Abstract
Alternative splicing has been recognized as a major mechanism for creating proteomic diversity from a limited number of genes. However, not all determinants regulating this process have been characterized. Using subviral human immunodeficiency virus (HIV) env constructs we observed an enhanced splicing of the RNA when expression was under control of the cytomegalovirus (CMV) promoter instead of the HIV long terminal repeat (LTR). We extended these observations to LTR- or CMV-driven murine leukemia proviruses, suggesting that retroviral LTRs are adapted to inefficient alternative splicing at most sites in order to maintain balanced gene expression.
The human proteome exceeds 100,000 isoforms, whereas the number of genes is significantly lower (30,000; reviewed in reference 18). Alternative splicing has been recognized as playing a major role in proteomic diversity related to the ability to generate several different mRNAs from one primary transcript (13). The same applies to retroviruses. Due to their genomic organization only one polycistronic transcript is made, and this encodes up to nine open reading frames (ORFs) in the case of human immunodeficiency virus (HIV) (6). Alternative splicing ensures regulated expression of several of these gene products (20), and mutations that disturb the balance of alternatively spliced transcripts result in severe attenuation (3, 16). For all retroviruses alternative splicing is regulated via the interplay of cis-acting sequences on the RNA and cellular splicing factors (7, 17, 25). This regulation involves the presence of both exonic and intronic splicing silencers as well as enhancers. Whereas HIV modulates mostly its 3′ splice sites (ss) (10), murine leukemia virus (MLV) uses sequences upstream of the 5′ ss to regulate alternative splicing (14). Transcription and 3′-end processing are closely connected to splicing (12), thus adding one more level to the complex regulation of gene expression. In this report we show that replacing the retroviral long terminal repeat (LTR) with the cytomegalovirus (CMV) promoter shifts the balance of alternatively spliced transcripts, resulting in higher levels of spliced RNA.
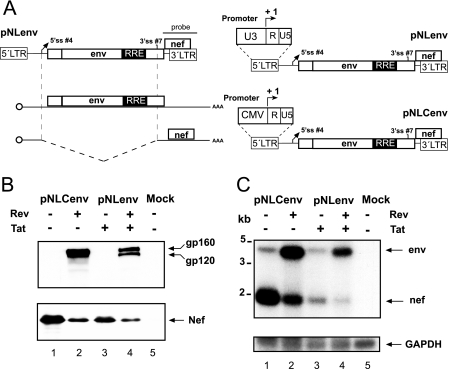
In order to study the effects of promoters on alternative splicing in retroviruses we used the previously described NLenv system (Fig. 1A) (2). This system is based on the HIV-1 proviral clone NL4-3 and generates an mRNA which is identical in sequence to the wild-type env mRNA by removing the sequence between the major 5′ ss and the env 3′ ss and restoring the natural exon junction. This RNA can undergo one splicing event, resulting in the nef mRNA (Fig. 1A, left panel). We exchanged the HIV U3 region with the CMV immediate-early promoter, leaving the transcriptional start site unchanged (Fig. 1A, right panel). Transfection of these constructs into HelaP4 cells and Western blot analysis showed Rev-dependent Env expression and Rev-independent Nef expression, as expected (Fig. 1B, lanes 1 and 2 and lanes 3 and 4). Northern blot analysis of total RNA probed with a 3′ LTR probe detected the unspliced transcript coding for env and the spliced RNA coding for nef (Fig. 1C, lane 3). The addition of Rev shifted the ratio towards unspliced RNA due to its nuclear export and translation, leading to stabilization of the RNA as an indirect consequence (Fig. 1C, lane 4) (2). Replacing the U3 region with the CMV promoter led to enhanced splicing of the primary transcript (Fig. 1C, lanes 1 and 3), although the sequences of the two RNAs are identical and differ only in the nontranscribed promoter region. Interestingly, the CMV promoter seems to function Tat independently in contrast to that of the viral LTR (Fig. 1C, lanes 1 and 2 and lanes 3 and 4), as reported previously (4, 22).
FIG. 1.
The CMV promoter enhanced splicing of a subviral HIV env RNA. (A) Schematic drawing of the NLenv system. The sequence between the major 5′ ss and the env 3′ ss was removed from the proviral clone NL4-3 by cloning, thereby mimicking the natural exon ligation. The plasmid contains both LTRs, the Env and the Nef ORF, and the Rev-responsive element (RRE). The remaining 5′ ss #4 can splice to 3′ ss #7 to generate the nef RNA. The right panel illustrates the promoter exchange from the viral LTR to the CMV promoter in the NLenv context. The probe used for detection in Northern blot analysis is indicated on the left panel. (B) Western blot analysis of HeLa P4 cell lysates corresponding to 20 μg of protein from transfections performed using the indicated constructs and a combination of anti-Env and anti-Nef antibodies. Cells were transfected with 10 μg of plasmid plus 5 μg of Tat- or Rev-encoding plasmid as indicated and collected 48 h after transfection. The Env products (gp160 and gp120) and the Nef protein are marked with arrows. (C) Northern blot using total RNA from the same transfection as described for panel B. RNA was separated on a denaturing agarose gel, transferred to nylon membrane, and probed with a labeled cDNA fragment as indicated in panel A. The left side shows molecular size markers, and the RNAs are marked on the right. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a loading control.
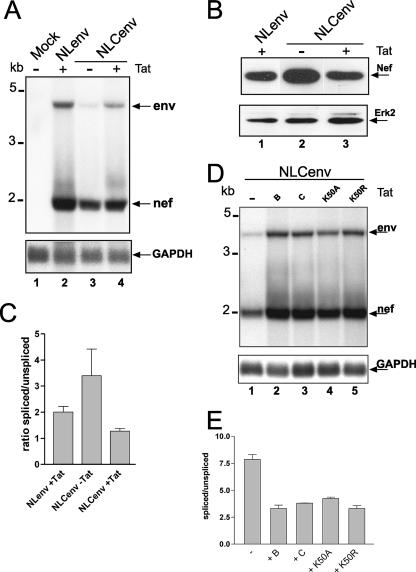
Since the transactivation by Tat is the major difference between the two promoters we looked at CMV transfections in the presence or absence of Tat. Figure 2A reveals that cotransfection of Tat led to the wild-type splicing pattern (lanes 2 and 4). To obtain transcript levels that were more comparable, the amount of NLCenv plasmid was reduced from 10 to 2 μg per 10-cm dish. Still, the CMV promoter was upregulated between two- and fourfold by Tat (Fig. 2A, lanes 3 and 4, and 2D, lanes 1 and 2) in agreement with previous findings (4, 22). Since the efficiency of splicing correlates with the amount of nef mRNA and Nef protein, we did Western blot analysis and detected elevated levels of Nef protein in the case of NLCenv compared to that for the wild-type construct and reduced levels upon Tat cotransfection (Fig. 2B). Quantification of the Northern blot data (Fig. 2A) by phosphorimager analysis again illustrated a role for Tat in alternative splicing, namely, that Tat shifts the ratio of spliced versus unspliced RNA back towards wild-type levels (Fig. 2C).
FIG. 2.
Cotransfection of Tat led to reversal of enhanced splicing. (A) LTR- or CMV-containing NLenv plasmids were transfected into HeLa P4 cells in the presence or absence of an HIV-Tat-encoding plasmid. Northern blot analysis was performed as described for Fig. 1C. GAPDH serves as a loading control. Molecular size markers are shown on the left, and the RNAs are named on the right. (B) Western blot analysis of cell lysates from the transfection performed as described for panel A. An anti-Nef antibody was used, and the blot was reprobed with an anti-extracellular signal-regulated kinase 2 (Erk2) antibody for a loading control. (C) Quantification of the Northern blot data using a PhosphorImager (Amersham). Mean values represent the results of four independent experiments. (D) The NLCenv construct was transfected together with different Tat plasmids as indicated or left untransfected (−). + B and + C specify the HIV Tat subtypes and K50A and K50R point mutations based on subtype C Tat. GAPDH served as a loading control. Molecular markers and RNA species are indicated as described for panel A. (E) Quantification of the Northern blot data as described for panel C.
We extended these observations to a complete proviral HIV clone (NL4-3) driven by the CMV promoter. Here, reduced infectivity (2.4-fold) is measurable (data not shown), but this clone can still produce Tat. Chang and Zhang looked at RNA levels of Tat minus proviral clones driven by hybrid promoters and found only slight difference in RNA levels (5), but the promoter construct differed from the ones reported here. Effects of Tat on alternative splicing were described in a recent report by Berro and colleagues, who established an interaction of acetylated Tat and the splicing inhibitor p32 leading to more unspliced RNA (1). Alternatively, it has also been shown that p32 can bind Rev (15) and that this interaction leads to rescue of excessive splicing in murine cells (26). We tried to test the Tat hypothesis in our system by using Tat mutants that have little effect on transactivation and should still exert an effect on splicing (1, 19). The mutants are based on subtype C Tat, whereas we used subtype B Tat (Fig. 2D). Transfection of NLCenv and different Tat mutants revealed that all of them lead to more unspliced RNA in comparison to the CMV promoter results in the absence of Tat (Fig. 2D and E). In particular, the K50A mutant showed no defect in transactivation compared to the parental construct (Fig. 2D, lanes 3 and 4) and only a very minor effect on splicing. As a control the K50R mutant behaved exactly like wild-type Tat (Fig. 2D, lanes 3 and 5).
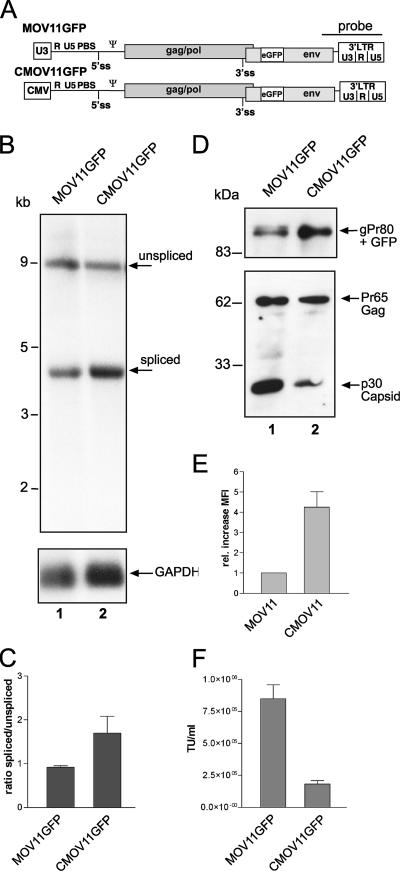
In order to extend our observations to other retroviruses, we cloned a CMV-driven murine leukemia virus (Fig. 3A). The construct is based on the MLV clone MOVGFP, which contains eGFP in the proline-rich region of the Env ORF (11). We exchanged the promoter-leader region to the CMV promoter and sequences from a retroviral vector (SCS11) (24). As a result both plasmids produced RNAs with identical sequences. Transfection into 293T cells revealed enhanced splicing when transcription was directed via the CMV (Fig. 3B, lanes 1 and 2). Whereas the parental MLV clone showed an equal ratio of unspliced and spliced RNAs, the CMV-driven construct showed an almost twofold enhancement of splicing (Fig. 3C). To evaluate this effect on the protein level, we probed cell lysates with anticapsid or anti-green fluorescent protein (GFP) antibodies (Fig. 2D), since GFP is encoded within the Env ORF (Fig. 3A). CMOV11GFP displayed less Gag expression and enhanced Env expression in comparison to the wild-type MLV (Fig. 3D) in agreement with the Northern blot data (Fig. 3B). This effect can also be measured via flow cytometry detecting the GFP. Here, a fourfold increase in the mean fluorescence intensity in the case of CMOV11GFP also indicates enhanced splicing, leading to more of the GFP Env fusion protein (Fig. 3E). To determine whether this change in gene expression has any impact on viral titer and therefore more biological relevance, we took supernatants of transient transfections and determined their titers on murine fibroblasts by use of the GFP as a reporter. The enhanced Env expression and reduced Gag expression led to a significant drop in titer, showing that a proper balance of Gag and Env indeed determines infectivity (Fig. 3F). It will be interesting to look for revertants in an MLV construct that carries the CMV in both LTRs.
FIG. 3.
The CMV promoter enhanced splicing in murine leukemia provirus. (A) Depiction of the MLV constructs used. Drawings show the genomic organization marking the gag-pol gene and the env gene. The eGFP coding sequence was inserted in frame into the proline-rich region of Env. The 3′ LTR is shown as well as the 5′ LTR. Here, only the U3 region is boxed to illustrate the exchange to the CMV promoter. The 5′ and 3′ ss are indicated. (B) Northern blot analysis of 293T transfected with 10 μg of the indicated proviral plasmids. The cells were harvested 48 h posttransfection and analyzed as described for Fig. 1C using an env/3′ LTR probe as shown. GAPDH served as a loading control. Molecular size markers are indicated on the left, and the RNA species are named on the right. (C) Quantification of the Northern blot data using a PhosphorImager. Mean values represent the results of three independent experiments. (D) Western blot analysis of transfected 293T cell lysates performed using an anti-GFP antibody and an anti-p30 antibody. Molecular size markers are indicated on the left, and the protein products are named on the right. (E) Enhanced GFP (eGFP) (Env expression) was measured using flow cytometry. The means for eGFP expression were calculated as a ratio to the parental construct MOV11GFP set to 1. Standard deviations represent the results of five independent experiments. (F) Titer analysis of the MLV constructs. Supernatants of transfected 293T cells were taken, and titers were determined according to their eGFP expression on murine fibroblasts. The titer is given in transducing units (TU) per milliliter of supernatant, and standard deviation values represent the results of three independent experiments.
Effects of the promoter type on alternative splicing events have been reported for the cellular fibronectin gene (8). Here, the CMV promoter also allowed enhanced inclusion of an alternative exon as found in our NLenv system, where the sequence from the weak nef 3′ ss to the poly(A) signal can be viewed as the terminal exon (Fig. 1A). Over the last several years it has become clear that promoters exert their effect on splicing via the processivity or elongation rate of the initiating polymerase (9). It seems that transcription through regulatory elements surrounding alternatively spliced exons and their presentation to the spliceosome determine the outcome of the splicing reaction. These observations have been extended to other genes like alpha tropomyosin (21) and the fibroblast growth factor receptor, where elements that slow down transcription influence splicing ratios (23). Our findings mark the first report on retroviral promoters and their influence on alternative splicing of the viral transcripts. In both HIV and MLV the CMV promoter leads to excessive splicing, indicating that the viral LTRs are adapted to produce a certain amount of unspliced RNA. For HIV this has been assigned to the Tat protein (1), although an alternative explanation is possible: namely, that the elongation rate is accelerated in the presence of Tat and that weak splice sites are simply over read by the polymerase. This model is strengthened by the results obtained using Tat mutants and MLV, where no Tat protein was present. Here, we could also observe a reduction in titer and viral fitness, showing the importance of the retroviral promoter for balanced viral gene expression and a previously undiscovered way to regulate alternative splicing in retroviruses.
Acknowledgments
We thank Barbara Schnierle for the MOVGFP plasmid and Melanie Ott for the Tat mutants, Valerie Bosch and Carol Stocking for providing reagents, Christopher Baum, Harry Wodrich, and Hans-Georg Kräusslich for their support and ongoing discussion, and Mariano Garcia-Blanco for critical reading of the manuscript.
This work was supported by DFG grant 1837/Ba4.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Berro, R., K. Kehn, C. de la Fuente, A. Pumfery, R. Adair, J. Wade, A. M. Colberg-Poley, J. Hiscott, and F. Kashanchi. 2006. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 80:3189-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohne, J., H. Wodrich, and H. G. Krausslich. 2005. Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5′ end. Nucleic Acids Res. 33:825-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, L. J., E. McNulty, and M. Martin. 1993. Human immunodeficiency viruses containing heterologous enhancer/promoters are replication competent and exhibit different lymphocyte tropisms. J. Virol. 67:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, L. J., and C. Zhang. 1995. Infection and replication of Tat- human immunodeficiency viruses: genetic analyses of LTR and tat mutations in primary and long-term human lymphoid cells. Virology 211:157-169. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 7.Cook, C. R., and M. T. McNally. 1999. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J. Virol. 73:2394-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Mata, M., C. R. Alonso, S. Kadener, J. P. Fededa, M. Blaustein, F. Pelisch, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525-532. [DOI] [PubMed] [Google Scholar]
- 10.Domsic, J. K., Y. Wang, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol. Cell. Biol. 23:8762-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erlwein, O., C. J. Buchholz, and B. S. Schnierle. 2003. The proline-rich region of the ecotropic Moloney murine leukaemia virus envelope protein tolerates the insertion of the green fluorescent protein and allows the generation of replication-competent virus. J. Gen. Virol. 84:369-373. [DOI] [PubMed] [Google Scholar]
- 12.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 14.Kraunus, J., D. H. Schaumann, J. Meyer, U. Modlich, B. Fehse, G. Brandenburg, D. Von Laer, H. Klump, A. Schambach, J. Bohne, and C. Baum. 2004. Self-inactivating retroviral vectors with improved RNA processing. Gene Ther. 11:1568-1578. [DOI] [PubMed] [Google Scholar]
- 15.Luo, Y., H. Yu, and B. M. Peterlin. 1994. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol. 68:3850-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen, J. M., and C. M. Stoltzfus. 2005. An exonic splicing silencer downstream of the 3′ splice site A2 is required for efficient human immunodeficiency virus type 1 replication. J. Virol. 79:10478-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchand, V., A. Mereau, S. Jacquenet, D. Thomas, A. Mougin, R. Gattoni, J. Stevenin, and C. Branlant. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323:629-652. [DOI] [PubMed] [Google Scholar]
- 18.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 30:13-19. [DOI] [PubMed] [Google Scholar]
- 19.Ott, M., M. Schnolzer, J. Garnica, W. Fischle, S. Emiliani, H. R. Rackwitz, and E. Verdin. 1999. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol. 9:1489-1492. [DOI] [PubMed] [Google Scholar]
- 20.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson, D., J. F. Elliott, and L. J. Chang. 1995. Retroviral vector with a CMV-IE/HIV-TAR hybrid LTR gives high basal expression levels and is up-regulated by HIV-1 Tat. Gene Ther. 2:269-278. [PubMed] [Google Scholar]
- 23.Robson-Dixon, N. D., and M. A. Garcia-Blanco. 2004. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 279:29075-29084. [DOI] [PubMed] [Google Scholar]
- 24.Schambach, A., D. Mueller, M. Galla, M. M. Verstegen, G. Wagemaker, R. Loew, C. Baum, and J. Bohne. 2006. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 13:1524-1533. [DOI] [PubMed] [Google Scholar]
- 25.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5:611-618. [DOI] [PubMed] [Google Scholar]