Abstract
The transmembrane (TM) domains of hepatitis C virus (HCV) envelope glycoproteins E1 and E2 have been shown to play multiple roles during the biogenesis of the E1E2 heterodimer. By using alanine scanning insertion mutagenesis within the TM domains of HCV envelope glycoproteins, we have previously shown that the central regions of these domains as well as the N-terminal part of the TM domain of E1 are involved in heterodimerization. Here, we used a tryptophan replacement scan of these regions to identify individual residues that participate in those interactions. Our mutagenesis study identified at least four residues involved in heterodimerization: Gly 354, Gly 358, Lys 370, and Asp 728. Interestingly, Gly 354 and Gly 358 belong to a GXXXG oligomerization motif. Our tryptophan mutants were also used to generate retrovirus-based, HCV-pseudotyped particles (HCVpp) in order to analyze the effects of these mutations on virus entry. Surprisingly, two mutants consistently displayed higher infectivity compared to that of the wild type. In contrast, HCVpp infectivity was strongly affected for many mutants, despite normal E1E2 heterodimerization and normal levels of incorporation of HCV glycoproteins into HCVpp. The characterization of some of these HCVpp mutants in the recently developed in vitro fusion assay using fluorescent-labeled liposomes indicated that mutations reducing HCVpp infectivity without altering E1E2 heterodimerization affected the fusion properties of HCV envelope glycoproteins. In conclusion, this mutational analysis identified residues involved in E1E2 heterodimerization and revealed that the TM domains of HCV envelope glycoproteins play a major role in the fusion properties of these proteins.
Hepatitis C virus (HCV) is an enveloped virus that belongs to the Hepacivirus genus in the Flaviviridae family (34). Its genome encodes a single polyprotein precursor of ∼3,010 amino acid residues. HCV polyprotein is synthesized on endoplasmic reticulum (ER)-associated ribosomes and is cleaved co- and posttranslationally by cellular and viral proteases to yield at least 10 mature products (reviewed in reference 34). The two envelope glycoproteins E1 and E2 are released from the polyprotein by signal peptidase cleavages (19).
Due to difficulties in propagating HCV in cell culture, many gaps remain in our understanding of the functions of HCV envelope glycoproteins. A major advance in the investigation of the functions of these proteins was the development of HCV pseudoparticles (HCVpp) consisting of native HCV envelope glycoproteins E1 and E2 assembled onto retroviral core particles (3, 16, 26). Furthermore, data obtained with HCVpp can now also be confirmed with the help of the recently developed cell culture system that allows efficient amplification of HCV (HCVcc) (33, 56, 58). Studies with HCVpp and HCVcc systems have shown that the envelope glycoprotein complex E1E2 is essential for HCV entry (3, 26, 33, 56, 58). However, detailed analyses of functional domains are still lacking.
After their syntheses, HCV glycoproteins E1 and E2 assemble as a noncovalent heterodimer, which is retained in the ER (15). This glycoprotein complex is the viral component present at the surface of HCV particles, and it is therefore the obvious candidate ligand for a cellular receptor(s) (13). In addition, after endocytosis by a clathrin-mediated pathway (5), HCV envelope glycoproteins are involved in the fusion process between the viral envelope and a membrane of an internal compartment of the host cell (5, 30). This fusion process has been demonstrated to be pH dependent (4, 5, 26, 29, 30, 54).
The transmembrane (TM) domains of HCV glycoproteins exhibit unusual features (41). These domains are less than 30 amino acid residues long and are composed of two hydrophobic stretches separated by a short segment containing one or two fully conserved charged residues (14). A study of the topology of the TM domains of HCV envelope glycoproteins has shown a reorientation of the C termini of these domains, leading to a single membrane-spanning topology (12). Interestingly, studies of HCV envelope glycoproteins with heterologous expression systems have shown that the TM domains of these proteins play a major role in the assembly of the E1E2 heterodimer (42) and its subcellular localization (9, 11). Alanine insertions within the TM domains of HCV envelope glycoproteins have identified the central regions of these domains as well as the N-terminal part of the TM domain of E1 as sequences involved in heterodimerization (42). To identify individual residues that participate in those interactions, a tryptophan replacement scan of these regions was carried out. Tryptophan has previously been used to obtain structural information on helix-helix interactions in membrane proteins (6, 51, 52). Indeed, tryptophan has a large, bulky hydrophobic side chain and so should be tolerated at positions that interact with lipids, but not at positions involved in close protein-protein interactions.
Our mutagenesis study identified residues that play a role in E1E2 heterodimerization. In addition, many mutants were defective in HCVpp entry due mainly to an alteration in their fusion properties. Furthermore, two mutations in E1 led to the production of more infectious HCVpp. Together, these data indicate that the TM domains of HCV envelope glycoproteins play a major role in virus entry.
MATERIALS AND METHODS
Cell culture.
Huh-7 human hepatocarcinoma cells (39) and 293T human embryo kidney cells (293tsA1609neo) obtained from the American Type Culture Collection (Manassas, VA) were grown in Dulbecco's modified essential medium (Invitrogen) supplemented with 10% fetal bovine serum.
Antibodies.
Monoclonal antibodies (MAbs) A4 (anti-E1) (18), H53 (anti-E2) (11), H47 (12), 3/11 (anti-E2; kindly provided by J. McKeating) (23), and R187 (anti-capsid of murine leukemia virus [MLV]) (ATCC CRL1912) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer.
Site-directed mutagenesis.
The DNA sequence used to construct the mutants was derived from the H strain (genotype 1a) (24). Plasmids phCMV-E1E2, phCMV-E1, and phCMV-E2 (3), expressing HCV envelope glycoproteins, were kindly provided by B. Bartosch and F. L. Cosset (INSERM U758, Lyon, France). Mutations in the transmembrane domain of E1 were introduced into phCMV-E1, a plasmid encoding E1 alone. Mutations in the transmembrane domain of E2 were introduced in the context of phCMV-E1E2, a plasmid expressing E1 and E2 as a polyprotein. The mutations were constructed by sequential PCR steps as described previously (2) using the high-fidelity Deep Vent DNA polymerase (New England Biolabs). The mutants were then assembled by a second PCR amplification. The sequences of all the constructs were verified.
Production of HCVpp and infection assays.
The production of HCVpp and infection assays have been described previously (3, 43). In this work, we used the Gag-Pol MLV packaging construct containing the MLV gag and pol genes and the MLV-Luc plasmid encoding an MLV-based transfer vector containing a cytomegalovirus-Luc internal transcriptional unit. For E1 mutants, HCV envelope glycoproteins were expressed from two different plasmids, phCMV-E1 and phCMV-E2, whereas for E2 mutants, HCV envelope glycoproteins were expressed from a single plasmid, phCMV-E1E2. In the case of E1 mutants, this should avoid any potential interference with the processing of E1E2 due to the function of reinitiation of translocation present at the C terminus of E1 (19). Pseudotyped particles produced in the absence of envelope proteins were used as controls (Delta env). To analyze the incorporation of HCV envelope glycoproteins into pseudotyped particles, HCVpp were pelleted by centrifugation through a 20% sucrose cushion and analyzed by Western blotting. Within a given preparation of virions, similar amounts of virion-associated MLV capsid proteins were detected for HCVpp generated with the different mutants. However, as previously observed, important differences in the absolute quantities of virion-associated capsid can be noticed when two independent preparations of HCVpp are compared (25, 31). Thus, to minimize variations due to differences in the quality of preparations, each experiment was conducted using HCVpp generated concurrently.
Western blotting.
After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), protein preparations were transferred to nitrocellulose membranes (Hybond-ECL, Amersham) by using a Trans-Blot apparatus (Bio-Rad) and revealed with a specific MAb, followed by rabbit anti-mouse immunoglobulin conjugated to peroxidase (Dako; dilution 1/1,000). The proteins of interest were revealed by enhanced chemiluminescence detection (ECL; Amersham) as recommended by the manufacturer.
Immunoprecipitation.
Monolayers of 293T cells grown in six-well plates were transfected with phCMV plasmids expressing E1 and E2 using a PEI ExGen 500-based protocol (Euromedex). At 16 h posttransfection, cells were washed and incubated in Dulbecco's modified medium without l-methionine and l-cysteine (Invitrogen) for 20 min. Transfected cells were pulse labeled for 15 min with 100 μCi per ml of Promix ([35S]methionine-cysteine; Amersham) as described previously (18). Cells were washed twice with medium containing a 10-fold excess of methionine and cysteine, followed by a 4-h chase. Cells were lysed with 1 ml of 0.5% Igepal CA-630 in phosphate-buffered saline (PBS) in the presence of 20 mM iodoacetamide, and immunoprecipitations were carried out as previously described (20). Immunoprecipitates were eluted from protein A-Sepharose beads in 25 μl of 4× nonreducing Laemmli sample buffer by heating for 10 min at 70°C and run on 10% SDS-PAGE gels. After electrophoresis, the gels were treated with sodium salicylate, dried, and exposed to preflashed Hyperfilm-MP (Amersham) at −70°C.
CD81 pull-down assay.
CD81 pull-down experiments were performed as previously described (10). Recombinant fusion proteins containing the large extracellular loop of human CD81 fused to glutathione S-transferase (GST) were preadsorbed onto glutathione-Sepharose 4B beads according to the manufacturer's recommendations (Pharmacia Biotech, Uppsala, Sweden). Precipitates were separated by SDS-PAGE (10% polyacrylamide), followed by Western blotting with anti-E2 (3/11) MAb.
Fusion assays.
The lipid mixing assay was described previously by Lavillette et al. (30). Liposomes were large unilamellar vesicles (100 nm) consisting of phosphatidylcholine and cholesterol (egg yolk phosphatidylcholine, 99% pure, and cholesterol; Sigma). Briefly, R18-labeled liposomes were obtained by mixing R18 (octadecyl rhodamine B chloride; Molecular Probes) and lipids as ethanol and chloroform solutions, respectively (5 mol% R18, final concentration). After the evaporation of solvents under a stream of nitrogen, liposomes were prepared by extrusion over polycarbonate filters in PBS at pH 7.4. Lipid mixing between HCVpp and liposomes was monitored as the dequenching of R18 as a function of time. R18-labeled liposomes (final lipid concentration 15 μM) were added to a 37°C-thermostated cuvette containing HCVpp in PBS, pH 7.4. After temperature equilibration and a decrease in pH to 5, fusion kinetics were recorded on an SLM Aminco 8000 spectrofluorometer over a 30-min time period, with excitation set at 560 nm and emission set at 590 nm. Maximal R18 dequenching was measured after the addition of 0.1% Triton X-100 (final concentration, vol/vol) to the suspension.
RESULTS
Identification of TM residues involved in heterodimerization.
To identify individual residues that participate in E1E2 interactions, a tryptophan replacement scan of these regions was carried out. Tryptophan has a large bulky hydrophobic side chain and so should be tolerated at positions that interact with lipids, but not at positions involved in close protein-protein interactions (6, 51, 52). The TM domains of E1 and E2 are present at the C termini of these proteins (Fig. 1). The N-terminal amino acids of these domains have not been precisely determined, but it has been predicted that they could start at positions 353 and 718 for E1 and E2, respectively (9, 11). A series of mutated proteins were obtained by site-directed mutagenesis in the TM domain of E1 or E2, in the regions previously identified as being involved in heterodimer formation (Fig. 1). The mutations in E2 were introduced in the context of an E1E2 polyprotein. To avoid any potential interference with the processing of E1E2 due to the function of reinitiation of translocation present at the C terminus of E1 (19), mutations in the TM domain of E1 were introduced in the context of a plasmid encoding E1 alone. For these mutants, the assembly of E1E2 was therefore analyzed by cotransfecting cells with two separate plasmids encoding E1 and E2.
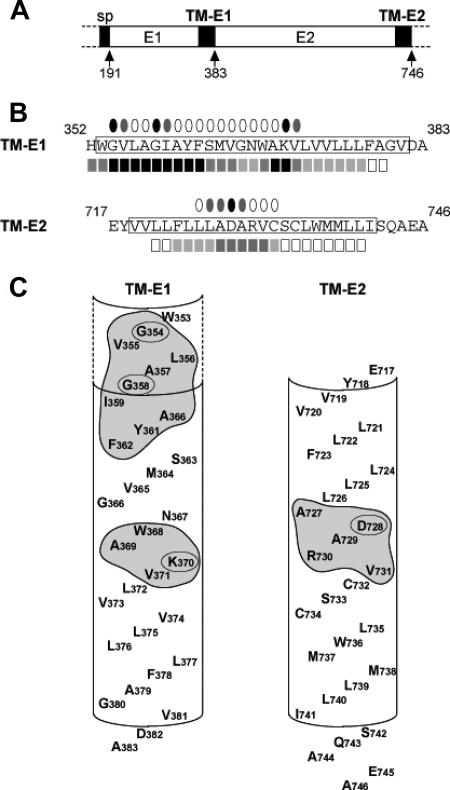
FIG. 1.
Presentation of the TM domains of HCV envelope glycoproteins. (A) Schematic representation of the HCV polyprotein in the region comprising the envelope glycoproteins. The TM domains of E1 and E2 as well as the signal peptide of E1 (sp) are indicated by black boxes. Numbers indicate the positions of the last residues of C, E1, and E2. (B) Summary of the effect of alanine insertion (square symbols) (42) and substitution mutations (this study) (oval symbols) in the TM domains of E1 and E2 on E1E2 heterodimerization. The effects of alanine insertion (square symbols below TM-E1 and TM-E2 sequences) were deduced from Fig. 2 and 3 of an article by Op De Beeck et al. (see reference 42) for TM-E1 and TM-E2, respectively. The intensity of the inhibition of E1E2 heterodimerization was color coded as follows: black, ≥90% inhibition; dark gray, 70 to 90% inhibition; light gray, 30 to 70% inhibition; white squares, <30% inhibition. The mutated residues in this study are indicated by ovals above the TM-E1 and TM-E2 sequences and the intensity of the inhibition of E1E2 heterodimerization is color-coded as follows: black, ≥60% inhibition; dark gray, 30 to 60% inhibition; white ovals, <30% inhibition. The putative TM sequences are boxed. (C) Schematic representation of TM-E1 and TM-E2 as helical structures. The shapes of predicted transmembrane helices are schematically indicated; the dotted section indicates the glycine-rich N-terminal part of TM-E1, which is assumed to be helical (42). The boxed areas underline the main residues deduced to be involved in E1E2 heterodimerization from an alanine insertion study (42). Residues that were shown to be essential for E1E2 heterodimerization by substitution mutations (this study) are highlighted by oval boxes.
The ability of the mutants to form a noncovalent E1E2 complex was analyzed by immunoprecipitation with a conformation-sensitive, E2-specific MAb (H53) that has been shown to specifically precipitate E2 as well as the native E1E2 complex (11, 21). Cells expressing mutated E1E2 polyproteins were pulse labeled for 10 min and chased for 4 h. These conditions have been shown to be appropriate to detect the peak of heterodimer formation (15). To evaluate the level of expression of E1, a control Western blot analysis, with a conformation-insensitive anti-E1 MAb (A4), was performed. E1 expression was found to be constant whatever the tryptophan replacement (data not shown). Since MAb H53 is E2 specific and because H53 epitope can be formed independently of E1 (11), the amount of E1 coprecipitated by MAb H53 is a good indicator of the assembly of the noncovalent heterodimer. To evaluate the percentage of heterodimerization, E1/E2 ratios were measured for each mutant and compared to the ratio obtained with wild-type (WT) proteins.
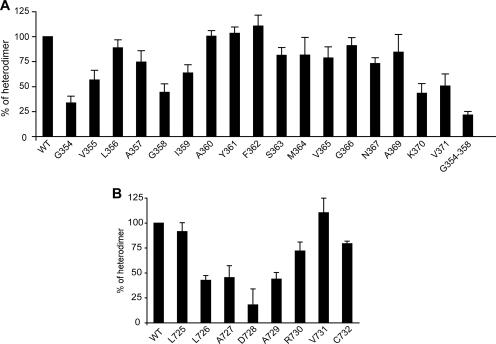
For E1 mutants, a decrease in heterodimer formation was observed when Gly 354, Gly 358, or Lys 370 was replaced by a tryptophan (Fig. 2A). Only ca. 35 to 45% of E1E2 heterodimers were detected for these mutants compared to that for wild-type proteins. In addition, the mutation of the residues located immediately downstream of positions 354, 358, and 370 (V355, I359, and V371, respectively) also had some effect on E1E2 heterodimerization, suggesting that they might also participate in the contact with the TM domain of E2. Indeed, these mutants exhibited 50 to 65% of E1E2 heterodimers relative to wild-type proteins. Interestingly, our structural model predicts that Gly 354 and Gly 358 are located on the same side of the TM domain of E1 (42), suggesting that these residues together would contribute to a continuous region involved in contact with the TM domain of E2. It is worth noting that when both Gly 354 and Gly 358 were mutated, a further reduction in E1E2 heterodimerization was observed (Fig. 2A). Indeed, heterodimerization was reduced to approximately 20% for the double mutant relative to ca. 35 and 45% for the G354 and G358 mutants, respectively. We cannot exclude that the residual heterodimerization is due to interaction between ectodomains (17). The observation that the mutation of the Lys 370 altered heterodimerization is in agreement with previously reported data showing that charged residues within the TM domains of HCV envelope glycoproteins play a role in E1E2 heterodimerization (14). Together, these results indicate that Gly 354, Gly 358, and Lys 370 play major roles in E1E2 heterodimerization.
FIG. 2.
Effect of tryptophan mutagenesis in E1 and E2 on the assembly of the noncovalent E1E2 heterodimer. (A) Effect of tryptophan mutagenesis in E1. 293T cells transfected with a plasmid expressing E2 and a plasmid expressing wild-type or mutated E1 were pulse labeled for 10 min and chased for 4 h. Cell lysates were immunoprecipitated with a conformation-sensitive, E2-specific MAb (H53) which recognizes the noncovalent E1E2 heterodimer. Proteins were separated by SDS-PAGE (10% acrylamide gels) under nonreducing conditions. The intensities of the bands corresponding to E1 and E2 proteins were measured by phosphorimaging for three independent experiments, and the mean percentage of noncovalent complex was calculated as follows for each mutant: (E1/E2 ratio from mutated E2 protein)/(E1/E2 ratio from wild-type proteins). Control Western blot analyses with conformation-insensitive MAbs A4 (anti-E1) and H47 (anti-E2) were performed to confirm that the levels of expression of these proteins were comparable for the different mutants (data not shown). (B) Effect of tryptophan mutagenesis in E2. 293T cells transfected with a plasmid expressing an E1E2 polyprotein, containing or not containing a mutation in E2, were pulse labeled for 10 min and chased for 4 h. Cell lysates were immunoprecipitated and processed as described for panel A. These data represent the mean values of three independent experiments. Error bars indicate standard deviations.
For E2 mutants, a strong reduction in heterodimer formation was observed when Asp 728 was replaced by a tryptophan (Fig. 2B). Indeed, less than 20% of E1E2 heterodimers were detected for this mutant. These data are in agreement with previously reported data showing that Asp 728 plays a major role in E1E2 heterodimerization (7). In addition, mutations of the residues located at positions upstream (726 and 727) and downstream (729) of Asp 728 also led to impaired E1E2 heterodimerization. Approximately 45% of E1E2 heterodimers were detected for these mutants compared to that for wild-type proteins. This suggests that residues close to Asp 728 might also contribute to E1E2 heterodimerization through their interaction with the TM domain of E1.
Altogether, our results point to major roles for Gly 354, Gly 358, and Lys 370 in the TM domain of E1 and for Asp 728 in the TM domain of E2 for E1E2 heterodimerization.
Effect of tryptophan mutagenesis on the infectivity of HCVpp.
A major advance in the investigation of the functions of HCV envelope glycoproteins was the development of HCVpp, consisting of native HCV envelope glycoproteins E1 and E2 assembled onto retroviral core particles (3, 16, 26). Therefore, to further characterize our tryptophan mutants, we analyzed whether these mutations would affect HCV entry in the context of HCVpp. The effects of the mutations on E1E2 heterodimerization and on HCVpp infectivity are summarized in Table 1. In addition, the incorporation of HCV envelope glycoprotein heterodimers into HCVpp was also determined by analyzing the relative level of incorporation of E1 associated with HCVpp compared to that of wild-type glycoproteins (Fig. 3B, 4B, and 5B). Indeed, since E1 needs to interact with E2 to be efficiently incorporated into HCVpp (47), the level of incorporation of E1 into HCVpp, as measured by Western blotting, is representative of the incorporation of E1E2 heterodimers. It is worth noting that slight differences in the migration of some mutated forms of E1 could be observed. A similar observation has been carried out with alanine insertion mutants (42). It is likely due to abnormal migration due to mutations in a hydrophobic domain.
TABLE 1.
Summary of the properties of the mutantsa
| Mutant or WTb | Groupc | Heterodimerization | HCVppd incorporation | Infectivity | Lipid mixing |
|---|---|---|---|---|---|
| WT | + | + | + | + | |
| E1 mutants | |||||
| G354 | II-b | +/− | +/− | + | NT |
| V355 | II-b | +/− | + | + | NT |
| L356 | III-a | + | + | +/− | +/− |
| A357 | I-a | + | + | ++ | ++ |
| G358 | IV-b | +/− | +/− | − | NT |
| I359 | III-b | +/− | +/− | +/− | NT |
| A360 | III-a | + | + | +/− | − |
| Y361 | III-a | + | + | +/− | NT |
| F362 | III-a | + | ++ | +/− | NT |
| S363 | IV-a | + | ++ | − | NT |
| M364 | IV-a | + | ++ | − | NT |
| V365 | I-a | + | + | ++ | NT |
| G366 | IV-a | + | + | − | NT |
| N367 | IV-a | + | + | − | NT |
| A369 | IV-a | + | + | − | NT |
| K370 | IV-b | +/− | +/− | − | NT |
| V371 | IV-b | +/− | +/− | − | NT |
| G354/358 | IV-b | − | − | − | NT |
| E2 mutants | |||||
| L725 | III-a | + | + | +/− | +/− |
| L726 | IV-b | +/− | +/− | − | +/− |
| A727 | IV-b | +/− | +/− | − | NT |
| D728 | IV-b | +/− | − | − | NT |
| A729 | IV-b | +/− | − | − | NT |
| R730 | III-a | + | +/− | +/− | +/− |
| V731 | III-a | + | + | +/− | NT |
| C732 | III-a | + | +/− | +/− | NT |
−, less than 10%; +/−, between 10% and 60%; +, between 60% and 130%; ++, more than 130%; NT, not tested.
Mutants for which both infectivity and heterodimerization were affected are shown in bold.
Group I, infectivity more than 130%; group II, infectivity between 60 and 130%; group III, infectivity between 10 and 60%; group IV, infectivity less than 10%; a, heterodimerization not affected; b, heterodimerization affected.
The incorporation of HCV envelope glycoprotein heterodimers into HCVpp was determined by Western blotting by analyzing the relative level of incorporation of E1 into HCVpp compared to wild-type glycoproteins.
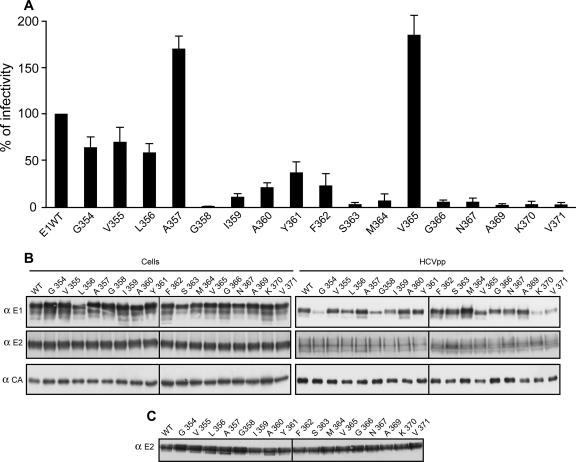
FIG. 3.
Effect of tryptophan mutagenesis in E1 on the infectivity of HCVpp. HCVpp were generated in 293T with a plasmid expressing E2 and a plasmid expressing wild-type or mutated E1. (A) Infectivity of HCVpp. Infection assays with the luciferase reporter gene were performed using target Huh-7 human hepatoma cells. Similar inputs of viral particles were used in each experiment, and this was confirmed by comparing the amounts of capsid protein incorporated into HCVpp. The results are expressed as percentages of infectivity. For each mutant, the percentage of infectivity was calculated as the luciferase activity of HCVpp produced with mutant glycoproteins divided by the luciferase activity of HCVpp produced with wild-type E1 and E2 (WT). Results are reported as the means ± standard deviations (error bars) of five independent experiments. Pseudotyped particles produced in the absence of envelope proteins were used as controls. The mean luciferase activity of such particles represented less than 0.5% of the activity measured for HCVpp (data not shown). (B) Incorporation of HCV envelope proteins into HCVpp. Particles were pelleted through a 20% sucrose cushion and analyzed by Western blotting. HCV glycoproteins and the capsid protein of MLV were revealed with specific MAbs anti (α)-E1 (A4), anti-E2 (H47), and anti-MLV capsid (CA). (C) Interaction of HCV envelope glycoproteins with human CD81LEL. HCVpp pelleted through a 20% sucrose cushion were lysed and analyzed in a CD81LEL-GST pull-down assay by using human CD81LEL fused to glutathione S-transferase. The presence of E2 glycoprotein in the precipitates was revealed by Western blotting with an anti-E2 MAb (3/11).
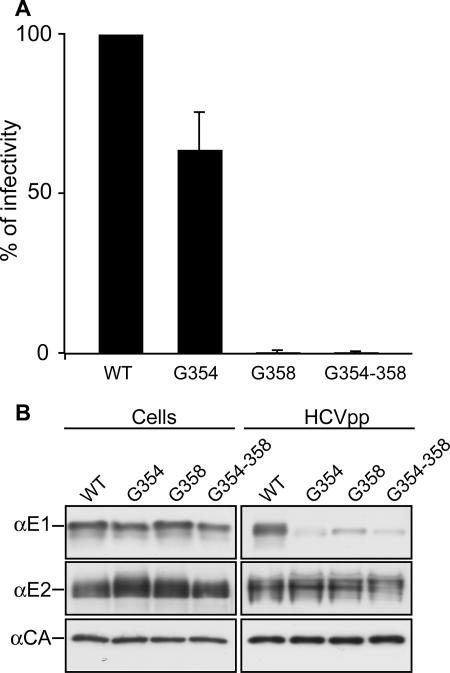
FIG. 4.
Effect of tryptophan mutagenesis of Gly 354 and Gly 358 on the infectivity of HCVpp. The mutants were expressed and processed as described in the legend for Fig. 3. α, anti.
FIG. 5.
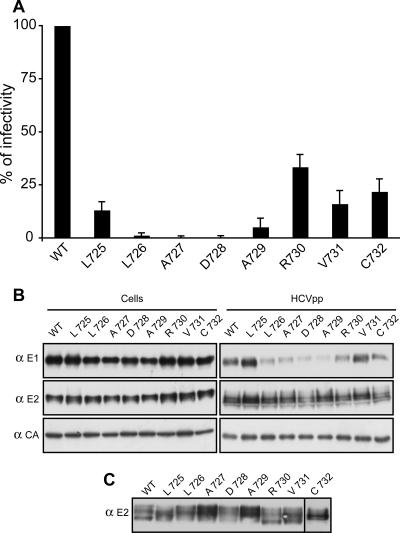
Effect of mutagenesis in E2 on the infectivity of HCVpp.HCVpp were generated in 293T with a plasmid expressing E1E2 polyprotein, containing or not containing a mutation in E2. The mutants were expressed and processed as described in the legend for Fig. 3. α, anti.
The characterization of E1 mutants allowed the definition of four infectivity phenotypes (Fig. 3 and Table 1). The first group (group I) contained mutants that were more infectious than was the wild type (A357 and V365). In the second group (group II), infectivity of the mutants was close to that of the wild type (mutants G354 and V355). The third group (group III) contained mutants that were still infectious, but their infectivity was reduced to less than 60% of that of the wild type (mutants L356, I359, A360, Y361, and F362). Finally, the fourth group (group IV) contained mutants that had almost totally lost their infectivity (mutants G358, S363, M364, G366, N367, A369, K370, and V371). These four groups were divided into two subgroups (subgroups a and b) depending on the effect the mutations had on heterodimerization. Mutants of subgroup a had normal levels of heterodimerization, whereas mutants of subgroup b showed reductions in heterodimerization.
Interestingly, the alteration in HCVpp infectivity observed in groups III-b and IV-b correlated with lower levels of incorporation of E1E2 heterodimers into HCVpp, which seems to be due to a decrease in heterodimerization. Indeed, it has been shown that E1 needs to interact with E2 for an efficient incorporation of E1E2 heterodimer into HCVpp (47). Importantly, the alteration in HCVpp infectivity observed in groups III-a and IV-a was not due to a problem of heterodimerization or incorporation into HCVpp, suggesting that these mutations had a direct effect on the entry function of HCV envelope glycoproteins. For mutants of group I, a slight increase in the incorporation of E1 into HCVpp was detected for mutant A357 (Fig. 3B). In addition, for mutant V365, a slight reduction in the level of particle production was observed, suggesting that after normalization, HCVpp should contain slightly more E1E2 heterodimers for this mutant (Fig. 3B). Whether this small increase in incorporation of HCV envelope glycoproteins into HCVpp is responsible for the increase in infectivity is not clear. Alternatively, these mutations might have a direct effect on the entry functions of HCV envelope glycoproteins. Surprisingly, mutants of group II-b displayed almost normal infectivity, despite some alteration in heterodimerization (Table 1). For such mutants, we cannot exclude the possibility that the reduction in heterodimerization is compensated by an increased efficiency of the entry function of HCV envelope glycoproteins, as was suggested for a mutation affecting glycosylation site N7 of E2 (25).
Despite the involvement of Gly 354 in E1E2 heterodimerization (Fig. 2), its replacement by a tryptophan had only a mild effect on pseudoparticle infectivity (Fig. 3A and Table 1). As discussed above, when both Gly 354 and Gly 358 were mutated, a higher reduction in E1E2 heterodimerization was observed (Fig. 2A). Therefore, we also analyzed the infectivity of pseudoparticles generated for this double mutant. As observed for the single mutant G358, the G354-G358 double mutant was no longer infectious (Fig. 4), indicating that the G358 mutation had a dominant effect on HCVpp infectivity.
E2 mutants could be classified into two groups (groups III-a and IV-b) (Table 1). As for E1, group III contained mutants that were still infectious, but their infectivity was reduced to less than 60% of that of the wild type (mutants L725, R730, V731, and C732) and group IV contained mutants that had almost totally lost their infectivity (mutants L726, A727, D728, and A729) (Fig. 5). The alteration in HCVpp infectivity observed in mutants of group IV-b correlated with a lower level of incorporation of E1E2 heterodimers into HCVpp, which seems to be due to a decrease in heterodimerization (Table 1). As for E1 mutants, the alteration in HCVpp infectivity observed in group III-a was not due to a defect in heterodimerization (Table 1), suggesting that some of these mutations had a direct effect on the entry function of HCV envelope glycoproteins.
Altogether, our data indicate that the TM domains of HCV envelope glycoproteins play a pivotal role in the entry functions of these proteins.
Effect of tryptophan mutagenesis on the fusion properties of HCV envelope glycoproteins.
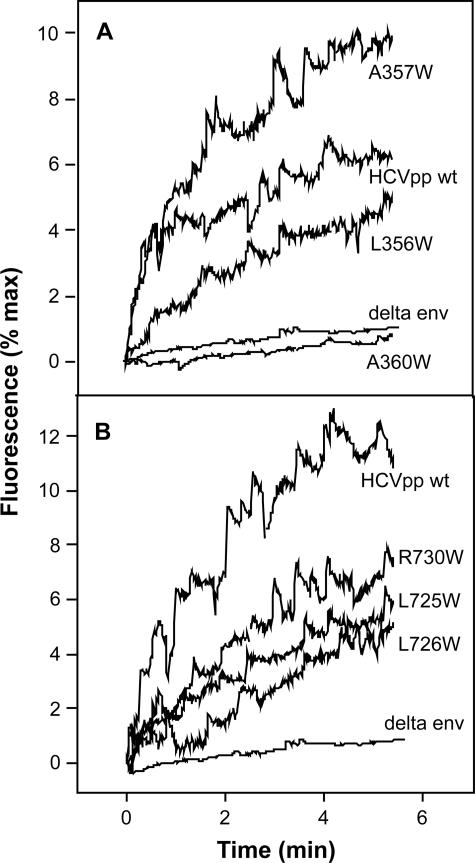
For some mutants, HCVpp infectivity was strongly reduced without displaying any alteration in the assembly of the E1E2 heterodimer. Therefore, we suspected that these mutants would be affected in their binding or fusion properties. The effect of the mutations on the binding properties of HCV envelope glycoproteins is not easy to measure in the context of HCVpp because the initial binding of retroviral particles to cells does not require specific interaction between a viral envelope protein and a receptor (44). However, the interaction between HCV envelope glycoproteins and CD81, a molecule involved in the early steps of HCV entry, can be tested. The binding of mutant HCVpp was analyzed in a GST pull-down assay using GST-CD81-LEL and was found not to be altered for all mutants tested (Fig. 3C and 5C). The fusion activity of some mutants was then tested in the recently developed in vitro fusion assay using fluorescently labeled liposomes and HCVpp (30). For E1, we selected mutants L356, A357, and A360 displaying normal or close to normal E1E2 heterodimerization (Fig. 2A) and normal incorporation of E1 and E2 onto HCVpp (Fig. 3B). For E2, the mutants chosen were L725, L726, and R730 displaying either reduced (L726) or close to normal heterodimer content (L725 and R730) (Fig. 2B) and reduced (L726 and R730) or normal (L725) HCVpp assembly properties (Fig. 5B).
As clearly shown in Fig. 6A, mutations in the TM domain of E1 that altered HCVpp infectivity affected HCVpp-mediated lipid mixing as well. Indeed, mutants L356 and A360 displayed low and negligible fusogenicity, respectively, compared to the fusion of wild-type HCVpp. The fusion properties of mutant A360 were comparable to those observed for our negative control for fusion, the Delta env pseudoparticles devoid of any envelope glycoproteins. Interestingly, mutants L356 and A360 were found to be poorly infectious in our cell infection assays (Fig. 3). The absence of fusogenicity of mutant A360, despite some residual infectivity in HCVpp, suggests that the presence of receptors on the cell surface might slightly help the fusogenicity of HCV envelope glycoproteins. Conversely, mutant A357, displaying a 1.5-fold increase in HCVpp infectivity compared to that of wild-type pseudoparticles (Fig. 3), exhibited a dramatic increase in HCVpp fusogenicity (Fig. 6A), both in terms of initial rate and final extent of lipid mixing. This observation suggests that the increase in HCVpp infectivity observed for mutant A357 is likely due to a better fusogenicity of the envelope glycoproteins. As for E1 mutants, a similar parallel evolution between infectivity and fusion held true for mutants in the TM of E2 as well (Fig. 6B). However, residual fusion activity was still observed for HCVpp bearing the L726 mutation in E2 in spite of a complete loss of infectivity (Fig. 5).
FIG. 6.
Effect of tryptophan mutagenesis on the fusion properties of HCV envelope glycoproteins. Wild-type (HCVpp wt) or mutant HCVpp fusion capacities were assayed by lipid mixing. Forty microliters of pseudoparticles were added to R18-labeled liposomes (final lipid concentration, 15 μM) in PBS buffer, pH 7.4, and after a 2-min equilibration at 37°C, fusion was initiated by decreasing the pH to 5 in the cuvette (time zero of the fusion kinetics; λexc 560 nm and λem 590 nm). Results are expressed as percentages of maximal fluorescence, obtained by the addition of Triton X-100 (final concentration, 0.1% vol/vol) to the pseudoparticle-liposome suspension. (A) Mutants in the TM domain of E1. (B) Mutants in the TM domain of E2. “Delta env” denotes pseudoparticles devoid of any envelope glycoprotein, used as a negative control for fusion. For both panels, the most representative curves are presented, and experiments were repeated four times.
Since the fusion assay is based on the use of plain lipid vesicles, which do not contain cell surface receptors for HCV, our results indicate that the mutations that reduced HCVpp infectivity without affecting E1E2 heterodimerization alter the fusion properties of HCV envelope glycoproteins.
DISCUSSION
The TM domains of HCV envelope glycoproteins are multifunctional. Indeed, besides their role as membrane anchor, they possess a signal of reinitiation of translocation in their C-terminal half and they are responsible for the ER retention of E1E2 heterodimer (19). In addition, even if they are probably not the only determinants for oligomerization (17), they are essential in E1E2 heterodimerization (42). Here, we identified individual residues that participate in E1E2 interactions. Our mutagenesis study identified Gly 354, Gly 358, Lys 370, and Asp 728 as residues playing a role in E1E2 heterodimerization. Our tryptophan mutants were also analyzed for their effects on the entry functions of HCV envelope glycoproteins. Importantly, HCVpp infectivity was strongly affected for many mutants, despite normal E1E2 heterodimerization and normal levels of incorporation of HCV envelope glycoproteins into the pseudotyped particles. In addition, some mutations were shown to affect the fusion functions of HCV envelope glycoproteins. In conclusion, this mutational analysis identified residues involved in E1E2 heterodimerization and revealed that the TM domains of HCV envelope glycoproteins play a major role in the fusion properties of these proteins.
Alanine scanning insertion mutagenesis (6, 38) within the TM domains of HCV envelope glycoproteins has previously identified the central regions of these domains as well as the N-terminal part of the TM domain of E1 as sequences involved in heterodimerization (42). This technique has been shown to be a powerful method to detect the dimerization of TM α-helices. Indeed, the insertion of a single amino acid into a TM helix displaces the residues on the N-terminal side of the insertion by 110° relative to those on the C-terminal side of the insertion, disrupting a helix-helix packing interface involving residues on both sides of the insertion. Two distinct segments of the TM domain of E1 and one of the TM domain of E2 were identified by this technique. Although alanine scanning insertion mutagenesis allowed the identification of segments involved in E1E2 heterodimerization through their TM domains, the very residues implicated in E1E2 interactions were not identified. Here, by tryptophan scanning mutagenesis, we identified at least four residues that are important for E1E2 heterodimerization: Gly 354, Gly 358, Lys 370, and Asp 728.
Importantly, the Gly 354 and Gly 358 residues belong to a GXXXG motif. The GXXXG motif has a strong propensity for TM helix interactions (46), as shown, for instance, in glycophorin A (32). Sequence analyses show that the GXXXG motif or GXXXG-like motifs, in which one or both glycine residues are replaced by other small residues, such as alanine or serine, occur more frequently in TM helices than their random expectation (1, 50). Glycine and, to a lesser extent, alanine and serine residues allow very close contact between TM helices. This proximity permits extensive interhelical van der Waals interactions (35). It has to be noted that additional sequence constraints as found in glycine zippers might also be necessary for the oligomerization mediated by the GXXXG motif (28, 37, 48). Although the GXXXG motif has been observed mainly in homooligomerization, its involvement in heterodimerization has also been suggested in some cases (49). In our study of HCV envelope glycoproteins, we confirm that this motif can be involved in heterodimerization.
The Lys 370 and Asp 728 residues have previously been shown to play a major role in the function of HCV envelope glycoproteins. Indeed, when Lys 370 was mutated together with Asn 368, the ER retention function of the TM domain of E1, as well as heterodimerization, was abolished (14). Similarly, when Asp 728 was mutated together with Arg 730, the ER retention function of the TM domain of E2 and heterodimerization were abolished (14). These mutations in E1 and E2 also affected the signal of reinitiation of translocation present in the C-terminal half of these TM domains. More recently, the mutagenesis in the TM domain of E2 has indicated that of the two charged residues of this domain, Asp 728 plays a major role in E1E2 heterodimerization (7). These observations as well as our tryptophan scanning mutagenesis indicate that, together with the GXXXG motif found in E1, the Lys 370 and Asp 728 residues play a major role in E1E2 heterodimerization. Although the presence of a positive and a negative charge in the TM domain of E1 and E2, respectively, would suggest that heterodimerization might be mediated by an ion pair, this hypothesis has not been confirmed experimentally (7). Due to their peculiar features, a high-resolution structure of the TM domains of HCV envelope glycoproteins in a hydrophobic environment will be necessary to better understand their organization within the membrane.
Some mutations affect the fusion function of HCV envelope glycoproteins without altering E1E2 heterodimerization and incorporation of HCV envelope glycoproteins into HCVpp (e.g., A360 and L725). The TM domains of fusion proteins are essential for their fusion activities. For instance, the hemagglutinin of influenza virus in which the TM and cytosolic domains have been replaced by a glycerylphosphatidylinositol moiety has been shown to lose the ability to mediate complete fusion but retain the ability to promote hemifusion (27, 36). Similarly, alterations in the TM domains of other fusion proteins led to a loss of fusion activity (8, 40, 45, 57). The TM domains of some fusion proteins have been shown to function at a late stage of the fusion process, subsequent to the formation of a hemifusion diaphragm (8, 27, 36). The rearrangement of this unstable reaction intermediate into a fusion pore constitutes a major late step in fusion, and this is likely to be potentiated by the TM domains. However, in the case of HCV envelope glycoproteins, the mutants were blocked at the lipid mixing stage, indicating that hemifusion was also blocked and that the mutations are altering an early step in the fusion process.
Although there is some controversy about the identity of the HCV fusion protein, HCV envelope glycoproteins are supposed to belong to class II fusion proteins based on the fact that HCV belongs to the Flaviviridae family (55). Since class II envelope proteins undergo rearrangements in their oligomeric structures at the budding and entry steps, it is likely that the mutant proteins are blocked in the organization and/or reorganization of their oligomeric form. Alternatively, we cannot exclude the possibility that anomalous or uncoordinated protein reorganization takes place, as observed for mutants of the TM domains of the Semliki Forest virus glycoproteins, because Semliki Forest virus also contains a class II fusion protein (53). Unfortunately, due to the high heterogeneity of the envelope glycoproteins associated with HCVpp (22), it is currently not possible to obtain biochemical information on HCV glycoprotein reorganization during the fusion process. In addition, we do not know the density of functional HCV glycoproteins on pseudoparticles that are required for infectivity.
In conclusion, our study shows that the GXXXG motif in the TM domain of E1, together with Lys 370 and Asp 728 residues, is involved in E1E2 heterodimerization. In addition, it also reveals that the TM domains of HCV envelope glycoproteins play a major role in the fusion properties of these proteins. Together, these data underline the essential role played by the TM domains of HCV envelope glycoproteins at the level of particle assembly and virus entry.
Acknowledgments
We thank Yves Rouillé for critical reading of the manuscript and Jennifer Molle, Sophie Desnoulez, André Pillez, and Sophana Ung for their technical assistance. We are grateful to J. McKeating, B. Bartosch, and F. L. Cosset for providing us with reagents. Fluorescence spectroscopy was performed by EIP on the platform “Physico-Chemical Analysis of Proteins” (PAP) at the Institut de Biologie et Chimie des Protéines (IBCP).
This work was supported by the Agence Nationale de Recherche sur le Sida et les Hépatites Virales (ANRS). J.D. is an international scholar of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Arkin, I. T., and A. T. Brunger. 1998. Statistical analysis of predicted transmembrane α-helices. Biochim. Biophys. Acta. 1429:113-128. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, A. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., New York, NY.
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C pseudo-particles containing functional E1E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, P., B. Persson, R. Kaback, and G. von Heijne. 1997. Alanine insertion scanning mutagenesis of lactose permease transmembrane helices. J. Biol. Chem. 272:29566-29571. [DOI] [PubMed] [Google Scholar]
- 7.Ciczora, Y., N. Callens, C. Montpellier, B. Bartosch, F. L. Cosset, A. Op De Beeck, and J. Dubuisson. 2005. Contribution of the charged residues of HCV glycoprotein E2 transmembrane domain to the functions of E1E2 heterodimer. J. Gen. Virol. 86:2793-2798. [DOI] [PubMed] [Google Scholar]
- 8.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocquerel, L., S. Duvet, J.-C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., C.-C. Kuo, J. Dubuisson, and S. Levy. 2003. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 77:10677-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocquerel, L., A. Op De Beeck, M. Lambot, J. Roussel, D. Delgrange, A. Pillez, C. Wychowski, F. Penin, and J. Dubuisson. 2002. Topologic changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerel, L., C. Voisset, and J. Dubuisson. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075-1085. [DOI] [PubMed] [Google Scholar]
- 14.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 17.Drummer, H. E., and P. Poumbourios. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066-30072. [DOI] [PubMed] [Google Scholar]
- 18.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 20.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 22.Flint, M., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fournillier Jacob, A., A. Cahour, N. Escriou, M. Girard, and C. Wychowski. 1996. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J. Gen. Virol. 77:1055-1064. [DOI] [PubMed] [Google Scholar]
- 25.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F. L. Cosset, C. Montpellier-Pala, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S., T. J. Jeon, A. Oberai, D. Yang, J. J. Schmidt, and J. U. Bowie. 2005. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. USA 102:14278-14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, M., M. C. Bennett, T. Bercot, and I. R. Singh. 2006. Functional analysis of hepatitis C virus envelope proteins, using a cell-cell fusion assay. J. Virol. 80:1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavillette, D., B. Bartosch, D. Nourrisson, G. Verney, F. L. Cosset, F. Penin, and E. I. Pecheur. 2006. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J. Biol. Chem. 281:3909-3917. [DOI] [PubMed] [Google Scholar]
- 31.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of hepatitis C virus of major genotypes and subtypes. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 32.Lemmon, M. A., J. M. Flanagan, H. R. Treutlein, J. Zhang, and D. M. Engelman. 1992. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry 31:12719-12725. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 35.MacKenzie, K. R., and D. M. Engelman. 1998. Structure-based prediction of the stability of transmembrane helix-helix interactions: the sequence dependence of glycophorin A dimerization. Proc. Natl. Acad. Sci. USA 95:3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melnyk, R. A., S. Kim, A. R. Curran, D. M. Engelman, J. U. Bowie, and C. M. Deber. 2004. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J. Biol. Chem. 279:16591-16597. [DOI] [PubMed] [Google Scholar]
- 38.Mingarro, I., P. Whitley, M. Lemmon, and G. von Heijne. 1996. Ala-insertion scanning mutagenesis of the glycophorin A transmembrane helix: a rapid way to map helix-helix interactions in integral membrane protein. Prot. Sci. 5:1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 40.Odell, D., E. Wanas, J. Yan, and H. P. Ghosh. 1997. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J. Virol. 71:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 42.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. Role of the transmembrane domains of hepatitis C virus envelope proteins E1 and E2 in the assembly of the noncovalent E1E2 heterodimer. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 43.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 73:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragheb, J. A., and W. F. Anderson. 1994. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J. Virol. 68:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 47.Sandrin, V., P. Boulanger, F. Penin, C. Granier, F. L. Cosset, and B. Bartosch. 2005. Assembly of functional hepatitis C virus glycoproteins on infectious pseudoparticles occurs intracellularly and requires concomitant incorporation of E1 and E2 glycoproteins. J. Gen. Virol. 86:3189-3199. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, D., and D. M. Engelman. 2004. Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 343:799-804. [DOI] [PubMed] [Google Scholar]
- 49.Senes, A., D. E. Engel, and W. F. DeGrado. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14:465-479. [DOI] [PubMed] [Google Scholar]
- 50.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol. 296:921-936. [DOI] [PubMed] [Google Scholar]
- 51.Sharp, L. L., J. Zhou, and D. F. Blair. 1995. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc. Natl. Acad. Sci. USA 92:7946-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp, L. L., J. Zhou, and D. F. Blair. 1995. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry 34:9166-9171. [DOI] [PubMed] [Google Scholar]
- 53.Sjöberg, M., and H. Garoff. 2003. Interactions between the transmembrane segments of the alphavirus E1 and E2 proteins play a role in virus budding and fusion. J. Virol. 77:3441-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voisset, C., and J. Dubuisson. 2004. Functional hepatitis C virus envelope glycoproteins. Biol. Cell 96:413-420. [DOI] [PubMed] [Google Scholar]
- 56.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss, C. D., and J. M. White. 1993. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 67:7060-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]