Abstract
Like other coronaviruses, severe acute respiratory syndrome coronavirus (SARS CoV) assembles at and buds into the lumen of the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC). Accumulation of the viral envelope proteins at this compartment is a prerequisite for virus assembly. Previously, we reported the identification of a dibasic motif (KxHxx) in the cytoplasmic tail of the SARS CoV spike (S) protein that was similar to a canonical dilysine ER retrieval signal. Here we demonstrate that this motif is a novel and functional ER retrieval signal which reduced the rate of traffic of the full-length S protein through the Golgi complex. The KxHxx motif also partially retained two different reporter proteins in the ERGIC region and reduced their rates of trafficking, although the motif was less potent than the canonical dilysine signal. The dibasic motif bound the coatomer complex I (COPI) in an in vitro binding assay, suggesting that ER retrieval may contribute to the accumulation of SARS CoV S protein near the virus assembly site for interaction with other viral structural proteins. In support of this, we found that the dibasic motif on the SARS S protein was required for its localization to the ERGIC/Golgi region when coexpressed with SARS membrane (M) protein. Thus, the cycling of SARS S through the ER-Golgi system may be required for its incorporation into assembling virions in the ERGIC.
An outbreak of severe acute respiratory syndrome (SARS) emerged in Guangdong Province of China around November 2002 and rapidly spread to 32 other countries in a short time. Shortly after the outbreak, a novel coronavirus (SARS-associated coronavirus [SARS CoV]) was identified as the causative agent of this lethal infectious disease (32, 56). This new virus likely originated from a coronavirus that had been circulating in animals and had acquired mutations that enabled it to infect humans (22, 30, 34, 39).
Coronaviruses are enveloped positive-strand RNA viruses with a genome of 27 to 31 kb. These viruses are classified into group 1, 2, or 3 based on their antigenicity and sequence homology (18); however, new data suggest that the addition of additional groups and subgroups may be necessary (64). Coronaviruses infect a wide range of vertebrate species and display variable cellular tropism ranging from the gastrointestinal and respiratory tracts to the nervous system. Coronaviruses are intriguing because they assemble at the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), bud into the lumen of the compartment, and then exit the cells by exocytosis (31). The advantage of assembly at this intracellular compartment is currently unknown. All known coronaviruses contain at least three envelope proteins, envelope (E), membrane (M), and spike (S), which are encoded in the last third of their genomes. The E protein is present at low levels in mature virions but may play a critical role in virus assembly (11, 33, 53). M is the most abundant protein in the viral envelope and, acting as a scaffold, orchestrates virus assembly. It interacts with E, S, and the nucleocapsid during assembly and is necessary for virus-like particle formation (2, 8, 15, 26, 45, 52, 67). The S protein is less abundant in virions; however, it is responsible for binding to receptors on target cells and determines the cell tropism of the virus (13). When expressed on the surface of infected cells, S proteins from some coronaviruses cause the fusion of infected cells with neighboring cells, resulting in the formation of syncytia (6).
Envelope proteins of viruses that assemble at intracellular compartments possess signals that direct them to the site of virus assembly (21). For instance, the CoV infectious bronchitis virus (IBV) E protein contains a Golgi-targeting signal within its cytoplasmic tail (7); the IBV M contains a Golgi-targeting signal in its first transmembrane domain (42, 63); and the IBV S contains a canonical dilysine ER retrieval signal in its cytoplasmic tail (41). The latter type of signal requires two lysine residues at the −3 and −4 (or −5) positions relative to the C terminus (65). Proteins with the dilysine signal bind to the coatomer complex I (COPI) and are recruited into vesicles that bud from the Golgi complex and travel back to the ER (9, 35). The efficiency of binding to COPI is influenced by the sequence surrounding the dilysine signal, resulting in an ER, ERGIC, or Golgi complex steady-state localization of proteins bearing this signal (65). Recombinant IBV containing a mutant S protein lacking the dilysine signal forms syncytia earlier than wild-type IBV but produces lower titers of infectious virus (71), suggesting a role for the dilysine signal in the pathogenesis and fitness of the virus.
SARS CoV has a genome organization similar to that of other coronaviruses (44, 56) and is distantly related to group 2 coronaviruses (61, 64). The virus has also been shown to assemble at and bud into the lumen of the Golgi/ERGIC region as well as into the ER (16, 46). However, as the sequence identity is only around 20% between proteins of SARS CoV and the analogous proteins from other coronaviruses (44, 56), it is possible that SARS CoV may be unique in some aspects of its biology. The SARS CoV S protein is a type I membrane protein and a class I fusion protein (5, 27, 40, 66) with 1,255 amino acids and 23 potential N-linked glycosylation sites (44, 56). It binds to the major SARS CoV receptor, angiotensin-converting enzyme 2 (38), and an alternative receptor, CD209L (28), mediates virus entry and cell-cell fusion (38, 60). Previously, we reported a dibasic motif (KxHxx-COOH) in the cytoplasmic tail of SARS S that resembles a canonical dilysine signal by demonstrating that the C-terminal 11 amino acids of SARS S retain a plasma membrane reporter protein in the ERGIC (41). In the present study, we explored the function of this motif in the context of the full-length SARS S protein and in the context of the entire cytoplasmic tail of the protein. We found that in transfected cells, SARS S was predominantly localized to the cell surface but the KxHxx dibasic motif decreased the extent of trafficking to the plasma membrane. The cytoplasmic tail of SARS S also decreased the extent of trafficking of two different plasma membrane reporter proteins to the cell surface, and this activity was dependent on the KxHxx dibasic motif. The KxHxx motif bound COPI, suggesting it could contribute to the retrieval of SARS S and collect it near the virus assembly site. Importantly, when coexpressed with SARS M, SARS S was localized to the Golgi compartment, and this localization was dependent upon the KxHxx motif. These results suggest a role for the KxHxx motif in efficient M-S interaction and virus assembly.
MATERIALS AND METHODS
Plasmids and expression vectors.
A cDNA clone encoding SARS S derived from the SARS CoV Tor 2 strain was obtained from the Institute for Genomic Research (Rockville, MD). Four fragments were amplified by PCR and subcloned in pBluescript (Stratagene, La Jolla, CA). After sequence verification, a full-length SARS S cDNA was assembled in pcDNA3.1/Hygro (Invitrogen, Grand Island, NY) utilizing BamHI and XhoI sites. The pcDNA3.1/SARS S2A construct was generated by mutagenizing the codons for Lys1251 and His1253 of pcDNA3.1/SARS S to alanines by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The SARS S and SARS S2A cDNA sequences were then transferred into another expression vector, pCAGGS-MCS (50), by using KpnI and XhoI sites. To make the constructs for expression of G-S and G-S2A, the DNA sequence encoding the SARS S or SARS S2A cytoplasmic tail (Cys1217-Thr1255) was amplified by PCR with a forward primer containing an introduced BamHI site and a reverse primer containing an XhoI site. The amplified fragment and a DNA fragment encoding the ectodomain and the transmembrane domain of vesicular stomatitis virus (VSV) G cut with EcoRI and BamHI from pBS-GTMB (54) were ligated into pCAGGS-MCS, using EcoRI and XhoI sites, generating pCAGGS/G-S or pCAGGS/G-S2A. To create the pCAGGS/Tac-S and pCAGGS/Tac-S2Aconstructs, the DNA fragment encoding the ectodomain and transmembrane domain of Tac was excised from pBluescript/Tac-VSV G (59) using KpnI and BamHI, and the fragment encoding the cytoplasmic tail of SARS S or SARS S2A was excised from pCAGGS/G-S or pCAGGS/G-S2A using BamHI and XhoI. The two fragments were assembled in pCAGGS-MCS digested with KpnI and XhoI. To create constructs for expressing glutathione S-transferase (GST) fusion proteins, the sequence encoding Leu1226 to Thr1255 of the SARS S C-terminal tail was PCR amplified from pCAGGS/SARS S. BamHI and EcoRI sites were introduced by primers to flank the amplified tail and were used for subcloning into the expression vector pGEX2T (Amersham Biosciences, Piscataway, NJ). QuikChange site-directed mutagenesis was used to change the SARS S dibasic signal (KLHYT) to alanines (ALAYT). A similar strategy was used to make pGEX/IBV S and pGEX/IBV S2A, using pcDNA3.1/IBV S (41) as the template. A cDNA clone encoding SARS M derived from the SARS CoV Tor 2 strain was obtained from the Institute for Genomic Research. The coding sequence was amplified by PCR and subcloned into pcDNA3.1/Hygro using BamHI and XhoI sites. After sequence verification, the cDNA encoding SARS M was transferred to pCAGGS-MCS by using KpnI and XhoI restriction sites.
Cells and transfection.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Grand Island, NY) containing 10% fetal calf serum (Atlanta Biologicals, Lawrenceville, GA) and 0.1 mg/ml Normocin (InvivoGen, San Diego, CA). CHO cells were maintained in alpha-modified minimum essential Eagle's medium (with sodium bicarbonate, without l-glutamine, ribonucleosides, deoxyribonucleosides) (Sigma, St. Louis, MO) supplemented with 10% fetal calf serum, 1% nonessential amino acids, 1% l-glutamine, and 0.1 mg/ml Normocin. Transient transfections were performed using a total of 1 to 2 μg of pCAGGS-based expression vectors encoding the appropriate cDNAs and FuGENE 6 transfection reagent (Roche, Indianapolis, IN), following the manufacturer's instructions. For vaccinia/T7 RNA polymerase-driven expression, HeLa cells were infected with vTF7-3 (12) at a multiplicity of infection of 5 for 1 h and then transfected with 1.5 μg of pcDNA3.1/Hygro or pcDNA3.1/SARS S and 6 μl of LT1 transfection reagent (Mirus, Madison, WI). Expression was analyzed at the indicated times posttransfection.
Antibodies.
The rabbit polyclonal antibody to SARS S (anti-SCT) was generated by immunizing rabbits with a conjugate of keyhole limpet hemocyanin and a peptide (CDSEPVLKGVKLHYT) derived from the C-terminal 14 amino acids of SARS S, with an N-terminal cysteine added for conjugation (Covance Research Products, Denver, PA). The rabbit polyclonal antibody to SARS M was made in a similar manner using a synthetic peptide corresponding to the C-terminal 14 amino acids, with a cysteine added for coupling (CDHAGSNDNIALLVQ). The mouse anti-VSV G monoclonal antibody (MAb) used in immunofluorescence recognizes the ectodomain of VSV G (36). A rabbit polyclonal anti-VSV antiserum (69) was used for immunoprecipitation. The mouse monoclonal and rabbit polyclonal anti-Tac antibodies were a generous gift from M. Marks (University of Pennsylvania School of Medicine, Philadelphia, PA). The mouse monoclonal anti-ERGIC-53 antibody was a kind gift from H. P. Hauri (Basel, Switzerland). The mouse anti-SARS immune serum was a generous gift from K. Subbarao (National Institute of Allergy and Infectious Diseases, Bethesda, MD). By indirect immunofluorescence, the anti-SARS mouse immune serum did not recognize the SARS M protein. The mouse monoclonal anti-GM130 antibody was from Transduction Labs (San Jose, CA). The golgin-160 antibody was previously described (24). Polyclonal rabbit anti-ɛ-COP antibody was a generous gift from M. Krieger (Massachusetts Institute of Technology, Cambridge, MA). Texas Red-conjugated donkey anti-rabbit immunoglobulin G (IgG) was obtained from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA), and Alexa 488-conjugated goat anti-mouse IgG was from Molecular Probes, Inc. (Eugene, OR). Horseradish peroxidase-conjugated goat anti-rabbit IgG was from Amersham.
GST fusion protein expression and purification.
Constructs encoding the GST fusion proteins were transformed into competent Escherichia coli BL21 cells, and expression and purification of fusion proteins were performed primarily following the manufacturer's instructions (GST Gene Fusion System; Amersham). Briefly, transformed E. coli cells were grown to an optical density at 600 nm (OD600) of approximately 0.7, and expression was induced with 0.1 mM isopropyl-β-d-thiogalactoside. After 3 h of induction, cells were harvested and sonicated in phosphate-buffered saline (PBS) binding buffer (10 mM EDTA, 5 mM benzamide, and protease inhibitor cocktail [Sigma, St. Louis, MO] in PBS). Glutathione-Sepharose 4B beads (Amersham) were added to the soluble protein fraction and incubated at 4°C for 1 h. After the beads were extensively washed in PBS binding buffer, the GST fusion proteins were eluted off the beads using increasing concentrations of GST elution buffer (10 to 50 mM glutathione, 50 mM Tris-HCl [pH 8.0]).
Indirect immunofluorescence microscopy.
HeLa cells were plated on coverslips in 35-mm-diameter dishes 1 day before transfection and transfected as described above. At the indicated times posttransfection, cells were fixed in 3% paraformaldehyde in PBS for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 3 min, and stained with the appropriate primary antibodies as previously described (63). Secondary antibodies were Texas Red-conjugated donkey anti-rabbit immunoglobulin G (IgG) and Alexa 488-conjugated goat anti-mouse IgG. To label surface SARS S proteins, intact cells were washed with ice-cold PBS and incubated with mouse anti-SARS serum for 10 min at 4°C. Cells were then washed twice with ice-cold PBS, fixed and permeabilized as described above, and stained for intracellular proteins with rabbit anti-SCT. A similar protocol was used to label surface G-S and Tac-S chimeric proteins, except that monoclonal anti-VSV and anti-Tac antibodies were used, respectively. For cells expressing both SARS S and M, surface localization was assessed by positive plasma membrane/rim staining. Images were acquired with a Zeiss Axioscop microscope (Thornwood, NY) equipped for epifluorescence with a Sensys charge-coupled-device camera (Photometrics, Tucson, AZ), using IPLab software (Scanalytics, Vienna, VA).
Metabolic labeling, immunoprecipitation, and glycosidase digestion.
HeLa cells were transfected with pCAGGS-based vectors encoding SARS S, SARS S2A, G-S, G-S2A, Tac S, or Tac S2A as described above and were starved for 20 min at 24 h or 18 h posttransfection with methionine- and cysteine-free medium at 37°C. The cells were then labeled with 100 to 200 μCi of [35S]methionine-cysteine ([35S]Promix; Amersham) per ml in methionine- and cysteine-free medium for 20 min at 37°C and chased in normal growth medium for various times. After a rinse in ice-cold PBS, the cells were lysed in a detergent solution (62.5 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.4% deoxycholate, 1% NP-40) containing a protease inhibitor cocktail. After the removal of nuclei and debris by centrifugation at 14,000 × g for 10 min, the lysate was adjusted to 0.2% sodium dodecyl sulfate (SDS) and incubated at room temperature for 6 h with 6 μl of rabbit anti-SCT antiserum (for full-length SARS S and S2A) for 1 h at room temperature or overnight at 4°C with 2 μl anti-VSV MAb or 200 μl of mouse anti-Tac MAb (for chimeric proteins). Antibody-antigen complexes were collected by incubation with fixed Staphylococcus aureus (Pansorbin; Calbiochem, San Diego, CA) for 20 min at room temperature. Immunoprecipitates were washed three times in radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 50 mM Tris-HCl [pH 8.0], 1% Na-deoxycholate, 150 mM NaCl, 1% Triton X-100) and subsequently eluted in 20 μl of 50 mM Tris-HCl (pH 6.8), 1% SDS by incubation at 95°C for 3 min. Ten microliters of eluate was mixed with 10 μl of 0.15 M sodium citrate (pH 5.5) without (mock) or with 0.1 mU of recombinant endoglycosidase H (endo H) (New England Biolabs, Beverly, MA) and incubated for 16 to 20 h at 37°C. The samples were then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using 6, 8, or 10% gels. Images were obtained by a Molecular Imager FX phosphorimager (Bio-Rad, Hercules, CA). Oligosaccharide processing was quantified by comparing band densities of endo H-sensitive and -resistant forms using Quantity One software (Bio-Rad).
COPI binding experiments.
CHO cells from a confluent 10-cm dish were lysed in 1 ml of lysis buffer [90 mM KCl, 0.5% Triton X-100, and 50 mM 2-(N-morpholino)ethanesulfonic acid (pH 6.5) or 50 mM HEPES (pH 7.4)] plus protease inhibitor cocktail. Binding experiments were similar to those described by Gomez et al. (17). Briefly, 10 μg of each purified GST fusion protein was preincubated with 20 μl of glutathione-Sepharose 4B beads for 1 h at room temperature in PBS binding buffer (10 mM EDTA, 5 mM benzamide, and protease inhibitor cocktail in PBS). The beads were washed once in PBS and mixed with 100 μl of cell lysate. After incubation for 2 h at room temperature, beads were washed three times in lysis buffer and once in PBS. Bound proteins were then eluted off the beads by boiling in 65 μl of sample buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 30% glycerol, 0.2% bromophenol blue, and 10% 2-mercaptoethanol) for 3 min at 95°C. Half of the eluate was then subjected to 15% SDS-PAGE and Western blotting analysis using anti-ɛ-COP antibody. The other half was run on 15% SDS-PAGE gels and stained with Coomassie blue for protein quantification.
Western blotting analysis.
Samples were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore Corporation, Bedford, MA). The membrane was blocked for 30 min at room temperature with 5% nonfat dry milk in TBST (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 0.05% Tween 20). The membrane was incubated overnight at 4°C with primary antiserum or purified antibodies diluted in TBST containing 0.02% NaN3 and 5% nonfat dry milk or 3% bovine albumin. After extensive washing, the membrane was incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit secondary antibody diluted in 5% nonfat dry milk-TBST. Antibody binding was detected with a chemiluminescent substrate (Amersham) and X-ray film.
In vitro transcription and translation (IVTT).
TNT Quick Coupled Transcription/Translation Systems (Promega Corporation, Madison, WI) was used to produce radiolabeled SARS M according to the manufacturer's protocol. Briefly, pcDNA3.1/SARS M was incubated in a TNT master mix containing [35S]methionine (Redivue; Amersham) in the presence of canine pancreatic microsomal membranes (Promega Corporation, Madison, WI) for 90 min at 30°C. The reaction was then diluted into IVTT binding buffer containing 50 mM HEPES (pH 7.1), 100 mM NaCl, 10 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40. Equal amounts of the IVTT reaction mixture were added to purified GST fusion proteins (GST, GST SARS S, or GST SARS S2A) prebound to glutathione-Sepharose 4B beads and incubated for 2 h at room temperature. After extensive washing in IVTT binding buffer, samples were eluted in sample buffer and subjected to 15% SDS-PAGE. Images were obtained by a Molecular Imager FX phosphorimager. Binding was quantified using Quantity One software, and total M protein bound to the GST fusion proteins was determined by comparison with 10% input of the IVTT reaction.
RESULTS
Full-length SARS S is localized to the cell surface.
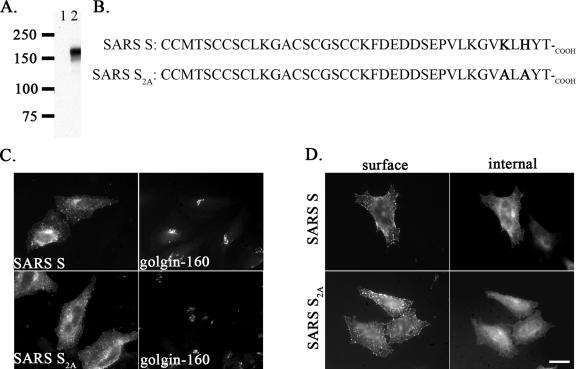
The localization of full-length SARS S expressed independently from cDNA has been controversial. While several groups reported localization on the plasma membrane (3, 25, 58, 60), another group suggested that it localizes to the ER and might escape to the cell surface when overexpressed (14). Many of these studies expressed epitope-tagged and/or codon-optimized SARS S. For studying the targeting of SARS S, we raised rabbit polyclonal antibodies (anti-SCT) against the C-terminal 14 amino acids of SARS S using a synthetic peptide. By immunoblotting, these antibodies specifically recognized a band of 180 kDa in the lysate of HeLa cells infected with a T7 RNA polymerase-encoding vaccinia virus (12) and transfected with pcDNA3.1/SARS S (Fig. 1A). This band was of the size predicted for N-glycosylated SARS S. To determine the subcellular localization of SARS S, we expressed this protein as an untagged construct using a cDNA with native codons to avoid overexpression and possible aberrant targeting. We used the expression vector pCAGGS, which expresses the transgene under the control of a chicken β-actin promoter (50). HeLa cells transiently transfected with pCAGGS/SARS S were double labeled with anti-SCT and an antibody recognizing an endogenous Golgi protein (golgin-160) and examined by indirect immunofluorescence microscopy. We examined transfected cells 24 h posttransfection since the staining with anti-SCT antibody was weak at earlier times. Untagged SARS S was located primarily on the surface of transfected cells, with staining throughout the secretory pathway; there was some overlap with the Golgi marker golgin-160 (Fig. 1C). Surface staining of intact cells with a mouse immune serum revealed that SARS S is expressed on the cell surface at steady state (Fig. 1D). To evaluate the contribution of the KxHxx motif to the localization of SARS S, KLHYT-COOH in SARS S was mutated to ALAYT-COOH, resulting in SARS S2A (Fig. 1B). This mutation did not dramatically change the steady-state localization of the full-length protein when expressed from pCAGGS/SARS S2A as determined by immunofluorescence microscopy, although more S was present in the Golgi region than S2A (Fig. 1C and D).
FIG. 1.
Localization of full-length SARS S. (A) Specificity of anti-SCT antibody. HeLa cells infected with vTF7-3 and transfected with pcDNA3.1/Hygro (lane 1) or pcDNA3.1/SARS S (lane 2) for 7 h were lysed and subjected to Western blotting analysis using a rabbit antiserum (anti-SCT, 1:5,000) raised against a peptide derived from the cytoplasmic tail of SARS S. Molecular masses in kDa are shown on the left. (B) Amino acid sequences of the cytoplasmic tails from SARS S and its mutant SARS S2A, with the lysine-histidine motif and alanine mutations in boldface type. (C) HeLa cells transfected with pCAGGS/SARS S or S2A for 24 h were fixed, permeabilized, and double labeled with mouse anti-SARS immune serum and rabbit anti-golgin-160 antibodies. (D) Cells transfected as in panel C were surface-labeled with SARS mouse immune serum at 4°C, fixed, permeabilized, and then stained with rabbit anti-SCT. Secondary antibodies were Alexa-488-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey anti-rabbit IgG. Bar, 10 μm.
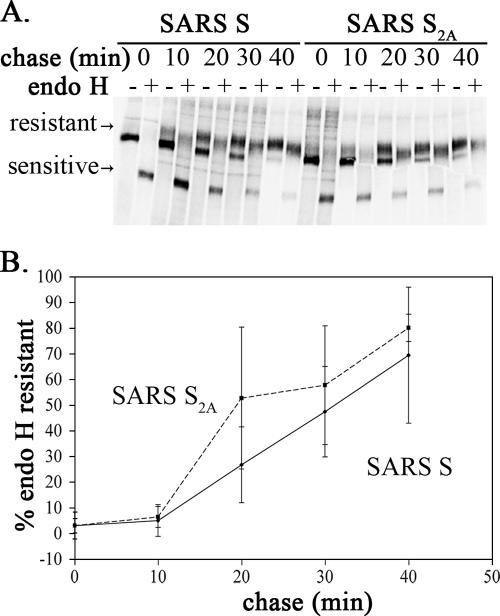
To determine if the rate of SARS S trafficking was influenced by the KxHxx motif, oligosaccharide processing was examined. Glycoproteins obtain their N-linked oligosaccharides in the ER during biosynthesis. When a protein moves through the Golgi complex, its oligosaccharides are processed in the medial Golgi compartment and become resistant to digestion with the endo H enzyme. Therefore, the rate of trafficking through the Golgi can be inferred from the rate of acquisition of endo H resistance. At 24 h posttransfection, HeLa cells expressing SARS S or S2A were pulse-labeled with [35S]methionine-cysteine for 20 min and chased for various times. SARS S was immunoprecipitated from cell lysates using anti-SCT antibodies and subjected to endo H digestion (Fig. 2A). The SARS S protein was efficiently processed to endo H resistance; however, the S2A protein was processed slightly more rapidly (Fig. 2B). Although the difference was small and not statistically significant, it was reproducible in each experiment. Since the differences in intra-Golgi trafficking were small, we went on to study the dibasic motif using reporter proteins, independent of full-length SARS S. Studying the dibasic motif using reporter proteins allowed the use of higher-titered antibodies, which permitted analysis at lower expression levels (earlier times posttransfection) and thus helped to quantify relatively small differences in trafficking rates.
FIG. 2.
Oligosaccharide processing of SARS S and SARS S2A. (A) HeLa cells transfected with pCAGGS/SARS S or pCAGGS/SARS S2A for 24 h were pulse-labeled with [35S]methionine-cysteine for 20 min, chased for 0, 10, 20, 30, or 40 min, lysed, and immunoprecipitated with rabbit anti-SCT antibody. The immunoprecipitates were mock treated (−) or treated with endo H (+) and subjected to 6% SDS-PAGE and autoradiography. The positions of the endo H-sensitive and -resistant forms of the proteins are indicated. (B) Endo H resistance was quantified, and the averages ± standard deviations from three independent experiments are shown.
The SARS S cytoplasmic tail slows the traffic of reporter proteins.
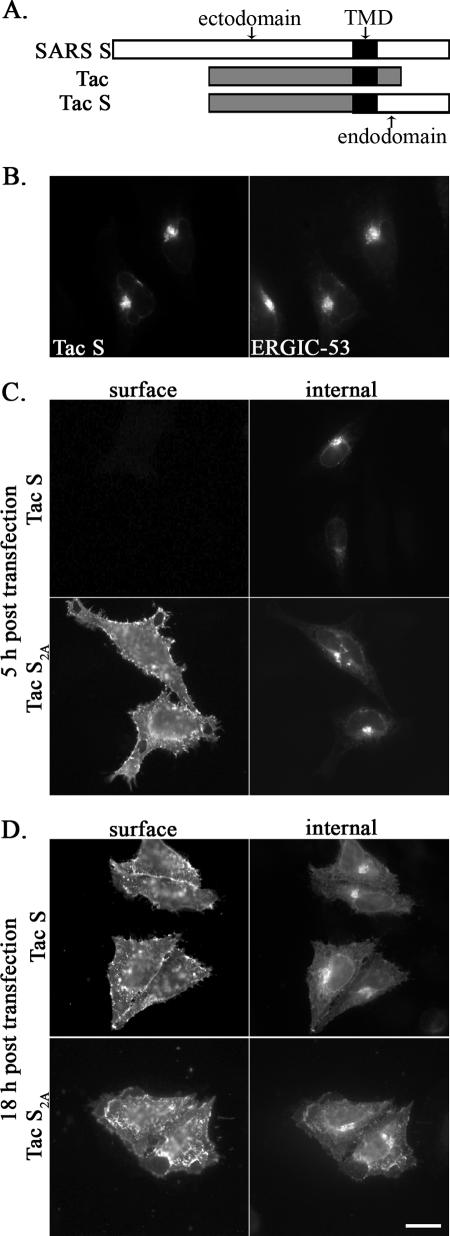
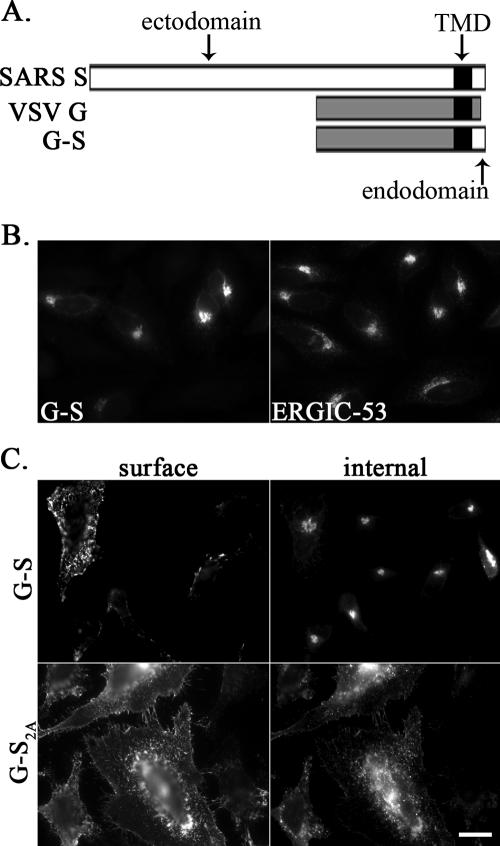
As mentioned above, only the last 11 amino acids of the SARS S cytoplasmic tail were appended to a chimeric protein in the previous study (41). To explore whether the KxHxx-COOH motif works in the context of the entire cytoplasmic tail of SARS S, we made a new construct, Tac-S, which contains the ectodomain and transmembrane domain of Tac antigen (human interleukin-2 receptor alpha chain) (49) and the entire cytoplasmic tail (Cys1217-Thr1255) of SARS S (Fig. 3A). To determine the localization of Tac-S, HeLa cells transiently transfected with pCAGGS/Tac-S were double labeled and examined by indirect immunofluorescence microscopy. We could detect expression of the chimeric protein at 5 h posttransfection. As shown in Fig. 3B, the distribution of Tac-S strongly overlapped with that of ERGIC-53, indicating that Tac-S accumulated in the ERGIC region.
FIG. 3.
Localization of Tac-S chimeric proteins. (A) Schematicrepresentation of Tac, SARS S, Tac-S, and Tac-S2A. TMD, transmembrane domain. (B) HeLa cells transfected with pCAGGS/Tac-S for 5 h were fixed, permeabilized and double labeled with rabbit anti-SCT and mouse anti-ERGIC-53 antibodies. (C and D) HeLa cells transfected with pCAGGS/Tac-S or pCAGGS/Tac-S2A for 5 h or 18 h were surface stained with a mouse monoclonal anti-Tac antibody at 4°C, fixed, permeabilized, and then stained with rabbit anti-Tac antibody for internal expression. Secondary antibodies were Alexa-488-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey anti-rabbit IgG. Bar, 10 μm.
To evaluate the role of the KxHxx-COOH dibasic motif in the SARS S cytoplasmic tail, we mutated the lysine and histidine residues in Tac-S to alanines, resulting in Tac-S2A. When examined at 5 h posttransfection by staining intact cells with a monoclonal antibody to the ectodomain of Tac, Tac-S was absent from the plasma membrane in a substantial population of cells; approximately 52% (54/104) of the Tac-S-expressing cells had low but detectable surface staining (Fig. 3C). By contrast, Tac-S2A, which contains the KLHYT-to-ALAYT mutation, was more strongly expressed on the cell surface (Fig. 3C); 100% (105/105) of the expressing cells were surface positive. However, at 18 h posttransfection, there were no obvious differences between the surface levels of Tac-S and those of Tac-S2A (Fig. 3D).
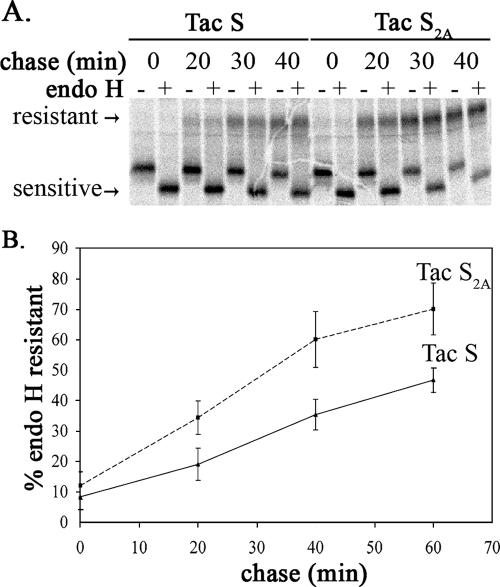
To further evaluate the contribution of the KxHxx motif to the retention of the reporter protein in the ERGIC, the rates of trafficking of Tac-S and Tac-S2A through the Golgi complex were compared. HeLa cells transfected with pCAGGS/Tac-S or pCAGGS/Tac-S2A for 18 h were pulse-labeled with [35S]methionine-cysteine for 20 min and chased for various times. The expressed proteins were immunoprecipitated and subjected to endo H digestion (Fig. 4A). Tac-S2A trafficked through the medial Golgi compartment significantly faster than Tac-S, as indicated by the results of pulse-chase labeling and endo H digestion experiments (Fig. 4B). Note that the endo H-resistant forms of the Tac chimeric proteins have much a slower mobility because Tac also acquires O glycosylation as it reaches the Golgi complex (37). After 60 min of chase, 45% of Tac-S acquired endo H resistance, while approximately 75% of Tac-S2A was resistant to endo H digestion. Taken together, the steady-state localization in the ERGIC/Golgi region at low expression levels and the slow processing of Tac-S compared with that of Tac-S2A suggest that the dibasic motif in the SARS S cytoplasmic tail plays a role in retrieval similar to that of the dilysine signal in the C terminus of IBV S.
FIG. 4.
Oligosaccharide processing of Tac-S and Tac-S2A. (A) HeLa cells expressing Tac-S or Tac-S2A for 18 h from pCAGGS-based vectors were pulse-labeled with [35S]methionine-cysteine for 20 min, chased for 0, 20, 40, or 60 min, lysed, and immunoprecipitated with mouse anti-Tac antibody. The immunoprecipitates were mock treated (−) or treated with endo H (+) and subjected to 10% SDS-PAGE and phosphorimaging. A representative gel is shown. The endo H-sensitive and -resistant forms are indicated. Note that the endo H-resistant forms of the Tac chimeric proteins have a much slower mobility because Tac also acquires O glycosylation (37). (B) Endo H resistance was quantified, and the averages ± standard deviations from three independent experiments are shown.
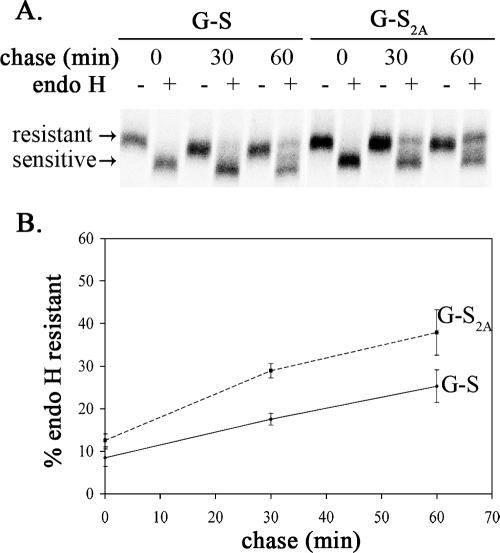
The Tac-S2A mutation was less efficient at restoring transport of the chimera compared to the analogous chimera containing the cytoplasmic tail of IBV S (41). However, Tac is a monomer when expressed from cDNA (29), whereas SARS S has been reported to form homotrimers (62). To determine if the KxHxx signal would be more efficient when presented as a trimer, we generated another chimeric protein using the ectodomain of VSV G, which mediates homotrimerization (10). The extracellular and transmembrane domains of VSV G were fused with the cytoplasmic tail of SARS S or SARS S2A, producing G-S and G-S2A, respectively (Fig. 5A). Similar to Tac-S, G-S also accumulated in the ERGIC region in HeLa cells transiently transfected with pCAGGS/G-S (Fig. 5B). At 8 h posttransfection, surface staining experiments showed that only 39% (82/208) of the cells expressing G-S had detectable surface staining, while this percentage was 100% (135/135) for G-S2A, and the staining was significantly brighter (Fig. 5C). G-S2A acquired endo H resistance more rapidly than G-S, although for both chimeric proteins only a portion (<50%) acquired endo H resistance by 60 min of chase (Fig. 6). This difference in oligosaccharide processing is consistent with the observed difference in surface expression described above. Thus, presenting the KxHxx signal as a trimer did not enhance the efficiency of ER retrieval. These results support the findings from the experiments with full-length SARS S and Tac-S, indicating that the dibasic motif is functional, although significantly less potent than the canonical dilysine signal.
FIG. 5.
Localization of G-S and G-S2A chimeric proteins. (A) Schematic representation of SARS S, VSV-G, G-S, and G-S2A chimeric proteins. TMD, transmembrane domain. (B) For the localization of G-S, HeLa cells transfected with pCAGGS/G-S were fixed, permeabilized, and double labeled with rabbit anti-SCT and mouseanti-ERGIC-53 antibodies. Secondary antibodies were Alexa-488-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey anti-rabbit IgG. (C) For surface and internal staining of G-S and G-S2A chimeric proteins, HeLa cells transfected with pCAGGS/G-S or pCAGGS/G-S2A for 8 h were incubated with mouse anti-VSV G antibody at 4°C, fixed, permeabilized, and then stained with rabbit anti-SCT for internal expression. Secondary antibodies were Alexa-488-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey anti-rabbit IgG. Bar, 10 μm.
FIG. 6.
Oligosaccharide processing of G-S and G-S2A. (A) HeLa cells expressing G-S or G-S2A from pCAGGS-based vectors for 18 h were pulse-labeled with [35S]methionine-cysteine for 20 min, chased for 0, 30, or 60 min, lysed, and immunoprecipitated with rabbit anti-SCT serum. The precipitates were mock treated (−) or treated with endo H (+) and subjected to 10% SDS-PAGE and phosphorimaging to determine the extent of endo H resistance. A representative image is shown. The endo H-resistant and -sensitive forms are indicated. (B) Quantification of oligosaccharide processing rates. The data represent the averages ± standard deviations from three independent experiments.
The SARS S dibasic motif binds COPI.
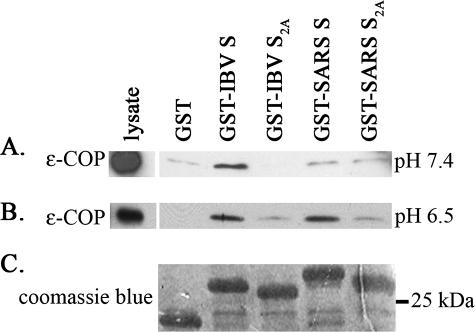
The results from the above experiments suggested that the KxHxx motif found at the SARS S C-terminal tail is a novel ER retrieval signal. It has been previously shown that the canonical dilysine ER retrieval signal binds to the COPI coat complex, the major component of COPI-coated vesicles that mediate the retrograde transport of proteins from the Golgi to the ER (9, 35). If the KxHxx motif is an ER retrieval signal, it should also bind COPI. To test this hypothesis, we performed an in vitro binding assay. GST or GST fusion proteins containing amino acids Leu1226 to Thr1255 of the SARS S C-terminal tail (named GST-SARS S) or the KLHYT-to-ALAYT mutation (named GST-SARS S2A) were expressed in bacteria and purified. As controls, GST-IBV S and GST-IBV S2A containing Gly1129 to Val1162 of the IBV S cytoplasmic tail (with KKSV-COOH) or its mutant (with AASV-COOH), respectively, were also prepared in the same way. The purified recombinant proteins were prebound to glutathione-Sepharose beads and incubated with cell lysate prepared at pH 7.4. Bound proteins were eluted and subjected to immunoblotting, and COPI binding was detected by an anti-ɛ-COP antibody. As shown in Fig. 7A, the IBV S GST fusion proteins behaved as expected: GST-IBV S bound COPI, whereas GST-IBV S2A did not. Neither GST-SARS S nor GST-SARS S2A bound COPI above the background levels bound by GST alone (Fig. 7A). However, when the cell lysate was prepared at pH 6.5, closer to the pKa of histidine, COPI specifically coprecipitated with GST-SARS S, indicating that GST-SARS S bound to COPI at this pH (Fig. 7B). Mutation of the KLHYT in GST-SARS S to ALAYT in GST-SARS S2A disrupted binding (Fig. 7B), indicating that the binding of GST-SARS S to COPI was indeed mediated by the dibasic motif. Given the results of steady-state localization and trafficking of the chimeric proteins, as well as COPI binding, we conclude that the dibasic motif (KxHxx-COOH) is an ER retrieval signal that plays a role similar to a canonical dilysine signal.
FIG. 7.
COPI binding to GST fusion proteins. CHO cell lysate (100 μl) was incubated at room temperature for 2 h with 10 μg of purified GST, GST-IBV S, GST-IBV S2A, GST-SARS S, or GST-SARS S2A prebound to glutathione-Sepharose 4B beads. After washing, bound materials were eluted in SDS sample buffer. One half of each eluate was subjected to Western blotting analysis by using rabbit anti-ɛ-COP antibody. As an input control, an aliquot of the cell lysate corresponding to 10% of the amount used in binding assays was also included on the gel. (A) A blot representative of two experiments is shown for binding at pH 7.4. (B) A blot representative of five independent experiments is shown for binding at pH 6.5. (C) The other half of each eluate was run on 15% SDS-PAGE gels and stained with Coomassie blue. A representative gel is shown for the samples from the same assay as in panel A. All the lanes from each experiment were from the same gel with the same exposure.
Coexpression with SARS M localizes SARS S to the Golgi region.
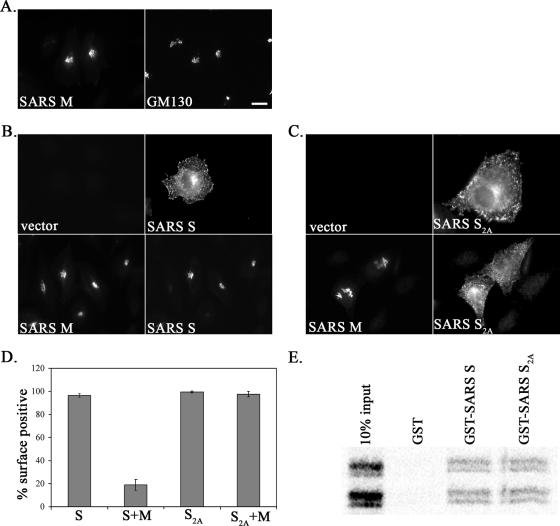
It has been shown for several coronaviruses that M protein interacts with S during assembly (15, 26, 47, 52). To determine if the cycling of SARS S through the ER-Golgi system mediated by the dibasic motif might allow more efficient interaction with SARS M, we evaluated the subcellular localization of SARS S when coexpressed with M. Cells were transfected with plasmids encoding SARS S or M or both and visualized by indirect immunofluorescence microscopy. When expressed alone, SARS M was localized to the Golgi region (Fig. 8A) and SARS S was transported to the cell surface (Fig. 8B, upper panel). However, when SARS S and M were coexpressed, S was retained intracellularly at the Golgi region and colocalized with M (Fig. 8B, lower panel). By contrast, when SARS S2A was coexpressed with SARS M, S2A was transported to the cell surface while M remained at the Golgi region (Fig. 8C, lower panel). The retention of S was nearly complete in the presence of M, whereas S2A was not retained (Fig. 8D). One possible explanation for these data is that the S2A mutation directly prevents the association with M protein. However, it has been previously shown that mouse hepatitis virus (MHV) S and M proteins interact via sequences in the juxtamembrane region of their cytoplasmic tails (4), suggesting that the mutation at the C terminus of SARS S should not disrupt interaction. To test this directly, we performed an in vitro binding assay with the cytoplasmic tails of SARS S or S2A fused to GST. SARS M was transcribed and translated in vitro with [35S]methionine in the presence of microsomes and solubilized with detergent. SARS M is partially N glycosylated, and both glycosylated and nonglycosylated forms of M run as a doublet (51, 68). Interestingly, this same pattern was seen for in vitro-translated protein (Fig. 8E). The solubilized M protein was incubated with equal amounts of purified GST, GST-SARS S, or GST-SARS S2A prebound to glutathione-Sepharose beads. Phosphorimaging and quantification showed that GST-SARS S and S2A interacted equally well with SARS M and that the interaction was at least 25-fold over background binding compared to that of GST (Fig. 8E and data not shown). Since the mutation in S2A does not prevent M-S interaction, our data suggest that ER retrieval via the dibasic signal in the S tail may be critical for S and M interaction during virus assembly in SARS CoV-infected cells. We propose that repeated cycling of SARS S through the ER-Golgi system allows sufficient opportunity for interaction with the M protein.
FIG. 8.
Relocalization of SARS S by SARS M and its dependence on the dibasic motif. (A) HeLa cells transfected with pCAGGS/SARS M for 24 h were fixed, permeabilized, and double labeled with rabbit anti-M and mouse anti-GM130 antibodies. (B) HeLa cells cotransfected for 24 h with plasmids encoding SARS S (along with vector alone) or SARS S and SARS M were fixed, permeabilized, and double labeled with rabbit anti-M and mouse anti-SARS immune serum. (C) HeLa cells cotransfected for 24 h with plasmids encoding SARS S2A (along with vector alone) or SARS S2A and SARS M were fixed, permeabilized, and double labeled with rabbit anti-M and mouse anti-SARS immune serum. (D) Cells were evaluated for the surface expression of S or S2A in the absence or presence of M as described in Materials and Methods. n ≥ 230 cells for each. The averages from four experiments ± standard deviations are shown. (E) SARS M interacts equally well with the cytoplasmic tails of SARS S and S2A. Equal amounts of in vitro-transcribed and -translated M were incubated with equal amounts of GST fusion proteins. Samples were washed, subjected to 15% SDS-PAGE, and visualized by phosphorimaging.
DISCUSSION
In the present study, we demonstrated that the cytoplasmic tail of the spike protein of SARS CoV contains a novel dibasic signal (KxHxx-COOH) that plays a role in the subcellular localization of the protein. The chimeric reporter proteins and COPI binding experiments indicated that the dibasic motif is an ER retrieval signal. To our knowledge, it is the first of its kind ever reported for a protein found in nature. The results of coexpression studies suggested that this novel motif aids the S protein in its interaction with the M protein.
The KxHxx motif present at the C terminus of SARS S is a novel functional ER retrieval signal.
Two types of ER localization signals have been well characterized for membrane proteins: dilysine and diarginine signals (65). Dilysine signals found in type I transmembrane proteins require two lysine residues at the −3 and −4 (or −5) positions relative to the C terminus (65). By contrast, diarginine motifs for membrane proteins are less strict in their position requirements: they can function at the N or C terminus of a membrane protein and do not require specific spacing relative to either terminus (57, 72). Proteins bearing a dilysine or a diarginine motif are retrieved from ER compartments back to the ER through direct interactions with the COPI complex, which forms the coat of COPI vesicles (35, 72). Although the COPI complex directly interacts with the dilysine or diarginine signal, the efficiency of binding is influenced by residues surrounding the motifs (65) and can affect the steady-state localization of a protein, as reported for ERGIC-53 (1).
It could be reasoned that the KxHxx motif found in group 1 coronaviruses and SARS coronavirus (41) can mimic the dilysine signal if the histidine residue is protonated. In fact, it has been shown that a histidine residue at the −3 position of the oligosaccharide transferase subunit OST48 can substitute for the lysine residue normally present at this position (23). Our analysis of the KxHxx motif found at the C terminus of SARS S showed that the dibasic motif is likely to function in ER retrieval. The KxHxx motif from SARS S retained reporter proteins in the ERGIC region at low expression levels and significantly reduced the rates of their trafficking to the cell surface. It is important to note that the acquisition of endo H resistance reports only the rate of the first passage through the medial Golgi; it cannot measure the transport of a cycling protein that has already been processed. Importantly, the results of in vitro binding experiments showed that the KxHxx motif binds the COPI complex at pH 6.5. Since histidine has a low pKa compared with those of lysine and arginine, it is likely to be less efficiently protonated in the neutral cytoplasm, and hence, the binding of the KxHxx motif to COPI in vivo might be weaker than that of a dilysine or a diarginine motif. This is reflected in the reduced efficiency of chimeric proteins with the SARS S tail to localize to the ERGIC region at high expression levels compared to those with the IBV S tail. This reduction in binding efficiency could explain why full-length SARS S is transported to the surface of cells while S proteins with a canonical dilysine signal, like those in IBV, are efficiently retained intracellularly. However, pH microenvironments can exist near membranes (43, 55), so it is possible that the histidine could be protonated during S trafficking. The Golgi complex has resident ion pumps and channels whose activities could alter the cytoplasmic pH near Golgi membranes (20). Also, the intracellular environment may change in infected cells. It has been previously shown that SARS infection can lead to apoptosis at late times postinfection (70). Interestingly, an event preceding apoptosis is acidification of the cytoplasm (19). This acidification could lead to an increase in the efficiency of S protein ER retrieval in infected cells as SARS CoV infection progresses. However, there is not yet enough information on the timing of apoptosis in SARS CoV-infected cells to predict the consequences for ER retrieval of SARS S on virus production.
Importantly, we showed that the SARS S KxHxx motif may be necessary for efficient interaction with the M protein. When S and M were coexpressed, S localization shifted dramatically from the plasma membrane to the Golgi region. A similar result was shown for MHV S: when expressed alone, it is transported to the cell surface, but when it is coexpressed with M, it is localized to the Golgi region (52). We showed that this dramatic change in SARS S localization can be disrupted by mutating the KxHxx motif. Since the cytoplasmic tails of these proteins can bind M equally well, we hypothesize that the cycling of SARS S (which is prevented by the mutations in S2A) is responsible for the differential localization of S and S2A when coexpressed with M. In this case, the S protein moves through the Golgi past the budding site but is retrieved back to the ER by COPI-coated vesicles, thus providing more opportunities to interact with M. When the KxHxx motif is mutated, the lack of retrieval would preclude efficient interaction with M. It will be important to determine the contribution of these signals to SARS CoV infection by using an infectious cDNA clone.
Implications of dibasic signals in the biology of coronaviruses.
Previously, we reported that the KxHxx-COOH motif found in the S protein of SARS CoV is also present in group 1 coronaviruses, including transmissible gastroenteritis virus (TGEV) (41). However, Schwegmann-Wessels and colleagues reported that a tyrosine-based motif in the cytoplasmic tail of TGEV S protein was responsible for intracellular localization (58). The difference between their conclusions and ours could be due to the different cell types and expression systems used and/or the fact that they analyzed only steady-state localization by immunofluorescence microscopy and long biosynthetic labeling periods. Thus, rapid internalization of their truncation mutants lacking the dibasic motif from the plasma membrane could have been missed. In addition to the KKxx-COOH motif found in the S proteins of group 3 CoV, we have found that IBV S contains a tyrosine-based endocytosis signal upstream of the dilysine signal in its cytoplasmic tail (41). However, no similar signals are observed in the S proteins from group 2 coronaviruses such as bovine coronavirus or mouse hepatitis virus (41). This raises interesting issues about the importance of these signals in the biology of coronaviruses.
Accumulation of the CoV envelope proteins at the virus assembly site is required for efficient virus production. An ER retrieval signal in the S protein is likely to contribute to this targeting. On the other hand, when expressed on the cell surface, the S proteins from coronaviruses, including SARS CoV, induce cell-cell fusion, resulting in the formation of syncytia and accompanying intracellular reorganization (6, 48). Syncytium formation may reduce the efficiency of virus replication or virion release, as suggested by our recent study with a mutant IBV lacking the KKxx motif (71). By limiting the level of surface S protein through ER retrieval or retention, the virus could have sufficient time for replication, maturation, and release. Late in infection, when the expression levels of S increase, the ER retrieval machinery could be saturated and the S protein would be transported to the plasma membrane. At this stage, endocytosis signals in some coronaviruses may further ensure that S proteins that reach the cell surface do not remain there for long. The group 2 coronaviruses, which lack ER retrieval and endocytosis signals in their S proteins, may depend on direct cell-to-cell spread of the infection through the formation of syncytia to a greater extent than coronaviruses of other groups. Alternatively, group 2 coronaviruses might have another as-yet-unidentified mechanism to control the levels of their S proteins on the cell surface. Further experiments are needed to test these possibilities.
Acknowledgments
This work was supported by National Institutes of Health grant GM64647.
We are grateful to Michael Marks for providing Tac cDNA and antibodies, Hans-Peter Hauri, Monty Krieger and Kanta Subbarao for antibodies, and Paul Bates for the expression plasmid pCAGGS-MCS. We also thank the members of the Machamer lab for helpful comments on the manuscript.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Andersson, H., F. Kappeler, and H. P. Hauri. 1999. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J. Biol. Chem. 274:15080-15084. [DOI] [PubMed] [Google Scholar]
- 2.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisht, H., A. Roberts, L. Vogel, A. Bukreyev, P. L. Collins, B. R. Murphy, K. Subbarao, and B. Moss. 2004. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA 101:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, B. J., C. A. de Haan, S. L. Smits, and P. J. Rottier. 2005. Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements. Virology 334:306-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-113. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, NY.
- 7.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629-1631. [DOI] [PubMed] [Google Scholar]
- 10.Doms, R. W., A. Ruusala, C. Machamer, J. Helenius, A. Helenius, and J. K. Rose. 1988. Differential effects of mutations in three domains on folding, quaternary structure, and intracellular transport of vesicular stomatitis virus G protein. J. Cell Biol. 107:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giroglou, T., J. Cinatl, Jr., H. Rabenau, C. Drosten, H. Schwalbe, H. W. Doerr, and D. von Laer. 2004. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 78:9007-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godeke, G.-J., C. A. M. de Haan, J. W. A. Rossen, H. Vennema, and P. J. M. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez, M., S. J. Scales, T. E. Kreis, and F. Perez. 2000. Membrane recruitment of coatomer and binding to dilysine signals are separate events. J. Biol. Chem. 275:29162-29169. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb, R. A., J. Nordberg, E. Skowronski, and B. M. Babior. 1996. Apoptosis induced in Jurkat cells by several agents is preceded by intracellular acidification. Proc. Natl. Acad. Sci. USA 93:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabe, M., and G. Oster. 2001. Regulation of organelle acidity. J. Gen. Physiol. 117:329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths, G., and P. Rottier. 1992. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 23.Hardt, B., and E. Bause. 2002. Lysine can be replaced by histidine but not by arginine as the ER retrieval motif for type I membrane proteins. Biochem. Biophys. Res. Commun. 291:751-757. [DOI] [PubMed] [Google Scholar]
- 24.Hicks, S. W., and C. E. Machamer. 2002. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J. Biol. Chem. 277:35833-35839. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, H., K. Hattermann, A. Marzi, T. Gramberg, M. Geier, M.Krumbiegel, S. Kuate, K. Überla, M. Niedrig, and S. Pöhlmann. 2004. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 78:6134-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., Z.-Y. Yang, W.-P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78:12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingallinella, P., E. Bianchi, M. Finotto, G. Cantoni, D. M. Eckert, V. M. Supekar, C. Bruckmann, A. Carfi, and A. Pessi. 2004. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. USA 101:8709-8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffers, S. A., S. M. Tusell, L. Gillim-Ross, E. M. Hemmila, J. E. Achenbach, G. J. Babcock, W. D. Thomas, Jr., L. B. Thackray, M. D. Young, R. J. Mason, D. M. Ambrosino, D. E. Wentworth, J. C. Demartini, and K. V. Holmes. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 101:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junghans, R. P., A. L. Stone, and M. S. Lewis. 1996. Biophysical characterization of a recombinant soluble interleukin 2 receptor (Tac). Evidence for a monomeric structure. J. Biol. Chem. 271:10453-10460. [DOI] [PubMed] [Google Scholar]
- 30.Kan, B., M. Wang, H. Jing, H. Xu, X. Jiang, M. Yan, W. Liang, H. Zheng, K. Wan, Q. Liu, B. Cui, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, X. Qi, K. Chen, L. Du, K. Gao, Y.-T. Zhao, X. Z. Zou, Y.-J. Feng, Y.-F. Gao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79:11892-11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. M. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, M. C., E. A. Miller, J. Goldberg, L. Orci, and R. Schekman. 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20:87-123. [DOI] [PubMed] [Google Scholar]
- 36.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121:168-174. [DOI] [PubMed] [Google Scholar]
- 37.Leonard, W. J., J. M. Depper, R. J. Robb, T. A. Waldmann, and W. C. Greene. 1983. Characterization of the human receptor for T-cell growth factor. Proc. Natl. Acad. Sci. USA 80:6957-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 40.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. R. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lontok, E., E. Corse, and C. E. Machamer. 2004. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 78:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 105:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maouyo, D., S. Chu, and M. H. Montrose. 2000. pH heterogeneity at intracellular and extracellular plasma membrane sites in HT29-C1 cell monolayers. Am. J. Physiol. Cell Physiol. 278:C973-C981. [DOI] [PubMed] [Google Scholar]
- 44.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 45.Mortola, E., and P. Roy. 2004. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 576:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng, M. L., S. H. Tan, E. E. See, E. E. Ooi, and A. E. Ling. 2003. Proliferative growth of SARS coronavirus in Vero E6 cells. J. Gen. Virol. 84:3291-3303. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen, V.-P., and B. G. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikaido, T., A. Shimizu, N. Ishida, H. Sabe, K. Teshigawara, M. Maeda, T. Uchiyama, J. Yodoi, and T. Honjo. 1984. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature 311:631-635. [DOI] [PubMed] [Google Scholar]
- 50.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 51.Oostra, M., C. A. M. de Haan, R. J. de Groot, and P. J. M. Rottier. 2006. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 80:2326-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puddington, L., C. E. Machamer, and J. K. Rose. 1986. Cytoplasmic domains of cellular and viral integral membrane proteins substitute for the cytoplasmic domain of the vesicular stomatitis virus glycoprotein in transport to the plasma membrane. J. Cell Biol. 102:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ro, H. A., and J. H. Carson. 2004. pH microdomains in oligodendrocytes. J. Biol. Chem. 279:37115-37123. [DOI] [PubMed] [Google Scholar]
- 56.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 57.Schutze, M. P., P. A. Peterson, and M. R. Jackson. 1994. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 13:1696-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwegmann-Wessels, C., M. Al-Falah, D. Escors, Z. Wang, G. Zimmer, H. Deng, L. Enjuanes, H. Y. Naim, and G. Herrler. 2004. A novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 279:43661-43666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song, H. C., M. Y. Seo, K. Stadler, B. J. Yoo, Q.-L. Choo, S. R. Coates, Y. Uematsu, T. Harada, C. E. Greer, J. M. Polo, P. Pileri, M. Eickmann, R. Rappuoli, S. Abrignani, M. Houghton, and J. H. Han. 2004. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 78:10328-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swift, A. M., and C. E. Machamer. 1991. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 115:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang, X. C., J. X. Zhang, S. Y. Zhang, P. Wang, X. H. Fan, L. F. Li, G. Li, B. Q. Dong, W. Liu, C. L. Cheung, K. M. Xu, W. J. Song, D. Vijaykrishna, L. L. Poon, J. S. Peiris, G. J. Smith, H. Chen, and Y. Guan. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 66.Tripet, B., M. W. Howard, M. Jobling, R. K. Holmes, K. V. Holmes, and R. S. Hodges. 2004. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 279:20836-20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voss, D., A. Kern, E. Traggiai, M. Eickmann, K. Stadler, A. Lanzavecchia, and S. Becker. 2006. Characterization of severe acute respiratory syndrome coronavirus membrane protein. FEBS Lett. 580:968-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weisz, O. A., A. M. Swift, and C. E. Machamer. 1993. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 122:1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan, H., G. Xiao, J. Zhang, Y. Hu, F. Yuan, D. K. Cole, C. Zheng, and G. F. Gao. 2004. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 73:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Youn, S., E. W. Collisson, and C. E. Machamer. 2005. Contribution of trafficking signals in the cytoplasmic tail of the infectious bronchitis virus spike protein to virus infection. J. Virol. 79:13209-13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zerangue, N., M. J. Malan, S. R. Fried, P. F. Dazin, Y. N. Jan, L. Y. Jan, and B. Schwappach. 2001. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc. Natl. Acad. Sci. USA 98:2431-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]