Abstract
The neutralist perspective on molecular evolution maintains that the vast majority of mutations affecting gene function are neutral or deleterious. After a gene duplication where both genes are retained, it predicts that original and duplicate genes diverge at clock-like rates. This prediction is usually tested for coding sequences, but can also be applied to another important aspect of gene function, the genes' expression pattern. Moreover, if both sequence and expression pattern diverge at clock-like rates, a correlation between divergence in sequence and divergence in expression patterns is expected. Duplicate gene pairs with more highly diverged sequences should also show more highly diverged expression patterns. This prediction is tested for a large sample of duplicated genes in the yeast Saccharomyces cerevisiae, using both genome sequence and microarray expression data. Only a weak correlation is observed, suggesting that coding sequence and mRNA expression patterns of duplicate gene pairs evolve independently and at vastly different rates. Implications of this finding for the neutralist–selectionist debate are discussed.
If most mutations in coding regions are neutral or deleterious, the number of amino acid substitutions in a protein should be roughly a linear function of time, as predicted by the neutral theory of molecular evolution (1). A “neutralist” position on molecular evolution holds that such clock-like substitution rates are the prevalent mode of molecular evolution, whereas the selectionist viewpoint holds that advantageous mutations occur at appreciable frequency in many genes. If they do, theory predicts (i) great rate variation in sequence evolution, caused by directional selection on advantageous mutations, and (ii) an elevated rate of nonsynonymous substitution in rapidly evolving genes (2, 3). Arguments for or against the neutralist position revolve around case studies on one or few genes that address these two issues in some way or another (2–10). Recent work in this area has increasingly focused on patterns of evolution in duplicated genes (8–12). Here, the neutral theory predicts that duplicated genes diversify in function mainly by means of neutral mutations, whose rate may be elevated as a consequence of “relaxed constraints” on gene function after duplication. An accelerated rate of sequence evolution is consistent with this view, as long as the rate of nonsynonymous substitution is greater than the rate of synonymous substitution. However, such rate elevation has been observed for a number of duplicated genes, and it is a strong argument against the neutral model for these genes (9–11).
Most contemporary tests of the neutralist position focus on sequence evolution. However, the neutralist position is primarily a postulate about how most mutations affect gene function, and only secondarily a statement about rates of sequence divergence. Clearly, the biochemical function of a gene product, as represented by its amino acid sequence, is only one aspect of gene function. Another important aspect is a gene's spatiotemporal expression pattern, which may be strongly influenced by selection. For instance, gene expression patterns can often not be altered without adversely affecting organismal development (13). Furthermore, expression patterns are sometimes so conserved that they serve as reliable indicators of key developmental events across a broad range of organisms (14, 15). Conversely, change in expression is often associated with change in function. For instance, an increasing number of genetic studies indicate that the divergent functions of many duplicate genes are the result of divergence in expression pattern rather than coding sequence (16–18). Take, for instance, the case of the mouse gene Myf5, whose knock-out mutation causes a defect in rib cage formation, which can be rescued if the coding region of its paralogue myogenin is placed under the control of the regulatory region of Myf5 (16–18). However, the two genes have diverged substantially in coding sequence since their duplication more than 400 million years ago (Mya) (19).
Although it is often stated that gene duplications and subsequent evolution of regulatory regions have driven most morphological evolution, it is not clear whether mutations causing such changes would be mostly advantageous or neutral. For instance, the expression of duplicated genes may become restricted to a part of a preduplication expression domain merely by mutations in enhancer sequences that abolish expression in part of this domain. Such mutations would have deleterious effects before a gene duplication, but might be neutral after duplication because of “relaxed constraints” (1).
Even in lower eukaryotes, the regulatory (enhancer) sequences that determine gene expression can contain many binding sites for different transcriptional regulators (20). They thus contain much DNA that might be useful to assess the importance of neutral evolution for regulatory regions. Although some attempts were made to study the molecular evolution of enhancers (21, 22), comparisons among anything but closely related species may often be difficult. This is because regulatory regions usually consist of much inert DNA with scattered islands of functionally important regions, whose location may shift without major functional consequences. Fortunately, microarray technology has made it possible to circumvent these problems by permitting the study of gene expression on the mRNA level (23).
Similar to the predicted gradual sequence divergence between original and duplicate genes, the similarity of spatiotemporal gene expression patterns between duplicated genes will also decline gradually if evolution of expression patterns is best approximated by a neutralist model. As an important corollary, a correlation between divergence in sequence and divergence in expression patterns is expected. The greater the sequence similarity between two duplicate genes, the greater should be the similarity in expression pattern between these genes. This prediction can be tested in two different ways. First, one could follow the divergence in sequence and expression pattern of one gene pair in multiple related organisms. Second, one could analyze a large sample of duplicated gene pairs with varying degrees of divergence within one organism. In the second case, the rate of divergence would depend on the age of the duplication and on the functional constraints on the duplicate genes. The availability of genome sequence data and large scale microarray expression data from various organisms has put the second approach within reach. It is pursued here for the yeast Saccharomyces cerevisiae, for which not only the complete genome sequence is known (24), but where also microarray expression data are available for all genes under multiple physiological conditions (23, 25–27). The results show that there is, at most, a very weak correlation between rates of divergence in sequence and expression pattern.
Methods
Two main sets of duplicated genes were analyzed in this study. The first consists of a large sample of conserved duplicate gene pairs of the yeast S. cerevisiae and was obtained in the following way. mRNA expression levels of 2467 yeast genes of known function, as published by Eisen et al. (25) and obtained by microarray experiments, were used to assess similarity among gene expression patterns. The expression data contains pooled information from 79 independent experiments in which the changes in expression level of these yeast genes were assessed at multiple time points during (i) the diauxic shift (7 time points; ref. 23), (ii) sporulation (11 time points; ref. 27), (iii) the cell cycle (47 time points from four different experiments; ref. 26), and (iv) temperature and reducing shocks (14 time points; ref. 25). This data set is ideal for the purpose of this study, because gene expression under many physiological conditions is represented. For each gene and each time point of an experiment, the data provides log2-transformed ratios of expression levels in an experimental population and a reference population of cells (25). For all (≈2 × 106) gene pairs, Pearson product-moment correlation coefficients r of expression levels were calculated. Gene pairs not encoding ribosomal proteins and with a value of r > 0.5 were retained for further analysis. Conventional tests to detect correlation coefficients significantly different from zero at some significance level P cannot be applied here, because the data are not sampled from a normal distribution. However, the following randomization was carried out to demonstrate that r = 0.5 is a statistically conservative cut-off value. Denote as x = (x1, … , xn) and y = (y1, … , yn) the expression time course for two genes x and y, as represented by log2-transformed expression ratios xi and yi. A permutation x(1) of one of the time series was generated by randomly shuffling all entries of the array x. Then, a Pearson correlation coefficient r(1) was calculated for the shuffled time series x(1) and y, and its magnitude was compared with r. This process was repeated k = 500 times. If the absolute value of r(i) exceeded the absolute value of r less than kP times (here, P = 0.01), r was considered significantly different from 0. When carried out for a sample of 1000 randomly chosen gene pairs, correlation coefficients with r > 0.35 were always accepted as significantly different from 0. Thus, the cut-off value of r = 0.5 is statistically conservative.
Next, the amino acid and DNA sequences, as obtained from the Saccharomyces genome database (ftp://genome-ftp.stanford.edu/pub/yeast/yeast_ORFs; January 1999), of each gene pair with r > 0.5 were compared. This was done by first aligning the amino acid sequences of the two genes with the software package clustalw (v1.7; ref. 28), and by then calculating protein distance based on percent accepted mutations (PAM) substitution matrices (29) by using phylip (v3.5c; ref. 30). Second, the third codon positions were extracted from the coding regions of each gene pair, aligned with clustalw (28), and their Kimura two-parameter distance was calculated by using phylip with a default transition to transversion ratio of 2:1 (30). For both distance measures, that of amino acid sequences, da, and that of third codon positions, d3, a value of d corresponds to the expected fraction of substitutions that occurred per amino acid or nucleotide position. Only gene pairs with both 0<da<1 and 0<d3<1 were retained for further analysis. Completely identical proteins were excluded, because they may be representatives of the small number of repeat regions in the yeast genome or the product of very recent gene conversion events. Because of the possibility of multiple substitutions at some sites, a value of d = 1 corresponds to a sequence divergence of approximately 50% in either amino acid or third codon position comparisons.
In sum, the first set of duplicated genes analyzed here contained all (n = 124) pairs of nonribosomal yeast genes of known function that were moderately to highly conserved in both sequence (0<da, d3<1) and expression pattern (r > 0.5). The second, smaller set of analyzed gene pairs contains genes duplicated in a genome duplication event approximately 100 Mya (31). This genome duplication event was identified by Wolfe and Shields (31) in the form of 55 syntenic pairs of gene clusters that comprise 376 pairs of highly similar genes with amino acid identities ranging from 24% to 100%. From this set of gene pairs, the subset of n = 20 nonribosomal gene pairs of known function with r > 0.5, and 0<da, d3<1 was analyzed. Ribosomal proteins are excluded here because of their unusual properties, such as very high expression levels and great similarity in expression levels, almost 100% conservation among duplicate genes, yet severe fitness effect of loss-of-function mutations (32, 33). The fraction of nonsynonymous (Ka) and synonymous substitutions (Ks) per nucleotide site was estimated by using the method of Comeron et al. (34), as implemented in the software package k-estimator (v4.4; ref. 34).
Results
To determine whether duplicated genes show correlated rate divergence in expression patterns and amino acid sequences, the remnants of a yeast genome duplication event were analyzed. These remnants consist of 376 gene pairs organized into 55 syntenic gene clusters (31). Of these 376 gene pairs, a subset of the 20 most highly conserved pairs was chosen for analysis. More specifically, this subset contains only pairs of nonribosomal genes of known function that showed (i) highly correlated expression under multiple physiological conditions, as measured by Pearson correlation coefficients r of their mRNA induction level in 79 different microarray experiments (25), and (ii) distances of their protein sequences da and their third codon positions ds within 0<da, d3<1. These sequence distances represent the expected number of substitutions per amino acid or nucleotide position. Because multiple substitutions may occur at each sequence position for large values of d, a value of d = 1 does not mean 100% sequence divergence, but a smaller amount of divergence whose exact value depends on the substitution model (2). For the data used here, d = 1 corresponded to an amino acid or nucleotide identity of approximately 50%.
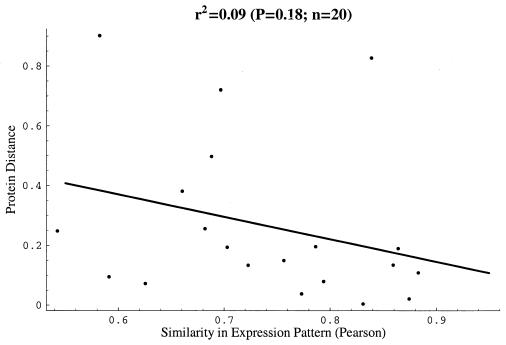
There is no statistically significant association (P = 0.18) between divergence in amino acid sequence and similarity in expression pattern among these 20 gene pairs (Fig. 1). The significance test used here was a randomization test identical to that carried out to determine significant similarity among expression patterns of two genes. Nonparametric tests of association between sequence similarity and similarity in expression patterns yield similar results (not shown). The advantage of analyzing this data set is that the duplications are likely to have occurred at the same time. However, the absence of a statistically significant correlation may simply be because of the small number of genes that are sufficiently highly conserved to be analyzed. A second, larger data set was thus generated that included all nonribosomal yeast genes of known function with highly correlated expression patterns (r > 0.5) and conserved sequences 0<da, d3<1. This data set of 124 gene pairs may contain duplications of different ages, such that the observed degree of divergence is a combination of duplication time and divergence rate.
Figure 1.
Similarity in gene expression pattern vs. protein distance da for 20 conserved gene pairs duplicated in a yeast genome duplication event. Significance P = 0.18 of the calculated coefficient of determination r2 = 0.09 was determined by a randomization assay.
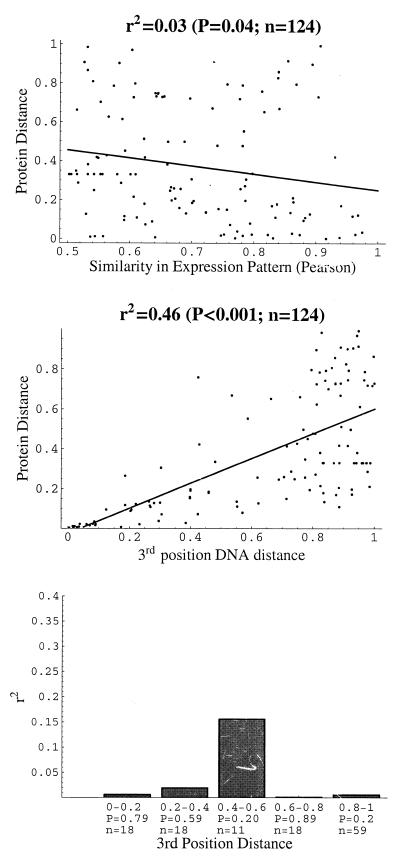
Statistical analysis of this larger data set yielded qualitatively identical results, in that there was only a very weak, albeit now marginally significant, statistical association between divergence in amino acid sequence and expression pattern (Fig. 2a; r2 = 0.03; P = 0.04). As has been argued above, if advantageous mutations are very rare, more recently duplicated genes may evolve under the influence of neutral (and deleterious) mutations for some time, until they are affected by advantageous mutations. It would thus be desirable to divide the set of 124 gene pairs into groups of different duplication age, and test these groups separately for associations between divergence in sequence and expression pattern. Amino acid divergence alone is clearly not a good indicator of duplication age, because different genes may be under vastly different functional constraints on sequence evolution (2). A better indicator may be the DNA distance d3 of two genes calculated from the third (wobble) nucleotides at each codon. This is because a large fraction of substitutions at third codon positions are synonymous, making them better approximate a model of neutral and clock-like evolution that could be used to calibrate duplication age. However, this indicator of duplication age is also far from perfect. First, differential codon usage bias may constrain evolution at the third position (35–38), and second, a still poorly understood correlation between the rate of nonsynonymous and synonymous substitutions is observed in a wide range of organisms (39–42). Thus, the strong correlation between the distance at third codon positions (d3) and amino acid distance (da) observed for the 124 gene pairs (Fig. 2b) will partially reflect a general correlation of amino acid sequence divergence with duplication age, and partially reflect the influence of these confounding factors. Although the relative age of gene duplications can thus not be determined without further phylogenetic information, third position distance will certainly be a better indicator of relative duplication age than amino acid based distance. The 124 gene pairs were thus classified into five groups according to their third position DNA distance, and statistical associations between amino acid divergence and divergence in expression patterns were assessed separately for each group. The results, summarized in Fig. 2c, demonstrate that neither of these groups showed a statistically significant association between sequence distance and distance in expression pattern. This holds also if nonparametric measures of association are used (not shown).
Figure 2.
(a) Similarity in gene expression pattern vs. amino acid distance da for 124 conserved gene pairs with protein distance 0<da<1, third position codon distance 0<d3<1, and Pearson correlation coefficient of mRNA induction level r > 0.5. Significance of the calculated coefficient of determination r2 = 0.03 was determined by a randomization assay. (b) Third position nucleotide distance d3 vs. protein distance da for the 124 genes pairs shown in a. (c) Coefficient of determination r2 between protein distances and similarity in expression pattern, calculated for groups of gene pairs with third position distances within a specified range, as shown on the x axis. Numbers of gene pairs in each distance group and P value indicating whether the estimated r2 is significantly different from 0 are shown on the x axis as well. P values were determined by a randomization assay.
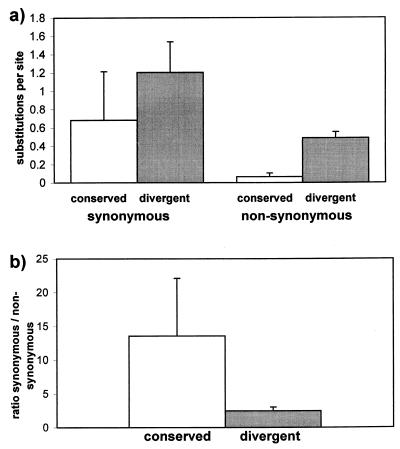
To evaluate whether differences in sequence divergence among gene pairs are associated with different patterns of synonymous and nonsynonymous substitutions, a subset of gene pairs with the most highly conserved expression patterns (r > 0.8) was analyzed. They were subdivided into two groups, a group of 7 gene pairs highly divergent in amino acid sequence (mean da = 0.85), and a group of 10 gene pairs with highly conserved amino acid sequence (mean da = 0.11). The estimated mean fraction of synonymous nucleotide substitutions per position differed only by a factor of 1.7 between the two groups, whereas the fraction of nonsynonymous substitutions differed by a factor of 7.6 (Fig. 3). The mean ratio of synonymous to nonsynonymous substitutions is 13.6 (s = 8.6) for the converged gene pairs, and 2.5 (s = 0.6) for the more diverged gene pairs. Thus, rapidly evolving gene pairs with highly conserved expression patterns show an elevated rate of nonsynonymous substitutions.
Figure 3.
(a) Mean and SD of the fraction of synonymous and nonsynonymous substitutions observed for two different groups of gene pairs with highly conserved (r > 0.8) mRNA induction levels. Values shown were determined for 10 “conserved” gene pairs with a protein distance 0<da<0.2, and 7 “diverged” gene pairs with a protein distance of 0.8<da<1. (b) The ratio of synonymous to nonsynonymous substitutions for these two groups of gene pairs.
Discussion
In sum, at best, a very weak statistical association is observed between divergence in amino acid sequence and temporal mRNA expression patterns of the moderately to highly conserved yeast gene pairs analyzed here. This holds both for a set of gene pairs that originated in an ancient yeast genome duplication event, and for a set of gene pairs identified only on the basis of their similarity in sequence and mRNA induction levels under a variety of physiological conditions. This second set of gene pairs was subdivided into groups of genes with different degrees of divergence at the third codon position, to attempt a rough classification according to different duplication ages. None of these groups showed the expected statistical association if neutrality is the dominant mode of evolution.
The most obvious interpretation of these results is that advantageous mutations occur at appreciable rates in either regulatory or coding regions of one of two duplicated genes, causing a variation in evolutionary rate so great that the expected correlation between sequence divergence in divergence in expression patterns erodes rapidly. However, there are some caveats to this interpretation.
Are Some Duplications Too Ancient, or Some Duplicates Too Divergent To Be Included?
The yeast genome duplication event in which the first set of gene pairs (Fig. 1) originated occurred of the order of 100 Mya (31), and some duplications in the second set of gene pairs (Fig. 2a) will be even older. One might argue that only the analysis of more recent duplications may allow meaningful conclusions.
In this regard, it is important to note two points. First, the validity of the neutralist position often does not depend on duplication age. Genes routinely used to infer phylogenies (an implicit assumption of evolution according to a neutral pattern) may evolve neutrally over significantly longer time spans (43). In a similar vein, many ancient gene duplicates (400 My; ref. 19) with redundant functions still exist in vertebrate genomes (18, 44–47). This great functional overlap among ancient duplicates suggests a largely neutralist pattern of (functional) evolution since the duplication.
Second, most gene pairs in the two data sets are highly conserved, high sequence conservation being one of the criteria by which these genes were originally identified (31). But what if the expected association would only hold for the most closely related genes? The data shown in Fig. 2 a and c address this question. They imply that even the most closely related duplicates do not show a strong association between divergence in sequence and expression pattern.
Data Quality.
The only data sets currently available to compare genome-scale gene expression patterns are temporal mRNA expression profiles. Among the shortcomings of the data are the considerable amount of noise and the neglect of spatial information, as well as of translational and posttranslational regulation. It is also not necessarily clear how to best quantify similarity among expression patterns. Two recent studies speak to perhaps the most troubling of these issues, the relation of mRNA to protein expression level (48, 49). Especially for genes with high codon bias, they suggest a strong correlation between mRNA and protein. Many of the duplicate gene pairs analyzed here do show strong codon bias (results not shown).
Do Sequence and Expression Patterns Diverge Neutrally, But at Very Different Rates?
This would be somewhat analogous to different rates of sequence evolution observed in different DNA sequences. For instance, because mitochondrial (mt) DNA evolves much faster than ribosomal (r) DNA (43), one might find only weak statistical associations between degrees of rDNA and mtDNA divergence on the broadest taxonomic levels. However, differential rates of evolution cannot explain the lack of correlation observed here, simply because there are many gene pairs with considerable sequence divergence (da > 0.8) that have highly conserved expression patterns (r > 0.8), and vice versa (Fig. 2a). This leaves the possibility that sequence is extremely constrained for some gene pairs, whereas evolution of their expression patterns is unconstrained, and vice versa, for other gene pairs. This is not supported by anecdotal observations, such as that duplicate gene pairs with highly conserved sequences (e.g., ribosomal genes and histone genes) often also show highly conserved expression patterns. In an attempt to exclude this possibility, the ratio of synonymous to nonsynonymous substitutions was determined for the set of gene pairs with the most highly conserved expression patterns (r > 0.8). If those members of the set with highly divergent sequence showed an increased rate of nonsynonymous substitutions relative to those members of the set with conserved sequence, one could conclude that the different rates in sequence evolution are the result of directional selection. The results (Fig. 3) are suggestive but not conclusive. The ratio of nonsynonymous to synonymous substitutions differs by a factor of five between the two groups. However, the fraction of nonsynonymous substitutions does not exceed that of synonymous substitutions for any gene pair analyzed.
Information relevant to resolve this last issue may also come from functional analysis of knock-out mutations in duplicated genes. If the neutralist view is correct, then one expects a correlation between the degree of divergence among duplicated genes, and the severity of the effect of loss-of-function mutations in one of the two genes. A recent study analyzing the results of a large-scale knock-out experiment in yeast (32) shows that there is no such correlation (33).
Strengths and Weaknesses of the Genome-Based Approach.
Most studies on the evolution of duplicate genes track one indicator of divergence (sequence) in a gene pair through multiple organisms. The perspective proposed here is radically different. It is based on the analysis of two indicators of divergence (sequence and expression pattern) in all duplicate gene pairs of one organism. Not surprisingly, it then also has unique strengths and weaknesses. First, by using a data set based only on criteria of similarity among duplicated genes, one may gain a representative picture of the forces driving molecular evolution on the whole-genome level, and thus avoid the potential (investigator) bias caused by focusing on genes of particular function. Second, proponents of the neutralist position have argued that variation in generation time or mutation rate across taxa may account for much of the observed variation in substitution rate that is in apparent conflict with their position (2, 3). By studying many genes in one organism, these problems are avoided altogether, because all studied genes share a common evolutionary history.
There are also disadvantages of this approach. First, using a criterion of conservation to select a set of duplicated genes might introduce a bias toward neutralist patterns of evolution. For if advantageous mutations are very rare, then one might observe their traces only in highly divergent genes. Second, unless one has access to a recently duplicated genome, the approach cannot easily disentangle conservation because of recent duplication and conservation because of functional constraints. Not even synonymous substitutions permit precise dating of gene duplications, because of a widely observed correlation between the rate of synonymous and nonsynonymous substitutions (39–42). Note, however, that from the neutralist view point, it should not matter whether two highly divergent genes differ because they were duplicated a long time ago, or because they are under few functional constraints. If the neutralist pattern holds for most genes, then even most anciently duplicated genes should diverge neutrally.
The first step from evolutionary genetics to evolutionary genomics is to study a large sample of genes in one organism, as opposed to tracking one gene pair through multiple organisms. The second step would consist in combining these two approaches by using whole genome information from multiple related organisms. Once this second step becomes feasible, the issues left open by either approach will be resolved, and a divisive and polarizing debate can then be put to rest.
Acknowledgments
I thank M. Lynch, D. Natvig, and M. Werner-Washburne for discussions on the subject. The financial and computational support of the Santa Fe Institute and of the Albuquerque High Performance Computing Center is gratefully acknowledged.
Abbreviation
- Mya
million years ago
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110147097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110147097
References
- 1.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 2.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 3.Gillespie J H. The Causes of Molecular Evolution. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 4.McDonald J H, Kreitman M. Nature (London) 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 5.Easteal S. Mol Biol Evol. 1991;8:115–127. doi: 10.1093/oxfordjournals.molbev.a040632. [DOI] [PubMed] [Google Scholar]
- 6.Easteal S, Collet C. Mol Biol Evol. 1994;11:643–647. doi: 10.1093/oxfordjournals.molbev.a040142. [DOI] [PubMed] [Google Scholar]
- 7.Kreitman M, Akashi H. Annu Rev Ecol Syst. 1995;26:403–422. [Google Scholar]
- 8.Ohta T. Genetics. 1994;138:1331–1337. doi: 10.1093/genetics/138.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirera S, Aguade M. Mol Biol Evol. 1998;15:988–996. doi: 10.1093/oxfordjournals.molbev.a026014. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J Z, Rosenberg H F, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long M Y, Langley C H. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 12.Li W H, Gojobori T. Mol Biol Evol. 1983;1:94–108. doi: 10.1093/oxfordjournals.molbev.a040306. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert S F. Developmental Biology. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 14.Patel N H, Martinblanco E, Coleman K G, Poole S J, Ellis M C, Kornberg T B, Goodman C S. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 15.Sordino P, Vanderhoeven F, Duboule D. Nature (London) 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- 16.Hanks M, Wurst W, Ansoncartwright L, Auerbach A B, Joyner A L. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 17.Li X L, Noll M. Nature (London) 1994;367:83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y K, Schnegelsberg P N J, Dausman J, Jaenisch R. Nature (London) 1996;379:823–825. doi: 10.1038/379823a0. [DOI] [PubMed] [Google Scholar]
- 19.Wray G A, Levinton J S, Shapiro L H. Science. 1996;274:568–573. [Google Scholar]
- 20.Breeden L, Nasmyth K. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang D G, Marsh J L, Ayala F J. Proc Natl Acad Sci USA. 1996;93:7103–7107. doi: 10.1073/pnas.93.14.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig M Z, Kreitman M. Mol Biol Evol. 1995;12:1002–1011. doi: 10.1093/oxfordjournals.molbev.a040277. [DOI] [PubMed] [Google Scholar]
- 23.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 24.Goffeau A, Aert R, Agostini-Carbone M L, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, et al. Nature (London) 1997;387:5. [Google Scholar]
- 25.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu S, Derisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dayhoff M, Schwartz R M, Orcutt B C. In: Atlas of Protein Sequence and Structure. Dayhoff M, editor. 5, Suppl. 3. Silver Spring, MD: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 30.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 31.Wolfe K H, Shields D C. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 32.Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Science. 1997;275:464–464. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 33.Wagner A. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 34.Comeron J M. J Mol Evol. 1995;41:1152–1159. doi: 10.1007/BF00173196. [DOI] [PubMed] [Google Scholar]
- 35.Sharp P M, Li W H. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- 36.Sharp P M, Tuohy T M F, Mosurski K R. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shields D C, Sharp P M, Higgins D G, Wright F. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 38.Moriyama E N, Hartl D L. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouchiroud D, Gautier C, Bernardi G. J Mol Evol. 1995;40:107–113. doi: 10.1007/BF00166602. [DOI] [PubMed] [Google Scholar]
- 40.Lipman D J, Wilbur W J. J Mol Evol. 1985;21:161–167. doi: 10.1007/BF02100090. [DOI] [PubMed] [Google Scholar]
- 41.Akashi H. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comeron J M, Kreitman M. Genetics. 1998;150:767–775. doi: 10.1093/genetics/150.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hillis D M, Moritz C, Mable B K. Molecular Systematics. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 44.Joyner A L, Herrup K, Auerbach B A, Davis C A, Rossant J. Science. 1991;251:1239–1243. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- 45.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dolle P, Chambon P. Development (Cambridge, UK) 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 46.Condie B G, Capecchi M R. Nature (London) 1994;370:304–307. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 47.Horan G S B, Kovacs E N, Behringer R R, Featherstone M S. Dev Biol. 1995;169:359–372. doi: 10.1006/dbio.1995.1150. [DOI] [PubMed] [Google Scholar]
- 48.Pavesi A. J Mol Evol. 1999;48:133–141. doi: 10.1007/pl00006451. [DOI] [PubMed] [Google Scholar]
- 49.Gygi S P, Rochon Y, Franza B R, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]