |
| MM70 | Back | CD*EO | 2.0 | A | 24; TJMEMDMGMK |
| MV29 | Urine | CDEO | 2.0 | A | 2; TJMBMDMGMK |
| LP62 | Wound | CD*EGOTTm | 2.0 | A | 2; TJMBMDMGMK |
| MV67 | Blood | CD*EO | 1.5 | A | 24; TJMEMDMGMK |
| MV48 | Urine | CDEIO | 3.0 | A | ND |
| LP66 | Blood | CDEGOT | 2.0 | 58; TJMAMGMK | |
| MM45 | Blood | CD*EO | 3.0 | 24; TJMEMDMGMK | |
| MM41 | Heel | CD*EO | 3.0 | ND | |
| MM22 | Knee fluid | CD*EO | 2.0 | E | 24; TJMEMDMGMK |
| MM30 | Urine | CEO | 1.5 | E | 2; TJMBMDMGMK |
| MM26 | Foot | CD*EO | 2.0 | E | 24; TJMEMDMGMK |
| MM21 | Foot | CEO | 3.0 | E | ND |
| MM29 | Sputum | CD*EO | 3.0 | E | ND |
| MM25 | Bronchial sample | CD*EO | 2.0 | E | 24; TJMEMDMGMK |
| MM4 | Sputum | CEO | 3.0 | D | ND |
| MM59 | Arm | CD*EO | 3.0 | D | ND |
| MM66 | Sputum | CDEO | 3.0 | D | ND |
| MV8 | Wound | DEO | 2.0 | 1; YHGFMBQBLO | |
| MM40 | Abdomen | CD*EO | 2.0 | 230; TMBMDMGMK | |
| MV47 | Surgical tissue | CD*EO | 2.0 | 303; TMEMDMGMK | |
| LP71 | Unknown | CDEIMO | 1.5 | 204; WGKAKAOMQQQQ |
C, ciprofloxacin; D, clindamycin; D*, inducible clindamycin resistance; E, erythromycin; G, gentamicin; I, imipenem; M, mupirocin; O, oxacillin; T, tetracycline; Tm, trimethoprim.
van, vancomycin. Vancomycin E-test MICs are boldface and underlined.
ND, not determined.
Five pulsed-field-type (PFT) groups containing 7 or more strains and 48 unrelated strains were identified (Fig. 1). PFT-A represented the largest PFT group (65 isolates, including 5 LP strains) and contained 13 strains of spa type 2 or 24, which diverge by only a single amino acid (B↔E) (Fig. 1; Table 1). PFT-B was the second-largest group (14 isolates, including 1 LP strain) (Fig. 1; Table 1) and was represented by two spa type 2 strains. spa types 2 and 24 are also represented in PFT-E, as well as four unrelated strains (MM19, MM45, MV20, and LP20), and overall these spa types represent 74% of the strains typed (Table 1). spa types 2 and 24 are representative of the multilocus sequence/SCCmec type ST5-MRSA-II, which identifies USA100 (16). LP57 is also spa type 2 (Table 1) and was previously identified as ST5-MRSA-II (12). PFT-C (nine strains) is represented by two spa type 1 strains, and MM2 at the PFT-C boundary and MV8 below PFT-D were also spa type 1 (Fig. 1, Table). spa type 1 is a marker for the U.S. community-associated MRSA clone USA300-0114 (17).
PFT-D and -E are made up entirely of MM strains, and MV and MM strains throughout tended to group together in PFGE (Fig. 1). This demonstrates that while the major clones spreading within Las Cruces medical centers are similar, the clones within the two hospitals have evolved independently following introduction and/or the initial clone introduction was unique to each hospital.
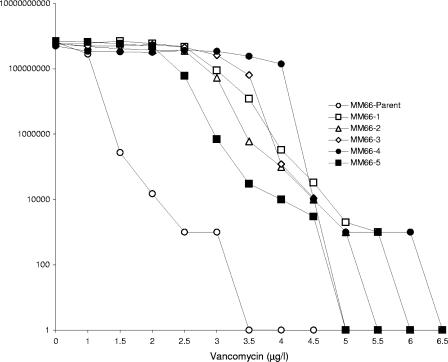
All strains investigated were susceptible to synercid, linezolid, and fusidic acid, and no vancomycin-resistant S. aureus strains were detected. Of the 152 MRSA strains analyzed, 123 were resistant to the drugs ciprofloxacin, clindamycin, and erythromycin. Ninety percent of the Las Cruces strains demonstrated vancomycin E-test MICs of ≥2.0, and 12 exhibited a MIC of 3.0 μg/ml (Table 1). All but one of the latter group of strains (MV48) came from one medical center, and five clustered within related PFT-D and -E (Fig. 1; Table 1). The agar dilution MIC of MM66 was 4 μg/ml, and initial vancomycin resistance population analyses on all strains with E-test MICS of 3 μg/ml revealed that only MM66 produced colonies on plates containing 3 μg/ml vancomycin. hVISA strain MM66 proved 96% identical by PFT to strain MM61, yet MM61 has an additional SmaI band of 80 kb (Fig. 1) and an E-test MIC of 2.0. MM66 colonies appeared at a frequency of 1.5 × 10−6 on 3 μg/ml vancomycin, and 5 MM66 VISA colonies were picked. The agar dilution/E-test vancomycin MICs (μg/ml) for these five VISA MM66 mutants were ≥4/≥4. Colonies of hVISA MM66 did not appear above 3.0 μg/ml, while all MM66 VISA mutants produced colonies above this vancomycin concentration (4.5 to 6.0 μg/ml) (Fig. 2).
FIG. 2.
Vancomycin resistance population analysis of MM66 (parent) and MM66 VISA mutants (MM66-1 through -5).
In conclusion, these findings demonstrate that USA100 clones are spreading within medical centers in two predominantly Hispanic Southwestern cities. The predominant community-associated clone USA300-0114 was also found in Las Cruces hospitals. Furthermore, we report a preponderance of MRSA with vancomycin MICs of ≥2 μg/ml in Las Cruces medical centers and have identified the first hVISA in this ethnically unique region.
Acknowledgments
We are indebted for help in determining spa types and to the training of Alejandro Delgado provided by Barry N. Kreiswirth, William Eisner, and Steve Naidich (Public Health Research Institute).
We acknowledge support from the National Institutes of Health: S06 GM008136-32 (J.E.G., NMSU SCORE Program), R25 GM07667-29 (NMSU-MARC Program), S06 GM61222-05 (A.D., NMSU-MBRS-RISE Program), and P20RR16480 from the NM-INBRE Program of the National Center for Research Resources.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Ariza, J., M. Pujol, J. Cabo, C. Pea, N. Fernandez, J. Linares, J. Ayats, and F. Gudiol. 1999. Vancomycin in surgical infections due to methicillin resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587-1588. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2001. NNIS system: national nosocomial infections surveillance (NNIS) system report, data summary from January 1992-April 2001, issued August 2001. Am. J. Infect. Control 29:400-421. [DOI] [PubMed] [Google Scholar]
- 3.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 4.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, F. C. Tenover, and the Vancomycin-Intermediate Staphylococcus aureus Epidemiology Study Group. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 6.Kazakova, S. V., J. C. Hageman, M. K. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468-475. [DOI] [PubMed] [Google Scholar]
- 7.Koreen, L. S., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Krieswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitani, N., M. Ohnishi, T. Masutani, K. Murakawa, and Y. Okamoto. 2002. Molecular typing of methicillin-resistant Staphylococcus aureus by protein A gene sequencing. Jpn. J. Infect. Dis. 55:179-180. [PubMed] [Google Scholar]
- 10.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Forshiem, L. K. McDougal, R. B. Carey, and D. A. Talan for the EMERGEny ID Net Study Group. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666-674. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing. M100-S13. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 12.O'Brien, F. G., T. T. Lim, D. C. Winnett, G. W. Coombs, J. C. Pearson, A. Delgado, M. J. Langevin, S. A. Cantore, L. Gonzalez, and J. E. Gustafson. 2005. Survey of El Paso methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:2969-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Leary, J. O., M. J. Langevin, C. T. D. Price, J. S. Blevins, M. S. Smeltzer, and J. E. Gustafson. 2004. Effects of sarA inactivation on intrinsic multidrug resistance mechanism of Staphylococcus aureus. FEMS Microbiol. Lett. 237:297-302. [DOI] [PubMed] [Google Scholar]
- 14.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in twelve New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 15.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 16.Soo Ko, K., K. R. Peck, W. Sup Oh, N. Y. Lee, K. Hiramatsu, and J. H. Song. 2005. Genetic differentiation of methicillin-resistant Staphylococcus aureus strains from Korea and Japan. Microb. Drug Resist. 11:279-286. [DOI] [PubMed] [Google Scholar]
- 17.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods, C. W., A. C. Cheng, V. G. Fowler, M. Moorefield, J. Frederick, G. Sakoulas, V. G. Meka, F. C. Tenover, P. Zwadyk, and K. H. Wilson. 2004. Endocarditis caused by Staphylococcus aureus with reduced susceptibility to vancomycin. Clin. Infect. Dis. 38:1188-1191. [DOI] [PubMed] [Google Scholar]
- 19.Wootton, M., T. R. Walsh, and A. P. MacGowan. 2005. Evidence for reduction in breakpoints used to determine vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3982-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]