Abstract
The Ras GTPase links extracellular signals to intracellular mechanisms that control cell growth, the cell cycle, and cell identity. An activated form of Drosophila Ras (RasV12) promotes these processes in the developing wing, but the effector pathways involved are unclear. Here, we present evidence indicating that RasV12 promotes cell growth and G1/S progression by increasing dMyc protein levels and activating dPI3K signaling, and that it does so via separate effector pathways. We also show that endogenous Ras is required to maintain normal levels of dMyc, but not dPI3K signaling during wing development. Finally, we show that induction of dMyc and regulation of cell identity are separable effects of Raf/MAPK signaling. These results suggest that Ras may only affect PI3K signaling when mutationally activated, such as in RasV12-transformed cells, and provide a basis for understanding the synergy between Ras and other growth-promoting oncogenes in cancer.
Keywords: Ras, Myc, PI3K, Drosophila, cell growth, cell cycle
Cancer is a genetic disease resulting from inappropriate activation or overexpression of proto-oncogenes, compounded by loss of tumor suppressor gene activity (Hanahan and Weinberg 2000). This process is often facilitated by mutation of so-called mutator genes, resulting in genomic instability and accelerated rates of genetic mutation. A great deal has been learned about interactions among various oncogenes including Ras, Myc, and members of the PI3K-signaling pathway that may contribute to oncogenesis. Most of these studies have been performed in vitro and in mammalian cell culture systems, however, and detailed in vivo analysis is lacking. It is also unclear whether these genes interact during normal development as they do when activated or overexpressed in tumors.
Ras is a membrane-associated guanine nucleotide-binding protein that is normally activated in response to the binding of extracellular signals, such as growth factors, to receptor tyrosine kinases. These receptors include the epidermal growth factor receptor (EGFR) in mammals and its Drosophila homologs dEGFR/DER, Sevenless, and Torso. Phosphotyrosine residues on activated receptors bind SH2 domains of adaptor proteins such as Drk/Grb2, which, via their SH3 domains, bind guanine-nucleotide exchange factors such as Sos. Sos catalyzes the conversion of Ras from a GDP-bound inactive state to a GTP-bound active state, resulting in conformational changes in two so-called switch domains (Boriack-Sjodin et al. 1998). GTP-bound mammalian Ras recruits several effector proteins including Raf, PI3K, and RalGDS to the cell membrane, in which they become activated by either Ras-induced conformational changes (Pacold et al. 2000) or other membrane-associated proteins (Morrison and Cutler 1997). Mutations in Ras that cause constitutive GTP binding result in constitutive signaling to downstream effector proteins and are frequently found in a range of human tumors (Barbacid 1987).
Ras signaling affects many cellular functions, including cell proliferation, apoptosis, migration, fate specification, and differentiation (for review, see Rommel and Hafen 1998). Some of these effects have been shown to require the combined activation of several Ras effector proteins. For example, studies in NIH3T3 fibroblasts showed that activation of both Raf and PI3K is required for Ras-dependent transformation (Rodriguez-Viciana et al. 1997) and contributes to cell cycle progression (Gille and Downward 1999). Specific Ras effectors have also been shown to perform distinct functions. For example, Ras stimulates membrane ruffling and inhibits apoptosis via PI3K in fibroblast cells (Kauffmann-Zeh et al. 1997; Rodriguez-Viciana et al. 1997). However, several studies contradict these findings. For example, stimulation of either Raf or PI3K signaling alone is sufficient to induce cell proliferation in cultured postmitotic chicken neuroretina cells (Peyssonnaux et al. 2000), and Raf has been implicated in both promoting and inhibiting apoptosis in several cell types (Kauffmann-Zeh et al. 1997; Zhong et al. 2001). Such discrepancies may reflect cell-type specificities or indicate shortcomings of studying signaling pathways in cell culture. To clarify the roles of effector proteins in mediating Ras signaling, it may be useful to study Ras and its downstream effectors in vivo during normal growth and development. Genetic analyses in vivo have already revealed novel functions for Ras, including the ability to regulate cellular growth (i.e., mass accumulation; Hunter et al. 1995; Heumann et al. 2000; Prober and Edgar 2000), although it is not known which effector proteins mediate this function.
PI3Ks are a family of intracellular signal transducers that are activated by receptor tyrosine kinases and G-protein-coupled receptors (for review, see Vanhaesebroeck et al. 2001). PI3Ks phosphorylate inositol lipids in the cell membrane, generating a range of second messengers including phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 recruits and activates downstream effectors such as Akt/PKB by binding to their pleckstrin homology (PH) domains. Studies in both mice and Drosophila have implicated this pathway in growth regulation, presumably due to its ability to promote glucose and lipid import, nutrient storage, and translation of RNAs involved in ribosome biogenesis (Vanhaesebroeck et al. 2001). Several lines of evidence implicate this pathway as a causal factor in cancer. For example, Akt/PKB was discovered as a viral oncogene, and PTEN, a lipid phosphatase that antagonizes PI3K signaling, is a tumor suppressor implicated in many human cancers and two cancer-associated inherited diseases (Cantley and Neel 1999).
Studies in cell culture have implicated the Myc family of transcription factors as regulators of the cell cycle that are rapidly induced in response to growth factors (for review, see Grandori et al. 2000). These studies are supported by the recent finding that mice with reduced Myc function have defects in cell proliferation (Trumpp et al. 2001). Several studies of Drosophila and mammalian Myc in vivo have also shown a critical function in regulating cell growth (Iritani and Eisenman 1999; Johnston et al. 1999; Kim et al. 2000; de Alboran et al. 2001; Douglas et al. 2001). These roles are supported by analyses of Myc target genes, which include regulators of both cell growth and the cell cycle (Coller et al. 2000; Boon et al. 2001). Myc is also a potent oncogene that is rearranged and overexpressed in a wide range of tumors.
The fact that Ras, Myc, and PI3K have all been implicated in regulating cell growth and the cell cycle suggests that they may interact with each other. In support of this idea, studies in mammalian cell culture have shown that Ras can stabilize Myc protein (Sears et al. 2000) and directly bind to and activate PI3K (Rodriguez-Viciana et al. 1994; Pacold et al. 2000). However, genetic evidence suggesting that Ras regulates Myc or PI3K in vivo is lacking. Ras signaling plays a major role in cell fate specification in Drosophila and Caenorhabiditis elegans (Rommel and Hafen 1998). In contrast, PI3K signaling does not regulate cell fate in C. elegans, and the Drosophila Myc and PI3K homologs regulate cell growth but not cell fate (for review, see Prober and Edgar 2001). The ability of Drosophila Ras to regulate both patterning and growth, however, raises the possibility that Ras might regulate growth via dMyc and/or dPI3K. Here we show that an activated form of Drosophila Ras is capable of driving growth via both dMyc and the dPI3K pathway, and that endogenous Ras is required to maintain normal levels of dMyc, but not dPI3K signaling, during wing development.
Results
Activated Ras increases dMyc levels and activates dPI3K signaling
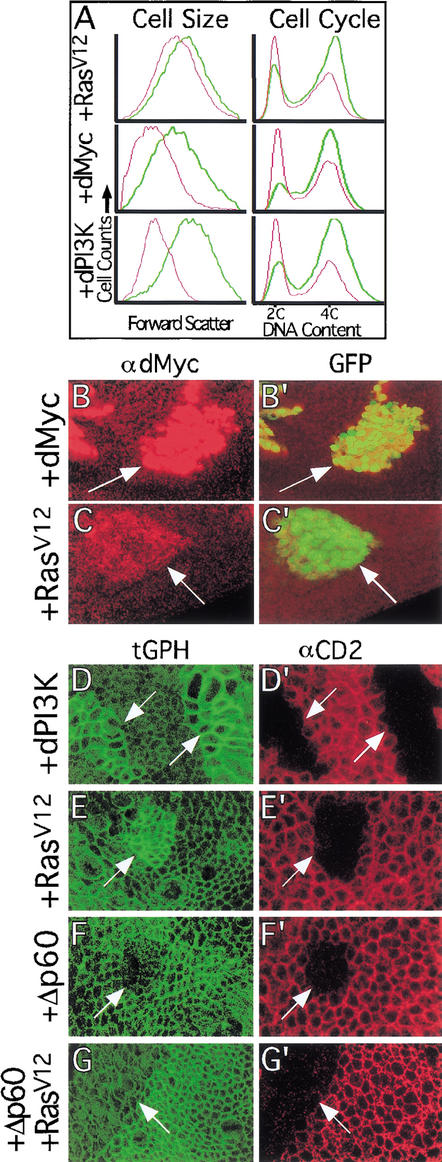
Substitution of valine for glycine at position 12 of mammalian Ras, which is frequently observed in tumors, inhibits GTP hydrolysis and results in constitutive activity (Barbacid 1987). We have shown previously that ectopic expression of a Drosophila Ras1 transgene containing this mutation (RasV12; Karim and Rubin 1998) accelerates the G1/S transition of the cell cycle, increases cell size, and increases clonal growth rates in the developing Drosophila wing (Prober and Edgar 2000). Because overexpression of the Drosophila homologs of Myc (dMyc) and PI3K (dPI3K) have similar but stronger effects (Johnston et al. 1999; Weinkove et al. 1999; Fig. 1A), we examined whether they mediate the effects of RasV12.
Figure 1.
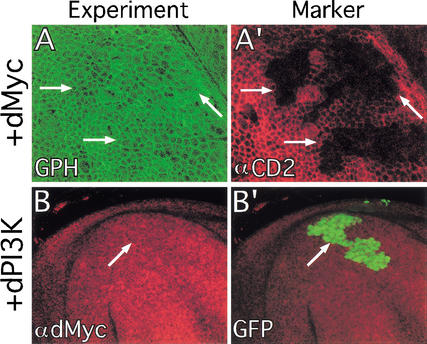
RasV12 expression increases dMyc protein levels and activates dPI3K signaling. (A) FACS analysis of wing disc cells overexpressing RasV12, dMyc, or dPI3K. Green and red traces represent GFP+ (experimental) and GFP− (control) cells, respectively. Each trace is normalized to fit the graph as the numbers of GFP+ and GFP− cells analyzed for each sample were not exactly equal. (Left) Cell size. Forward scatter data, which gives a relative measure of cell size, shows that expression of RasV12, dMyc, or dPI3K increases cell size. (Right) Cell cycle. DNA content data, which shows the proportion of cells in G1 (2C) and G2 (4C), indicates that expression of RasV12, dMyc, or dPI3K decreases the proportion of cells in G1. (B,C) Flp/Gal4 clones (marked with GFP in B‘ and C‘) expressing either dMyc (B,B‘) or RasV12 (C,C‘) show increased staining with a dMyc-specific antibody (B,C). (D–G) Flp/Gal4 clones (marked by loss of the CD2 cell membrane marker in D′,E′,F‘,G‘) expressing either dPI3K (D) or RasV12 (E) have increased cell membrane-associated tGPH, a PH-GFP fusion protein used as an indicator of dPI3K signaling. In contrast, clones expressing either Δp60 (F) or Δp60+RasV12 (G) have reduced cell membrane-associated tGPH. Note that apical tGPH and lateral αCD2 sections are shown, resulting in slight misalignment of tGPH and αCD2 in these images. Magnification: B,C, 40×; D–G, 100×.
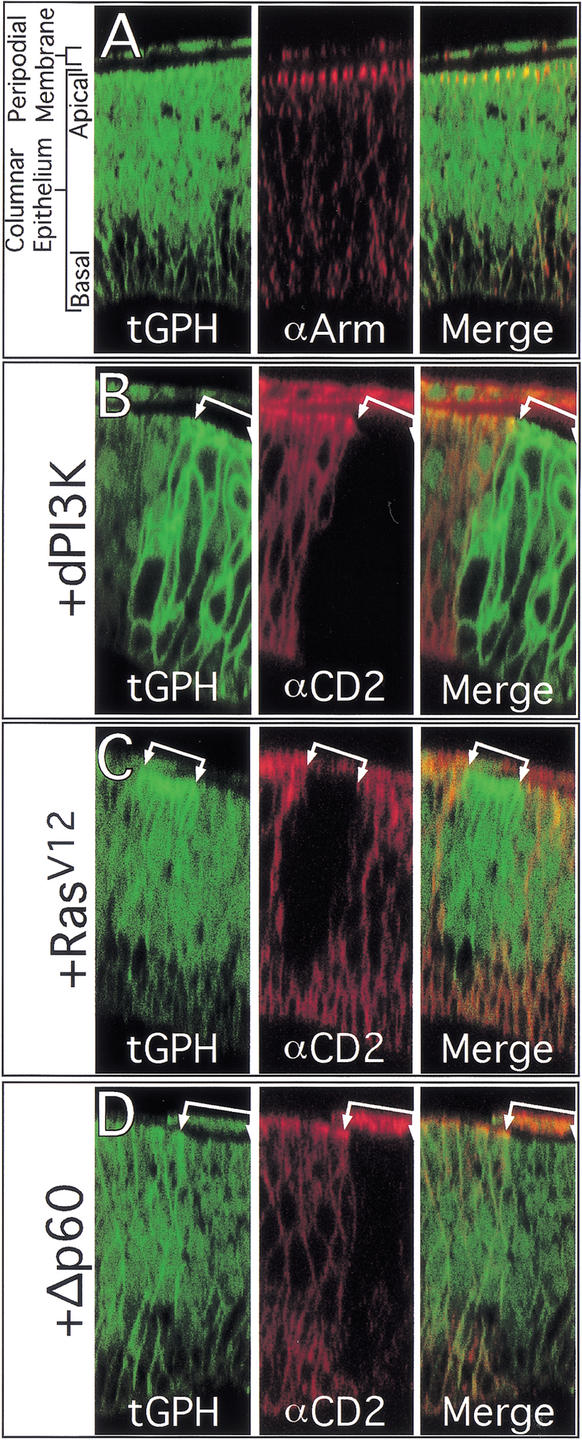
To examine the effect of RasV12 expression on dMyc and dPI3K, we generated clones of cells expressing RasV12 using the Flp/Gal4 technique (Struhl and Basler 1993; Pignoni and Zipursky 1997; Neufeld et al. 1998). This technique allows generation of permanent, heritable expression of UAS-regulated transgenes in random clones of cells in response to a heat shock. As we have shown previously, ectopic expression of RasV12 increases dMyc protein levels (Fig. 1C). To assay dPI3K signaling in vivo, we used a reporter stock that constitutively expresses a transgene encoding the pleckstrin-homology (PH) domain from the Drosophila homolog of general receptor for phosphoinositides-1 (GRP1) fused to GFP (tGPH; Britton et al. 2002). The PH domain of mammalian GRP1 specifically binds PIP3 (Ferguson et al. 2000; Lietzke et al. 2000), which is the second messenger generated by PI3K. Because PIP3 is generally located in lipid membranes, particularly the cell membrane, GRP1 is recruited to the cell membrane when PI3K activity increases cellular PIP3 levels. Fusion proteins containing the GRP1 PH domain are recruited similarly to cell membranes by binding PIP3, and thus serve as in situ reporters for PI3K activity (Gray et al. 1999; Oatey et al. 1999). The tGPH reporter binds PIP3 with high affinity in vitro (W. Lockwood and S. Cohen, pers. comm.) and responds to increases or decreases in dPI3K signaling in vivo by increasing or decreasing its localization at the cell membrane (Britton et al. 2002). In normal wing disc cells, we observed tGPH fluorescence throughout the entire cell membrane (Fig. 2A). Higher tGPH levels colocalized with Armadillo, however, which is located in adherens junctions at the apical region of the cell membrane (Fig. 2A). This suggests that dPI3K signaling is normally most active at the apical region of wing disc cells. To determine whether the observed tGPH fluorescence in wild-type cells reflects endogenous dPI3K activity, we inhibited dPI3K activity by expressing Δp60. p60 is an adapter protein that links the catalytic subunit of dPI3K (Dp110) to upstream activators such as the Drosophila Insulin receptor (dInr) and the IRS homolog Chico. Δp60 is a variant of p60 that lacks the Dp110 interaction domain and inhibits dPI3K activity, presumably by preventing endogenous Dp110 from binding to and becoming activated by dInr and Chico (Weinkove et al. 1999). Overexpression of Δp60 (Figs. 1F, 2D) reduced tGPH fluorescence throughout the cell membrane, indicating that much of the membrane-associated tGPH in wild-type discs results from the activity of endogenous dPI3K. Similar results were obtained upon overexpression of dPTEN, a lipid phosphatase that dephosphorylates PIP3 (data not shown).
Figure 2.
Subcellular localization of tGPH. (A) Optical cross-sections showing that tGPH is localized throughout the cell membranes, cytoplasm, and nuclei of cells in the columnar epithelium and peripodial membrane of imaginal wing discs. Intense GFP fluorescence is present at the apical region of the cell membrane, colocalizing with Armadillo, a component of apically localized adherens junctions. (B–D) Flp/Gal4 clones (marked by loss of CD2) overexpressing dPI3K have strong tGPH fluorescence throughout the entire cell membrane (B). Clones expressing RasV12 have increased tGPH fluorescence at the apical region of the cell membrane (C). Clones expressing Δp60 have reduced tGPH fluorescence throughout the cell membrane (D). Magnification, 100×.
Overexpression of dPI3K (Fig. 1D) resulted in strong recruitment of tGPH to the cell membrane. Expression of RasV12 also resulted in high tGPH levels at the cell membrane (Fig. 1E), suggesting that RasV12 activates dPI3K signaling, and thus increases PIP3 levels. To verify that these effects did not result simply from increased overall levels of the tGPH reporter, we performed Western blots on wing discs in which most cells overexpressed either dPI3K or RasV12. Overexpressed dPI3K or RasV12 did not increase tGPH levels relative to controls (Supplemental Fig. 1 in Supplementary Material at http://www.genesdev.org). This result is consistent with our interpretation that increased levels of membrane-associated tGPH in response to RasV12 and dPI3K results from relocalization and not increased levels of the reporter. These results suggest that RasV12 activates dPI3K, consistent with experiments in mammalian systems, showing that RasV12 can directly bind to and activate dPI3K (Rodriguez-Viciana et al. 1994; Pacold et al. 2000). However, we cannot rule out other potential mechanisms that may explain how RasV12 recruits tGPH to the cell membrane, such as inhibition of the lipid phosphatase dPTEN.
Overexpression of dPI3K increased tGPH fluorescence throughout the entire cell membrane, including both apical and basolateral regions (Fig. 2B). In contrast, RasV12 only recruited tGPH to the apical region of the cell membrane (Fig. 2C), in which tGPH colocalized with Armadillo (data not shown). This effect did not result simply from recruitment of overexpressed RasV12 to apical regions, as overexpressed RasV12 was distributed throughout the cell membrane (Supplemental Fig. 2 in Supplementary Material at http://www.genesdev.org). These results suggest that Ras may require other apically localized factors to activate dPI3K signaling, although other explanations are possible.
To verify that the ability of RasV12 to induce tGPH relocalization was dependent on dPI3K signaling, we coexpressed RasV12 and Δp60. Δp60 completely blocked the ability of coexpressed RasV12 to recruit tGPH to the cell membrane (Fig. 1G), indicating that the ability of RasV12 to relocalize tGPH is dependent upon dPI3K signaling. These experiments therefore show that RasV12 is capable of increasing dMyc levels and activating dPI3K signaling in the developing wing.
RasV12 effector loop mutants activate different effector pathways
Several mutations in the effector loop region of mammalian Ras have been identified that prevent Ras from binding and activating specific downstream effectors (White et al. 1995). For example, RasV12S35 specifically binds and activates mammalian Raf, but not RalGDS or PI3K (Rodriguez-Viciana et al. 1997; Kinashi et al. 2000; Pacold et al. 2000). RasV12G37 interacts specifically with RalGDS (resulting in activation of the Ral GTPase; Rodriguez-Viciana et al. 1997), PI3Kγ (Pacold et al. 2000), and PI3Kδ (Kinashi et al. 2000), but not with PI3Kα or Raf. A third variant, RasV12C40, specifically interacts with PI3Kα but not with PI3Kγ, PI3Kδ, Raf, or RalGDS. The amino acid sequence of the effector loop region of Drosophila Ras is identical to that of mammalian Ras, and transgenic flies containing UAS-regulated forms of these variants have been generated, all containing the activating V12 mutation (Karim and Rubin 1998).
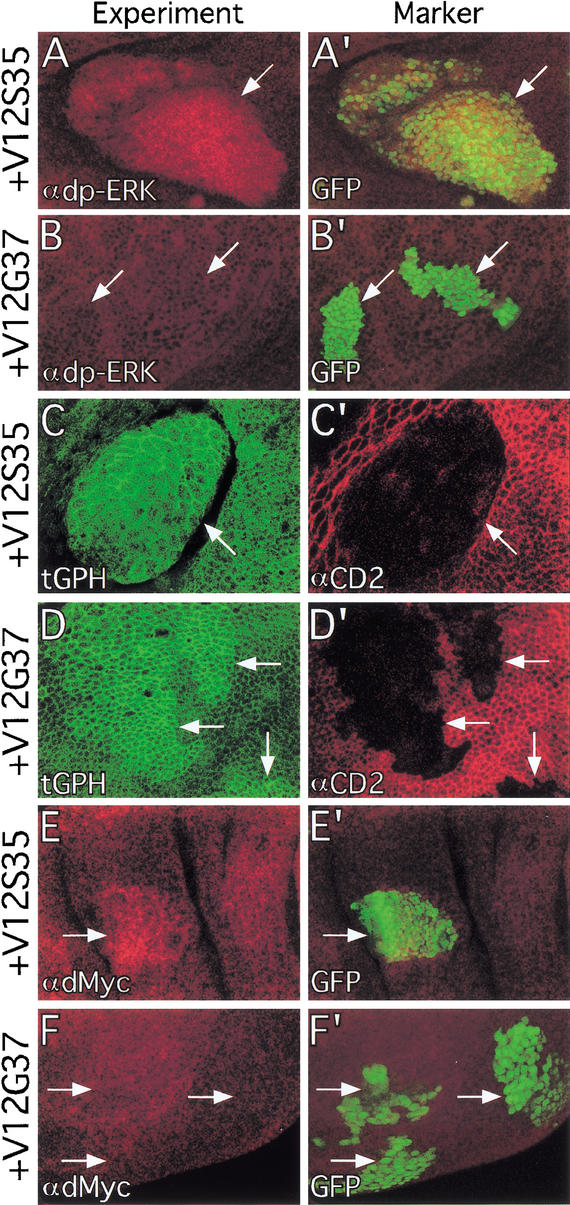
To determine whether the RasV12 effector loop mutants specifically activate different Ras effector pathways in vivo, we generated clones of cells expressing the RasV12 effector loop mutant transgenes in the developing wing and assayed their abilities to activate MAPK and dPI3K signaling. Staining with an antibody that recognizes the activated form of MAPK (dp-ERK) revealed that RasV12, an activated form of Raf (RafGOF), and RasV12S35 activated MAPK throughout the developing wing, whereas dPI3K and RasV12G37 did not (Fig. 3A,B; data not shown). Conversely, expression of RasV12 or RasV12G37, but not RafGOF, caused recruitment of tGPH to the cell membrane in all regions of the developing wing (Fig. 3; data not shown). We occasionally observed tGPH recruitment to the cell membrane in response to RasV12S35 expression, but at much lower levels than for RasV12 or RasV12G37 (Fig. 3C). Like RasV12, RasV12G37-dependent recruitment of tGPH was restricted to the apical region of the cell membrane (data not shown). Thus, at the sensitivity thresholds of our assays, RafGOF specifically activates MAPK but not dPI3K signaling, RasV12S35 primarily activates MAPK, and RasV12G37 specifically activates dPI3K signaling but not MAPK. Expression of RasV12C40 had no detectable effect on either MAPK activity or dPI3K signaling (data not shown).
Figure 3.
Ras effector loop mutants activate different effector pathways. (A,B) Flp/Gal4 clones (marked with GFP in A‘,B‘) expressing RasV12S35 have increased dp-ERK staining (A), whereas clones expressing RasV12G37 do not (B). (C,D) Flp/Gal4 clones (marked by loss of CD2 in C‘,D‘) expressing RasV12G37 have increased membrane-associated tGPH (D), whereas clones expressing RasV12S35 do not (C). (E,F) Flp/Gal4 clones (marked with GFP in E‘,F‘) expressing RasV12S35 have increased dMyc protein levels (E), whereas clones expressing RasV12G37 do not (F). Magnification: A,B,E,F, 40×; C,D, 100×.
RasV12 effector loop mutants that activate Raf or dPI3K signaling induce growth and G1/S progression
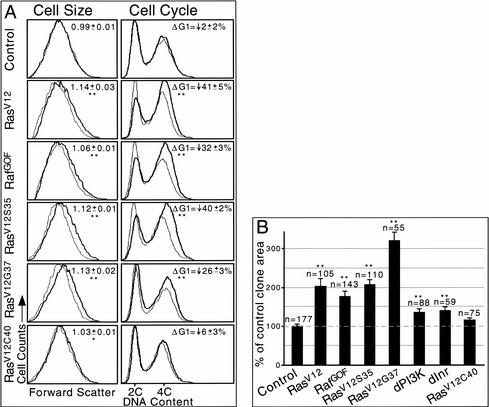
To test whether RasV12 promotes growth and G1/S progression via MAPK or dPI3K signaling, we generated GFP-marked clones of cells expressing the RasV12 effector loop mutant transgenes in the developing wing. We then performed flow cytometry and compared transgene-expressing cells with nonexpressing internal control cells (Neufeld et al. 1998). This technique provides forward scatter data, which gives a relative measure of cell size, as well as cell cycle phasing data, which indicates the proportion of cells in the G1, S, and G2 phases of the cell cycle. We also assayed the effects of the RasV12 effector loop mutant transgenes on clonal growth by measuring clone areas. Like RasV12, expression of RasV12S35, RasV12G37, or RafGOF truncated G1 (Fig. 4A, right), increased cell sizes (Fig, 4A, left), and increased clone areas (Fig. 4B). Because clones expressing RasV12, RafGOF, or RasV12S35 bulge out of the apical surface of the epithelium (see below), two-dimensional area measurements are underestimates of the amount of clonal growth produced. In fact, expression of RasV12G37, which does not cause apical bulging, showed a greater increase in two-dimensional clone areas (Fig. 4B) and likely provides the most accurate measure of clonal growth rates resulting from ectopic Ras signaling. Thus, the RasV12S35 and RasV12G37 effector loop mutants can both drive growth and G1/S progression to a similar extent as RasV12. In contrast, expression of Drosophila RasV12C40 had little effect in these assays (Fig. 4A,B). We also expressed an activated form of Drosophila Ral (dRal20V), a putative Ras target via RalGDS in mammals, that has been implicated in regulating morphogenesis but not growth in Drosophila (Sawamoto et al. 1999). Expression of dRal20V caused a decrease in cell size without affecting cell cycle phasing (data not shown), suggesting that Drosophila Ras does not promote growth or cell cycle progression via dRal.
Figure 4.
Ras effector loop mutants increase cell size, promote G1/S progression, and increase clonal growth. (A) Flow cytometry. Dark and light traces represent GFP+ (experimental) and GFP− (control) cells, respectively. Each trace is normalized to fit the graph as the numbers of GFP+ and GFP− cells analyzed for each sample were not exactly equal. Data shown are from an experimental set performed in parallel. Values significantly different from control are indicated (*, P < 0.05; **, P < 0.01). (Left) Cell size. Forward scatter data, which gives a relative measure of cell size, shows that expression of RasV12, RafGOF, RasV12S35, or RasV12G37 increases cell size. Numbers indicate the mean value of (GFP+ mean FSC)/(GFP− mean FSC) ± standard error of the mean for six experiments (except for RafGOF, which was repeated four times). (Right) Cell cycle. DNA content data, which shows the proportion of cells in G1 (2C) and G2 (4C), indicates that expression of RasV12, RafGOF, RasV12S35, or RasV12G37 decreases the proportion of cells in G1. Numbers indicate the mean percentage change of GFP+ cells compared with GFP− cells in the proportion of cells in G1 (ΔG1) ± standard error of the mean for six experiments (except for RafGOF, which was repeated four times). (B) Median areas of Flp/Gal4 clones expressing various transgenes are shown. Expression of any transgene except RasV12C40 significantly increased clone areas relative to control clones expressing GFP alone. Clones were induced at 48 h after egg deposition (AED; except for dInr, which was induced at 72 h AED) and analyzed at 120 h AED. n, number of clones measured. Error bars, standard error of the mean; **, P < 0.01 vs. control. Note that larvae overexpressing dPI3K or dInr under these conditions are developmentally delayed, and as a result the clone areas measured in this experiment underestimate dPI3K-and dInr-dependent clonal growth. Overexpression of dMyc also greatly increases clonal growth rates (Johnston et al. 1999).
Ras increases dMyc levels via the Raf/MAPK pathway
We next asked which effector pathway is responsible for the RasV12-mediated increase in dMyc protein levels. Staining with a dMyc-specific antibody revealed that expression of RasV12S35 or RafGOF increased dMyc levels, whereas expression of RasV12G37 did not (Fig. 3E,F; data not shown). This was observed in all regions of the wing disc, although it was more difficult to detect in regions containing high levels of endogenous dMyc, such as the wing pouch (see below). Thus, RasV12 regulates dMyc levels via the Raf/MAPK pathway and not via dPI3K signaling. The ability of RasV12 to affect dMyc and dPI3K signaling via separate pathways suggests that dMyc and dPI3K may not regulate each other. Accordingly, overexpression of dMyc had no effect on tGPH localization (Fig. 5A) and overexpression of dPI3K had no effect on dMyc levels (Fig. 5B). Thus, RasV12 is capable of activating two growth-promoting pathways, but these pathways are incapable of activating each other.
Figure 5.
Overexpressed dMyc and dPI3K do not affect each other. (A) Clones of cells overexpressing dMyc (marked by loss of CD2 in A‘) have normal tGPH localization. (B) Clones of cells overexpressing dPI3K (marked with GFP in B‘) have normal dMyc protein levels. Magnification: A, 100×; B 40×.
Ras is required to maintain normal levels of dMyc protein but not dPI3K signaling
Having shown that activated variants of Ras can increase dMyc protein levels and activate dPI3K signaling, we next examined whether endogenous Ras does so during normal development. Ras signaling is strongly activated in developing vein cells at the end of larval development, as determined by use of an antibody specific for the activated form of MAPK (Gabay et al. 1997; Guichard et al. 1999; Martin-Blanco et al. 1999). This pattern of activity is maintained until ∼30 h after pupation, at which point Ras activity is down-regulated in vein cells and up-regulated in intervein regions. Genetic data suggests that Ras/MAPK signaling is also active at low levels throughout the tissue (Diaz-Benjumea and Hafen 1994; Prober and Edgar 2000; Halfar et al. 2001). High levels of dmyc mRNA and protein are present in the developing wing pouch, except for several rows of cells along the dorsal-ventral boundary in which dmyc expression is repressed by Wingless (Supplemental Fig. 3A in Supplementary Material at http://www.genesdev.org; Johnston et al. 1999). High dMyc protein levels are also present in the notum, and low levels are observed throughout the rest of the tissue. Thus, dMyc appears to be expressed ubiquitously at low levels, with higher levels present in specific regions. In contrast, dPI3K-signaling levels, assayed using tGPH localization, appear uniform throughout the wing during larval and pupal development (Supplemental Fig. 4C in Supplementary Material at http://www.genesdev.org; data not shown). Whereas patterns of dMyc expression and dPI3K activity do not coincide with those of high MAPK activity, we reasoned that the ubiquitous low levels of Ras activity may play a role in regulating dMyc protein levels and dPI3K signaling.
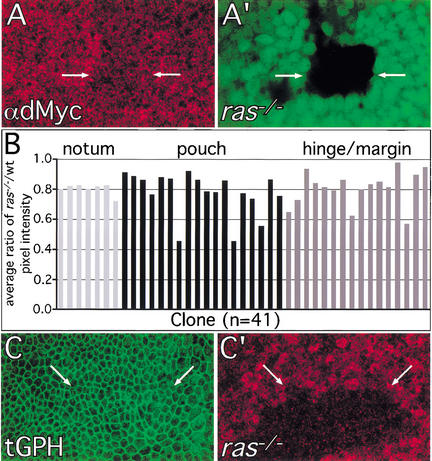
To test whether this is the case, we used FLP/FRT-mediated mitotic recombination to generate ras mutant clones (ras−/−) using the rasc40b allele, in which the entire ras ORF is deleted (Schnorr and Berg 1996). We quantified the requirement for ras to maintain normal dMyc protein levels by staining wing discs with a dMyc-specific antibody and comparing pixel intensities in neighboring regions inside and outside of ras−/− clones (see Materials and Methods). We found dMyc antibody staining intensity to be significantly lower in ras−/− cells in all areas of the developing wing, regardless of endogenous dMyc protein levels, although the effect was modest (Fig. 6A,B). Thus, Ras is required to maintain normal dMyc protein levels throughout the developing wing. In contrast, ras−/− clones had no detectable effect on tGPH intensity or localization (Fig. 6C), suggesting that Ras is not required to maintain normal PIP3 levels, and thus levels of dPI3K signaling.
Figure 6.
Endogenous Ras is required to maintain normal levels of dMyc protein but not dPI3K signaling. The FLP/FRT technique was used to generate ras−/− clones in ras+/− tissues. (A) ras−/− cells (marked by loss of GFP in A‘) have reduced levels of dMyc protein. Genotype: hs-FLP122; UAS-P35/en-Gal4; FRT (82B) rasc40b/FRT (82B) Ub-GFP. (B) Quantitation of dMyc antibody staining intensity in ras−/− clones compared with neighboring ras+/− and ras+/+ regions. Ratios of average pixel intensity ras−/−/average pixel intensity ras+/− or ras+/+ were calculated for individual clones. To minimize confounding bias due to patterned endogenous dMyc levels, each bar represents the average value of 10–15 ratios for a single clone (see Materials and Methods). Regions within ras−/− clones consistently had lower values than surrounding regions (average = 0.79 ± 0.02; P = 2 × 10−6 for 41 clones). dMyc antibody staining intensities in neighboring ras+/− and ras+/+ regions were not significantly different (average ratio of ras+/−/ras+/+ = 0.9 5 ± 0.05, P = 0.45 for 11 clones). ras−/− clones had similar effects on dMyc antibody staining intensities in regions of the wing with high (notum and pouch) and low (hinge/margin) endogenous dMyc levels. (C) ras−/− cells (marked by loss of the πmyc marker in C‘) have normal tGPH localization and intensity. Genotype: hs-FLP122; UAS-P35/tGPH; en– Gal4; FRT (82B) rasc40b/FRT (82B) hs-πmyc. Magnification: A,B, 100×.
Ras affects cell fate and cell adhesion via the Raf/MAPK pathway
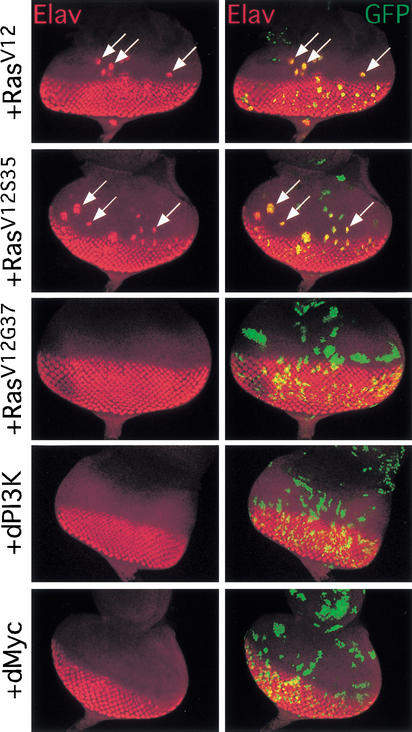
Ras signaling regulates cell identities in developing imaginal tissues (Rommel and Hafen 1998). To determine which effector pathway is utilized, we examined the abilities of the RasV12 effector loop mutants to affect cell differentiation and cell affinity, two aspects of cell identity. Ras signaling promotes specification of photoreceptor neurons in cells posterior to the morphogenetic furrow in the developing eye, which express the neuron-specific protein Elav. We found that transgenes that activated Raf/MAPK signaling (RasV12, RafGOF, and RasV12S35) induced precocious Elav expression anterior to the morphogenetic furrow, whereas transgenes that activated dPI3K signaling (RasV12G37 and dPI3K) did not (Fig. 7). Overexpressed dMyc and RasV12C40 also had no effect on Elav expression (Fig. 7; data not shown).
Figure 7.
Ras promotes precocious photoreceptor differentiation via Raf/MAPK signaling. Eye discs with Flp/Gal4 clones (marked with GFP at right) expressing various transgenes are shown. Expression of RasV12 or RasV12S35 induces precocious Elav staining (arrows), whereas expression of RasV12G37, dPI3K, or dMyc does not. Magnification, 20×.
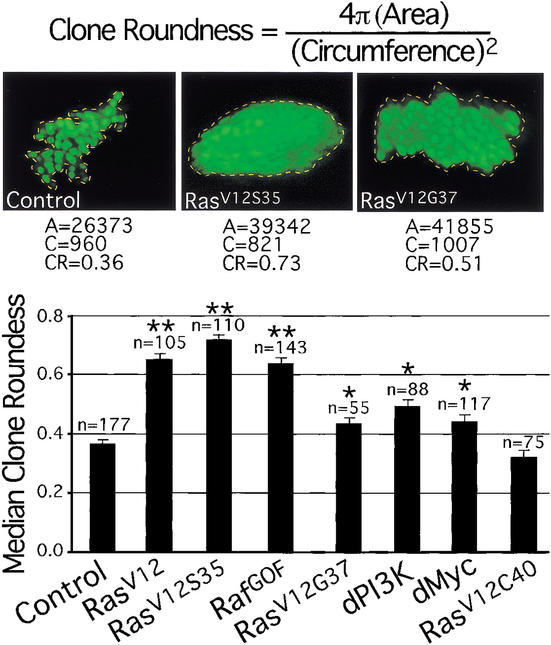
Clones expressing RasV12 are round with smooth borders (Prober and Edgar 2000), suggesting that they have altered cell affinities, and, as a result, minimize contact with neighboring wild-type cells. These clones also often generate ectopic folds in the tissue and bulge out of the apical surface of the epithelium. This contrasts with control clones expressing GFP alone, which are randomly shaped and intermingle with neighboring cells. To determine which Ras effector pathway is responsible for this phenotype, we measured clone roundness by comparing the areas of clones with their circumferences, using a formula that produces a value of 1 for a perfect circle (see Materials and Methods). Clones expressing RasV12, RafGOF, or RasV12S35 were all visibly round and had values of ∼0.7, whereas control clones expressing GFP alone and clones expressing RasV12C40 had significantly lower values of ∼0.35 (Fig. 8). Clones expressing either RasV12G37, dPI3K, or dMyc were not visibly round, did not generate ectopic folds, and did not bulge out of the epithelium. These clones had values of ∼0.45 (Fig. 8). This small, but significant increase in clone roundness values compared with controls resulted from a more compact arrangement of cells within the clones. These results indicate that the Raf/MAPK pathway is primarily responsible for the effect of RasV12 on cell adhesion. Furthermore, the inability of overexpressed dMyc to produce round clones or induce precocious Elav expression shows that regulation of dMyc levels and cell identity are separate effects of Raf/MAPK signaling. Interestingly, the round clone phenotype of RasV12, RafGOF, and RasV12S35 was associated with increased levels and mislocalization of DE-Cadherin/Shotgun (Shg) and β-catenin/Armadillo (Arm), whereas transgenes that did not produce round clones had no effect on Shg or Arm (D.A. Prober, unpubl.). Shg and Arm mediate intercellular adhesion, suggesting a potential mechanism for the effect of Ras/Raf/MAPK signaling on cell affinity. We therefore conclude that Ras affects cell identities via Raf/MAPK and not via dPI3K.
Figure 8.
Ras affects clone shape via the Raf/MAPK pathway. Median clone roundness values of Flp/Gal4 clones expressing various transgenes are shown. Expression of any transgene except RasV12C40 significantly increases clone roundness relative to control clones expressing GFP alone, although RasV12, RasV12S35, and RafGOF have significantly stronger effects than RasV12G37, dPI3K, and dMyc. Representative images and measurements are shown. Clones were induced at 48 h after egg deposition (AED) and analyzed at 120 h AED. A, clone area; C, clone circumference; CR, clone roundness; n, number of clones measured. Error bars: standard error of the mean; *, P < 0.001 vs. control; **, P ≪ 0.001 vs. control.
Lack of cross-talk between dEGFR- and dInr-dependent signaling
We next assayed cross-talk between the dEGFR/Ras and dInr/dPI3K pathways at the level of the receptors. Expression of an activated form of the Drosophila EGF receptor (dEGFRλtop) resulted in elevated levels of MAPK activity (data not shown) and dMyc protein (Supplemental Fig. 5A in Supplementary Material at http://www.genesdev.org) in wing discs, and induces precocious Elav expression in the eye (Dominguez et al. 1998). In contrast, dEGFRλtop had no detectable effect on dPI3K signaling (Supplemental Fig. 5B in Supplementary Material at http://www.genesdev.org). Overexpressed dInr strongly recruited tGPH throughout the cell membrane (Supplemental Fig. 5D in Supplementary Material at http://www.genesdev.org; data not shown), but did not activate MAPK (data not shown), increase dMyc levels (Supplemental Fig. 5C in Supplementary Material at http://www.genesdev.org), or affect Elav expression in the eye (data not shown). Thus, we found no evidence for cross-talk between these pathways at the receptor level.
Discussion
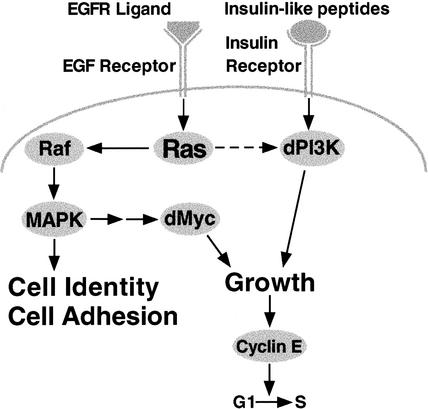
Recent studies using the developing Drosophila wing have shown that Ras, dMyc, and dPI3K regulate rates of cellular growth (i.e., mass accumulation) and progression through the G1/S transition of the cell cycle without affecting overall rates of cell division (Johnston et al. 1999; Weinkove et al. 1999; Prober and Edgar 2000). These studies concur with experiments in mice showing that Ras, Myc, and PI3K promote cell growth without affecting rates of cell division (Iritani and Eisenman 1999; Heumann et al. 2000; Kim et al. 2000; Shioi et al. 2000). Here, we show that an activated form of Drosophila Ras (RasV12) is capable of increasing dMyc protein levels as well as levels of dPI3K signaling, suggesting that RasV12 drives growth and G1/S progression via both of these mechanisms. We also use RasV12 effector loop mutants to show that RasV12 affects dMyc and dPI3K signaling via separate pathways, and we show that overexpressed dMyc and dPI3K do not cross-regulate each other. Thus, we have established a hierarchy for these growth-regulatory proteins (Fig. 9).
Figure 9.
Model for interactions between Ras, dMyc, and dPI3K in the developing Drosophila wing. Ectopic expression of RasV12 drives cell growth via at least two genetically separable pathways. RasV12 activates Raf/MAPK signaling, which increases levels of dMyc protein. RasV12 also independently activates dPI3K signaling. The resulting increased rate of cell growth increases cyclin E protein levels, thereby promoting G1/S progression (Prober and Edgar 2000). Ras is normally activated by the binding of ligands to the EGF receptor, and is required to maintain normal levels of dMyc protein but not dPI3K signaling (dashed arrow). dPI3K is likely normally regulated by the binding of insulin-like peptides to the Insulin receptor. RasV12 also affects cell identity and adhesion via Raf/MAPK signaling. Arrows indicate genetic interactions and do not imply direct molecular interactions.
Ras is required to maintain normal dMyc levels during development
Wing disc cells lacking ras had reduced levels of dMyc protein, indicating that Ras is required to maintain normal dMyc protein levels during wing development. ras−/− cells contained significant levels of dMyc protein, however, indicating that Ras is not absolutely necessary for dMyc expression, and suggesting that reduced dMyc levels may not fully explain the growth deficit of ras−/− cells. However, we found that dMyc antibody staining intensity was ∼40% lower for dmycP0 or dmycP1 homozygotes than for dmycP0 heterozygotes in regions of the wing disc that normally contain high dMyc levels (i.e., wing pouch and notum; Supplemental Table 1 in Supplementary Material at http://www.genesdev.org). Because dmycP0/P0 clones have severely reduced growth rates (Johnston et al. 1999), it seems reasonable to expect that the ∼20% reduction of dMyc levels in ras−/− clones will also reduce growth rates. We showed previously that RasV12 increases dMyc levels post-transcriptionally (Prober and Edgar 2000), and studies in mammalian cell culture showed that RasV12 stabilizes Myc protein (Sears et al. 2000). Therefore, it is likely that ras−/− cells still transcribe dmyc mRNA, but that following translation, dMyc protein is less stable. What other mechanisms may regulate dMyc levels? Wingless (Wg) signaling represses dmyc expression along the dorsal-ventral boundary of the developing wing (Johnston et al. 1999). In addition, expression of an activated version of the Decapentaplegic (Dpp) receptor Thickveins (TkvQ238D) can increase levels of dMyc protein in the wing, whereas loss of this same receptor suppresses dMyc levels (C. Martin-Castellanos and B.A. Edgar, unpubl.). Thus, Ras signaling may be one of many inputs affecting dMyc expression in the wing. Ras may stabilize the low levels of dMyc protein observed throughout the developing wing and/or refine the patterned dmyc expression regulated by other signals. The complex regulation of dMyc expression in vivo may account for the lack of a clear correspondence between patterns of high endogenous Ras activity and dMyc expression (Supplemental Fig. 4 in Supplemental Materials at http://www.genesdev.org).
Ras is not required to maintain normal dPI3K-signaling levels
We found that overexpressed Drosophila RasV12 recruited the tGPH reporter to the cell membrane, suggesting that RasV12 activates dPI3K signaling, and thereby increases PIP3 levels, in the developing wing. We infer that Drosophila RasV12 directly activates dPI3K, because mammalian studies have shown that RasV12 can directly bind and activate PI3K (Rodriguez-Viciana et al. 1994; Pacold et al. 2000). Alternatively, Drosophila RasV12 may activate dPI3K signaling via other mechanisms, such as by inhibiting the lipid phosphatase dPTEN. This possibility seems less likely, however, as direct interactions between Ras and PTEN have not been described. Contradicting the generally accepted idea that PI3K is normally an effector of Ras signaling (Rommel and Hafen 1998), we found that localization of the PI3K reporter tGPH was not detectably affected in ras−/− cells. Although our observations using the tGPH reporter were not quantitative, and small effects could have been missed, these results nevertheless indicate that Ras does not normally play a major role in regulating dPI3K in the developing wing. Consistent with this hypothesis, expression of an activated form of dEGFR (dEGFRλtop) had no effect on dPI3K signaling (Supplemental Fig. 5B in Supplementary Material at http://www.genesdev.org). Because the ability of dEGFRλtop to activate downstream pathways is limited by the amount of endogenous Ras, this result suggests that higher levels of Ras activity than can be generated in wild-type cells are required to activate dPI3K.
An alternative explanation for the discrepancy between the involvement of Drosophila and mammalian Ras in regulating PI3K signaling may relate to the evolution of ras genes. The Drosophila and C. elegans Ras homologs are more homologous to mammalian K-Ras than to H- or N-Ras (Neuman-Silberberg et al. 1984; Han and Sternberg 1990), suggesting that K-Ras may have an older, more general function than the other mammalian ras genes. In support of this idea, H- and N-Ras are dispensable, whereas K-Ras is essential, for normal mouse development (Umanoff et al. 1995; Johnson et al. 1997; Koera et al. 1997). It is also interesting to note that overexpressed K-Ras preferentially activates Raf over PI3K, whereas the opposite is true for H-Ras (Yan et al. 1998). Thus, K-Ras may play a more fundamental role in developmental processes dependent on Raf, but independent of PI3K, whereas H- and N-Ras may have evolved to perform less critical functions in which they regulate PI3K.
Regulation of dPI3K signaling in the developing wing
Using the tGPH reporter, we found that levels of dPI3K signaling are not patterned but rather are uniform throughout wing development (Supplemental Fig. 4 in Supplementary Material at http://www.genesdev.org). It is therefore unlikely that dPI3K signaling is regulated by localized patterning signals such as the morphogens Vein, Dpp, and Wg, which are secreted from the notum, anterior-posterior boundary, and dorsal-ventral boundary of the wing, respectively, and are thought to pattern growth and cell proliferation of the wing. Furthermore, cell-autonomous activation of Dpp signaling using an activated form of its receptor (TkvQ253D), which is a potent growth driver in the wing (Martin-Castellanos and Edgar 2002), has no effect on tGPH localization (C. Martin-Castellanos and B.A. Edgar, unpubl.). It may be that Dpp and Wg regulate cell growth rates by affecting the ability of cells to respond to ubiquitous dPI3K-dependent growth signals. They may do so by regulating the expression or activity of signaling proteins or transcription factors required for transducing dPI3K-dependent signals.
The tGPH reporter revealed that the polarized epithelial cells of Drosophila wing discs contain dense regions of tGPH colocalized with Armadillo at the apical region of the cell membrane, with lower tGPH levels present throughout the basolateral cell membrane (Fig. 2A). This does not simply reflect an apical accumulation of membrane microdomains enriched in PIP3 in polarized cells, because inhibiting dPI3K activity by expressing Δp60 dramatically reduced apical tGPH fluorescence (Figs. 1F, 2D). In contrast, tGPH is uniformly localized throughout the cell membranes of unpolarized Drosophila fat body cells in vivo and Drosophila S2 cells in culture (Britton et al. 2002). Similarly, mammalian PI3K is uniformly active throughout the cell membrane of cultured HEK 293 cells (Gray et al. 1999). Thus, the dynamics of dPI3K signaling are dependent on the cellular context, which is likely disturbed when tissues are dissociated into single cells that are studied in culture. This process may allow signaling interactions not normally occurring in vivo (Simons and Toomre 2000). In support of this idea, overexpression of the Drosophila Insulin receptor homolog (dInr) did not activate MAPK in the developing wing (Table 1) or affect Ras-mediated cell fate specification in the developing eye (Table 1; Brogiolo et al. 2001), whereas addition of insulin to cultured Drosophila or mammalian cells does activate Ras/MAPK signaling (Biggs and Zipursky 1992; Saltiel and Kahn 2001). Alternatively, our failure to detect activation of Ras signaling in response to overexpressed dInr may reflect a cell-type specificity for this interaction or insufficient sensitivity of our assays. It will therefore be interesting to compare the subcellular localization of PI3K signaling complexes in cultured mammalian cells with the tissues from which they are derived.
Table 1.
Summary of effects resulting from expression of UAS-regulated transgenes
| UAS-transgene
|
Increased cell size
|
Truncated G1
|
Increased clone area
|
Active MAPK
|
Increased dMyc
|
Active dPI3K
|
Round clones
|
Ectopic elav
|
|---|---|---|---|---|---|---|---|---|
| RasV12 | + | + | + | + | + | + | + | + |
| RasV12S35 (MAPK-specific) | + | + | + | + | + | ± | + | + |
| RafGOF (MAPK-specific) | + | + | + | + | + | − | + | + |
| RasV12G37 (dPI3K-specific) | + | + | + | − | − | + | ± | − |
| dPI3K | + | + | + | − | − | + | ± | − |
| dInr | +a | +a | +a | − | − | + | ±a | − |
| dMyc | + | + | + | − | + | − | ± | − |
| dEGFRλtop | +b | +b | N.D. | + | + | − | + | +c |
| RasV12C40 | − | − | − | − | − | − | − | − |
A. de la Cruz and B.A. Edgar, unpubl.
K. Plow and B.A. Edgar, unpubl.
N.D., Not done.
dPI3K signaling is thought to be regulated by a family of secreted Drosophila insulin-like peptides (dilps) that bind and activate dInr (Brogiolo et al. 2001; Rulifson et al. 2002). Brogiolo et al. (2001) reported that dilp2 is ubiquitously expressed in imaginal tissues, whereas dilp2 and other dilp family members are expressed in a variety of larval tissues including the gut and neurosecretory cells in the brain. dilp2 that is expressed in imaginal tissues is likely secreted apically into the lumen between cells of the columnar epithelium and the overlying peripodial membrane. This would result in preferential binding of dIlp2 to dInr at the apical region of the cell, which could account for the high levels of apically localized tGPH that we observed (Fig. 2A). Alternatively, apical dPI3K signaling may reflect a local concentration of dPI3K-signaling complexes. Consistent with the latter possibility, RasV12 only recruited tGPH to apical regions of the cell membrane. This result suggests that RasV12 may require other apically localized factors to activate dPI3K signaling. This possibility is supported by our finding that coexpressed Δp60, which prevents dPI3K from interacting with upstream activators, blocked RasV12-mediated activation of dPI3K signaling, as it does in mammals (Rodriguez-Viciana et al. 1997). Because mammalian Ras can directly bind the catalytic subunit of PI3K (Rodriguez-Viciana et al. 1994; Pacold et al. 2000), we infer that coexpressed Δp60 should not affect the ability of Drosophila Ras to activate dPI3K signaling unless Ras-dependent activation requires other apically localized factors that bind Δp60. These factors may include the Insulin receptor substrate Chico, dInr itself or other receptor tyrosine kinases, G-protein coupled receptors, or components of signaling complexes that are recruited upon activation of these receptors. Several receptor tyrosine kinases, including dEGFR, as well as phosphotyrosine-containing proteins, are concentrated at the apical cell surface (Woods et al. 1997; C. Delorenzo and P. Bryant, pers. comm.), although dInr is distributed throughout the cell membrane (Supplemental Fig. 6A in Supplementary Material at http://www.genesdev.org). The Drosophila homolog of the heterotrimeric G-protein subunit Gαi (Schaefer et al. 2001), which presumably transduces signals from a large family of associated receptors, is also concentrated apically in wing disc cells (Supplemental Fig. 6B,C in Supplementary Material at http://www.genesdev.org). This is consistent with the possibility that heterotrimeric G-proteins may regulate dPI3K signaling in Drosophila, as they do in mammals (Vanhaesebroeck et al. 2001).
Implications for cancer
Much of our understanding of Ras function, and that of most oncogenes, derives from studies in homogenous cell culture systems. These studies have focused primarily on cell-autonomous effects of oncogenes rather than upon the roles of interactions among cells within tissues in tumor development. Tissue homeostasis is maintained by a continuous exchange of signals between cells, the extracellular matrix, and the local environment. An important feature of tumor development is escape from this regulation, initially allowing the autonomous growth and proliferation of tumor cells, and eventually resulting in altered adhesion and migration of tumor cells away from their site of origin. The behavior of clones of cells with elevated Raf/MAPK signaling levels in developing Drosophila epithelia is strikingly similar to that of tumor cells within mammalian tissues. These cells have altered adhesive properties and cell identities, and as a result minimize contact with neighboring wild-type cells. In contrast, dPI3K and dMyc do not regulate cell identity or adhesion (Figs. 7, 8). Studies in Drosophila and vertebrates have also suggested that even though both dMyc and dPI3K stimulate growth, they appear to do so via different mechanisms. dPI3K signaling promotes nutrient import and storage (Saltiel and Kahn 2001), whereas dMyc promotes nucleolar growth and protein synthesis (Iritani and Eisenman 1999; Britton, 2000; Coller et al. 2000; Guo et al. 2000; Boon et al. 2001). Thus, the ability of RasV12 to up-regulate both of these pathways may generate a more robust and balanced growth response than activation of either dMyc or dPI3K alone. Furthermore, the ability of RasV12 to deregulate cell identity and adhesion may underlie the strong synergy between Ras and other growth-promoting oncogenes in vivo (Land et al. 1983; Sinn et al. 1987).
Materials and methods
Fly strains
All transgenes are P[+] in w− strains. hs-FLP122; +; UAS-RasV12 (Karim and Rubin 1998); hs-FLP122; +; UAS-RasV12S35 (Karim and Rubin 1998); hs-FLP122; +; UAS-RasV12G37 (Karim and Rubin 1998); hs-FLP122; +; UAS-RasV12C40 (Karim and Rubin 1998); hs- FLP122; +; UAS-RafGOF (Brand and Perrimon 1994); hs-FLP122; +; UAS-dMyc (Zaffran et al. 1998); hs-FLP122; +; UAS-dPI3K (Dp110; Leevers et al. 1996); hs-FLP122; +; UAS-Δp60 (Weinkove et al. 1999); hs-FLP122; +; UAS-dInr (Huang et al. 1999); hs- FLP122; +; UAS-dRal20V (Sawamoto et al. 1999); UAS-EGFRλtop; +; + (Queenan et al. 1997); w; +; Actin 5c>CD2>Gal4, UAS-GFPNLS (Pignoni and Zipursky 1997; Neufeld et al. 1998); w; tGPH, Actin 5c>CD2>Gal4 (Britton et al. 2002); w; UAS-P35; + (Hay et al. 1994); w; +; FRT (82B) rasc40b/TM6B (Schnorr and Berg 1996); w; +; FRT (82B) M(3)95A Ub-GFP/TM6B (Andersson et al. 1994); w; +; FRT (82B) hs-πmyc; w; hs-FLP7A; +; and w; en-Gal4; +.
Flow cytometry and clonal growth rate analysis
Clones of cells overexpressing various UAS-regulated transgenes were generated using the Flp/Gal4 method (Struhl and Basler 1993; Pignoni and Zipursky 1997; Neufeld et al. 1998). Larvae were staged from hatching and raised at a density of 50 per vial at 25°C. For FACS analysis (Neufeld et al. 1998; Prober and Edgar 2000), Flp/Gal4 clones were induced by a 1-h heat shock at 37°C at 72 h after egg deposition (AED), resulting in ∼50% GFP+ cells, and wing discs were analyzed at 120 h AED. Approximately 20,000 GFP+ and GFP− cells were analyzed for each genotype in each experiment. For clone area measurements, ∼5–10 clones/disc were induced by a 30-min heat shock at 37°C at 48 AED and measured at 120 h AED. Samples were blinded for analysis. Clone areas were measured using the histogram function of Adobe Photoshop. P values were calculated using a two-tailed student's t-test. All samples and controls were tested in parallel.
Clone shape measurements
Clone shapes were measured using the formula 4πA/L2, in which A = clone area and L = clone circumference (Lawrence et al. 1999). By use of this formula, a perfect circle has a value of 1 and more irregular shapes have values <1. Samples were blinded for analysis, which was performed using NIH Image. P values were calculated using a two-tailed student's t-test.
Mitotic recombination
Mitotic recombination was induced using the FLP/FRT method (Xu and Rubin 1993) by a 45-min heat shock at 48 h AED, and wing discs were analyzed at 120 h AED. Larvae were heat shocked for 1 h immediately prior to dissection when the πmyc clonal marker was used. en–Gal4 was used to express P35 in posterior wing compartments to increase the size of rasc40b clones (Prober and Edgar 2000).
Immunocytochemistry
Imaginal discs were fixed in 4% paraformaldehyde/PBS for 20 min at 37°C. All washes and antibody incubations were performed in 0.1% Tween-20/PBS. Primary antibodies used were mouse anti-dMyc (P4C4 B10, Prober and Edgar 2000, undiluted), mouse anti-CD2 (Serotec, 1:500), mouse anti-Armadillo (N2 7A1, Developmental Studies Hybridoma Bank, 1:100), mouse anti-dpERK (Sigma, 1:200), mouse anti-c-Myc (Calbiochem, 1:50), and rat anti-Elav (DSHB, 1:30). Alexa 488, 568, and 660-conjugated secondary antibodies (Molecular Probes) were used at 1:1500. Tissues were permeabilized for 30 min in 0.3% Triton X-100/PBS prior to incubation with dMyc- and Elav-specific antibodies. Discs were mounted in Fluoroguard (Bio-Rad) and imaged on a Leica TCS SP confocal microscope. Cross-section images were obtained using double-stick tape as a spacer between the slide and coverslip and the XZ function of the Leica software.
dMyc antibody quantitation
We measured pixel intensities in 10–15 small regions within each of 41 clones and compared intensities in each ras−/− region to a neighboring wild-type region. The ratio of average pixel intensity ras−/−/average pixel intensity ras+/− or ras+/+ was calculated for each region, and the average ratio was calculated for each clone. The regions were selected to evenly sample pixel intensities throughout each clone. Because each region was relatively small, endogenous dMyc levels and tissue folds were usually similar for a region both inside and outside of the clone, thereby minimizing confounding bias due to these variables. There was no significant difference between ras+/− and ras+/+ average pixel intensities. Pixel intensities were measured using the histogram function of Adobe Photoshop.
Acknowledgments
We thank Wendy Lockwood and Steve Cohen for providing us with the tGPH stock prior to publication. We also thank Celeste Berg, Ulrike Gaul, Ernst Hafen, Felix Karim, Sally Leevers, Hideyuki Okano, and Trudy Schupbach for fly stocks; Robert Eisenman, Rafael Fernandez, Peter Gallant, Juergen Knoblich, and Cynthia Yost for antibodies; Aida de la Cruz, Cristina Martin-Castellanos, Lenora Loo, Cecilia De Lorenzo, Amir Oryan, Kelly Plow, and Cynthia Yost for sharing unpublished data; Celeste Berg, Robert Eisenman, Savraj Grewal, and Tom Neufeld for comments on the manuscript; and members of the Edgar laboratory for helpful discussions during the course of this work. D.A.P. was a predoctoral fellow of the National Science Foundation and the Molecular Training Program in Cancer Research. Supported by NIH GMS R01–51186.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bedgar@fhcrc.org; FAX (206) 667-3308.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.991102.
References
- Andersson S, Saeboe-Larssen S, Lambertsson A, Merriam J, Jacobs-Lorena M. A Drosophila third chromosome Minute locus encodes a ribosomal protein. Genetics. 1994;137:513–520. doi: 10.1093/genetics/137.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Biggs WH, III, Zipursky SL. Primary structure, expression, and signal-dependent tyrosine phosphorylation of a Drosophila homolog of extracellular signal-regulated kinase. Proc Natl Acad Sci. 1992;89:6295–6299. doi: 10.1073/pnas.89.14.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes & Dev. 1994;8:629–639. doi: 10.1101/gad.8.5.629. [DOI] [PubMed] [Google Scholar]
- Britton JS. “Genetic and environmental control of growth and the cell cycle during larval development of Drosophila melanogaster.” Ph.D. Thesis. Seattle, WA: University of Washington; 2000. [Google Scholar]
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gray A, Van Der Kaay J, Downes CP. The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem J. 1999;344 Pt 3:929–936. [PMC free article] [PubMed] [Google Scholar]
- Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]
- Han M, Sternberg PW. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Heumann R, Goemans C, Bartsch D, Lingenhohl K, Waldmeier PC, Hengerer B, Allegrini PR, Schellander K, Wagner EF, Arendt T, et al. Transgenic activation of Ras in neurons promotes hypertrophy and protects from lesion-induced degeneration. J Cell Biol. 2000;151:1537–1548. doi: 10.1083/jcb.151.7.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- Hunter JJ, Tanaka N, Rockman HA, Ross J, Jr, Chien KR. Ventricular expression of a MLC-2v-ras fusion gene induces cardiac hypertrophy and selective diastolic dysfunction in transgenic mice. J Biol Chem. 1995;270:23173–23178. doi: 10.1074/jbc.270.39.23173. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes & Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Rodriguez-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus- mediated expression of c-Myc in vivo. Proc Natl Acad Sci. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi T, Katagiri K, Watanabe S, Vanhaesebroeck B, Downward J, Takatsu K. Distinct mechanisms of alpha 5β 1 integrin activation by Ha-Ras and R-Ras. J Biol Chem. 2000;275:22590–22596. doi: 10.1074/jbc.M000633200. [DOI] [PubMed] [Google Scholar]
- Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Casal J, Struhl G. The hedgehog morphogen and gradients of cell affinity in the abdomen of Drosophila. Development. 1999;126:2441–2449. doi: 10.1242/dev.126.11.2441. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development. 1999;126:5739–5747. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Morrison DK, Cutler RE. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schejter E, Hoffmann FM, Shilo BZ. The Drosophila ras oncogenes: Structure and nucleotide sequence. Cell. 1984;37:1027–1033. doi: 10.1016/0092-8674(84)90437-9. [DOI] [PubMed] [Google Scholar]
- Oatey PB, Venkateswarlu K, Williams AG, Fletcher LM, Foulstone EJ, Cullen PJ, Tavare JM. Confocal imaging of the subcellular distribution of phosphatidylinositol 3,4,5-trisphosphate in insulin- and PDGF-stimulated 3T3-L1 adipocytes. Biochem J. 1999;344 Pt 2:511–518. [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Provot S, Felder-Schmittbuhl MP, Calothy G, Eychene A. Induction of postmitotic neuroretina cell proliferation by distinct Ras downstream signaling pathways. Mol Cell Biol. 2000;20:7068–7079. doi: 10.1128/mcb.20.19.7068-7079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Prober DA, Edgar BA. Ras1 promotes cellular growth in the Drosophila wing. Cell. 2000;100:435–446. doi: 10.1016/s0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- ————— Growth regulation by oncogenes–new insights from model organisms. Curr Opin Genet Dev. 2001;11:19–26. doi: 10.1016/s0959-437x(00)00151-9. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Rommel C, Hafen E. Ras–a versatile cellular switch. Curr Opin Genet Dev. 1998;8:412–418. doi: 10.1016/s0959-437x(98)80111-1. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Winge P, Koyama S, Hirota Y, Yamada C, Miyao S, Yoshikawa S, Jin MH, Kikuchi A, Okano H. The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH(2)-terminal kinase pathway. J Cell Biol. 1999;146:361–372. doi: 10.1083/jcb.146.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schnorr JD, Berg CA. Differential activity of Ras1 during patterning of the Drosophila dorsoventral axis. Genetics. 1996;144:1545–1557. doi: 10.1093/genetics/144.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T, Kang PM, Douglas PS, Hampe J, Yballe CM, Lawitts J, Cantley LC, Izumo S. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 2000;19:2537–2548. doi: 10.1093/emboj/19.11.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: Synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Umanoff H, Edelmann W, Pellicer A, Kucherlapati R. The murine N-ras gene is not essential for growth and development. Proc Natl Acad Sci. 1995;92:1709–1713. doi: 10.1073/pnas.92.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- Woods DF, Wu JW, Bryant PJ. Localization of proteins to the apico-lateral junctions of Drosophila epithelia. Dev Genet. 1997;20:111–118. doi: 10.1002/(SICI)1520-6408(1997)20:2<111::AID-DVG4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Chartier A, Gallant P, Astier M, Arquier N, Doherty D, Gratecos D, Semeriva M. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development. 1998;125:3571–3584. doi: 10.1242/dev.125.18.3571. [DOI] [PubMed] [Google Scholar]
- Zhong J, Troppmair J, Rapp UR. Independent control of cell survival by Raf-1 and Bcl-2 at the mitochondria. Oncogene. 2001;20:4807–4816. doi: 10.1038/sj.onc.1204614. [DOI] [PubMed] [Google Scholar]