Abstract
The basal eudicot Aquilegia (columbine) has an unusual floral structure that includes two morphologically distinct whorls of petaloid organs and a clearly differentiated fifth organ type, the staminodium. In this study, we have sought to determine how Aquilegia homologs of the B class genes APETALA3 (AP3) and PISTILLATA (PI) contribute to these novel forms of organ identity. Detailed expression analyses of the three AP3 paralogs and one PI homolog in wild-type and floral homeotic mutant lines reveal complex patterns that suggest that canonical B class function has been elaborated in Aquilegia. Yeast two-hybrid studies demonstrate that the protein products of Aquilegia's AP3 and PI homologs can form heterodimers, much like what has been observed for their core eudicot homologs. Downregulation of AqvPI using virus-induced gene silencing indicates that in addition to petal and stamen identity, this locus is essential to staminodial identity but may not control the identity of the petaloid sepals. Our findings show that preexisting floral organ identity programs can be partitioned and modified to produce additional organ types. In addition, they indicate that some types of petaloid organs are not entirely dependent on AP3/PI homologs for their identity.
INTRODUCTION
Among the most striking features of the ABC model of floral organ identity is its streamlined simplicity: three functionally overlapping classes of gene activity establish the identity of four organ types. The basic formula holds that A function alone determines sepal identity, A+B determines petal identity, B+C determines stamen identity, and C alone determines carpel identity (Coen and Meyerowitz, 1991). Many aspects of this model have been shown to be evolutionarily conserved, particularly the functions of the B and C gene homologs (reviewed in Becker and Theissen, 2003). To some degree, these findings reflect the fact that variation in most aspects of floral morphology, such as organ number, color, size, and symmetry, are likely to be due to genetic changes downstream of or in parallel to the establishment of organ identity (reviewed in Kramer, 2005a). There are some flowers, however, that appear to exhibit variation in fundamental aspects of floral organ identity, suggesting that their ABC programs may be more complex. In particular, although true petals occur by definition in the second whorl of a bipartite perianth, the general phenomenon of petaloidy can occur in many different positions within the flower (Albert et al., 1998; Kramer and Jaramillo, 2005). Complicating this spatial flexibility, no single diagnostic character can be used to universally define petaloidy. Features often associated with petaloidy include bright coloration and the common presence of papillated epidermal cells, which are important for color reflectance (Endress, 1994; Christensen and Hansen, 1998; Martin et al., 2002). Perhaps the most straightforward candidate for spatial redeployment of petal identity is the entirely petaloid perianth of many monocots or magnoliid dicots, which could potentially be explained by a simple outward shift in the B gene expression domain (Kanno et al., 2003; Nakamura et al., 2005). Flowers such as those of Lilium have two whorls of petaloid organs that are highly similar to one another at maturity and could conceivably be produced by expression of the same identity program in multiple whorls of the flower. By contrast, flowers like those of Aquilegia (columbine) or Fuschia, for example, have petaloid sepals in the first whorl that are morphologically distinct from the true petals that arise in the second whorl. Furthermore, these different types of petaloid organs are found in actinomorphic (radially symmetric) flowers and are not due to variation in floral symmetry, as in Antirrhinum majus, where the dorsal petal is quite different in shape and size from the lateral and ventral petals (all of which are positioned in the second whorl) (Luo et al., 1999). It would appear, then, that flowers with two whorls of morphologically distinct petaloid organs must possess a genetic mechanism, presumably acting at the level of organ identity, whereby these types can be distinguished.
An even more dramatic example of novelty in floral organ identity is the evolution of fifth organ types. One commonly observed type of fifth organ is staminodia, which represent stamens that have become sterilized and otherwise modified. The term staminodia is applied to a wide variety of organ types, ranging from simple aborted stamens (e.g., in the context of bilateral symmetry or sex determination) to organs that have acquired elaborate novel functions and morphologies (reviewed in Walker-Larsen and Harder, 2000). Although the presence of staminodia is relatively rare, they are broadly distributed across the angiosperms and reflect what is considered to be a general tendency to transform parts of the androecial whorl into novel organ types (Takhtajan, 1991). In many cases, staminodia are involved in pollination-related processes and are well represented in several major angiosperm lineages, including gingers and orchids. These types of staminodia can be found in the inner or outer whorls of the androecium and sometimes represent morphological grades, such as the transitional organ types that can be found between petals and fertile stamens (Buzgo et al., 2004). In other instances, they are completely distinct in their morphology. When such staminodia appear in the context of zygomorphy and do not occupy an entire whorl, it is possible that the new morphology reflects the activity of a genetic pathway acting in parallel to the ABC program (e.g., CYCLOIDEA in Antirrhinum; reviewed in Cubas, 2002). However, in several cases, morphologically distinct staminodia do occupy an entire whorl of a radially symmetric flower, which would seem to indicate a modification of the ABC program itself via the evolution of a new identity program. In this regard, the lodicule of grasses, which represents a modified petal, stands as precedent for the derivation of a novel organ type from a preexisting organ identity program (Ambrose et al., 2000; Nagasawa et al., 2003). In that example, however, all of the preexisting petals are transformed into lodicules, while a conversion to staminodial identity typically affects only part of the androecium, resulting in five organ types.
The B class genes are implicated in cases of both morphologically distinct petaloid sepals and staminodia. In the core eudicots and several grass species, homologs of the Arabidopsis thaliana type II MADS box genes APETALA3 (AP3) and PISTILLATA (PI) have been shown to contribute to the identity of petals (or organs thought to be petal derivatives) and stamens, suggesting that their functions are generally conserved (reviewed in Kramer and Jaramillo, 2005). As a rule, the protein products function as obligate heterodimers between the AP3- and PI-like proteins (Zachgo et al., 1995; Riechmann et al., 1996; Vandenbussche et al., 2004; Whipple et al., 2004), although there are a few potential exceptions (Tzeng and Yang, 2001; Winter et al., 2002; Kanno et al., 2003). Across the angiosperms, all developing stamens and almost all types of petaloid organs express homologs of AP3 and PI (reviewed in Kim et al., 2005; Zahn et al., 2005). Within the core eudicots, expression in petaloid organs tends to be fairly homogeneous and constitutive, while outside of this clade, expression patterns show more variation in both their temporal and spatial aspects (reviewed in Kramer and Irish, 2000). The evolutionary history of the gene lineages is similarly complex, being marked by multiple gene duplication events at many phylogenetic levels (Kramer et al., 1998, 2003; Aoki et al., 2004; Kim et al., 2004; Stellari et al., 2004; Aagaard et al., 2005). There is some evidence that these frequent gene duplication events have played a role in the functional evolution of the AP3 and PI lineages. In the orchid Phalaenopsis, for instance, there are four AP3 paralogs, each of which exhibits a distinct expression pattern that could play a role in distinguishing between the different types of petaloid organs seen in these flowers (Tsai et al., 2004). A series of experiments in the core eudicot Petunia hybrida revealed a more traditional case of subfunctionalization, where a pair of AP3/PI paralogs became stamen-specific in their function (Vandenbussche et al., 2004; Rijpkema et al., 2006). Thus, it is intriguing to consider whether there is any relationship between the common occurrence of AP3/PI gene duplication and the evolutionary lability observed in petaloid and staminoid organs.
In this study, we utilized the emerging genetic model Aquilegia (columbine; Ranunculaceae) to investigate the role that AP3 and PI homologs may play in the development of petaloid sepals and novel staminodia. Aquilegia is the subject of genetic and genomic studies focusing on both ecological and evolutionary questions, and a number of tools are available or under development. These include a large EST data set (The Institute for Genomic Research Aquilegia Gene Index; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=aquilegia), a physical genome map (Clemson University Genomics Institute Aquilegia Physical Map; www.genome.clemson.edu/projects/aquilegia/), genome sequencing (Joint Genome Institute, Community Sequencing Program FY2007; www.jgi.doe.gov/sequencing/why/CSP2007/aquilegia.html), and virus-induced gene silencing (VIGS) approaches (B. Gould, K. Fifer, and E.M. Kramer, unpublished data). Among the many interesting features of Aquilegia is its distinct floral morphology with five types of floral organs: petaloid sepals, second whorl petals distinguished by large nectar spurs, stamens, staminodia, and carpels (Figure 1). Previous studies of AP3 and PI homologs in the Ranunculaceae revealed that the lineages experienced multiple duplication events both predating and during the diversification of the family (Kramer et al., 2003). Most notably, before the radiation of the Ranunculaceae, the AP3 lineage underwent two duplication events that gave rise to three distinct AP3 types. It is important to note that these events are independent of the AP3 duplication that occurred close to the base of the core eudicots; thus, all of the Ranunculaceae AP3-like genes are members of what is termed the paleo-AP3 lineage (Kramer et al., 1998). Aquilegia possesses all three of these AP3 paralogs as well as one PI homolog. RT-PCR of dissected floral organs in Aquilegia found that two of the AP3 paralogs and the PI homolog are expressed in the petaloid sepals as well as in the petals and stamens, while the third AP3 is restricted to the petals and stamens. In order to better understand these patterns, we have used in situ hybridization to obtain a detailed picture of AP3 and PI expression throughout wild-type floral development. Furthermore, we have characterized the function of these genes using yeast two-hybrid analysis, studies of an Aquilegia floral homeotic mutant, and VIGS of AqvPI. Collectively, these findings demonstrate that B function in Aquilegia encompasses the novel staminodia as well as the petals and stamens. In the case of the petaloid sepals, it does not appear that AP3/PI homologs play a role in their identity, although they may contribute to late aspects of organ maturation.
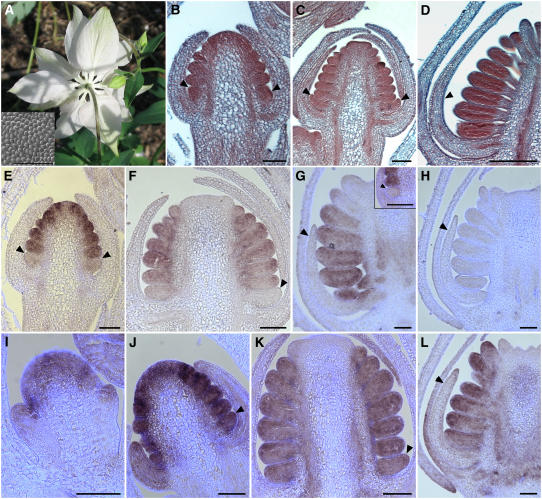
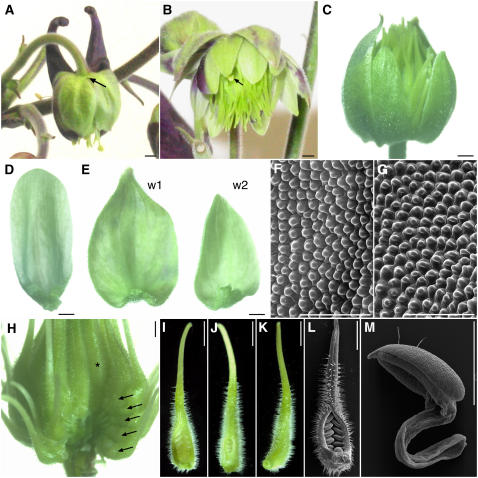
Figure 1.
Floral Morphology of Wild-Type A. vulgaris.
(A), (B), and (D) to (H) Light micrographs.
(I) to (K), (M) to (P), and (R) Scanning electron micrographs.
(L) and (Q) Environmental scanning electron micrographs.
(A) Side view of a flower at anthesis. Two sepals, one petal, and several whorls of stamens were removed to expose the staminodia. c, carpel; p, petal; s, sepal; sta, stamen; std, staminodium.
(B) Head-on view of a flower at anthesis.
(C) Floral diagram showing relative positions of five organ types. Whorls one and two are composed of 5 sepals and 5 petals, respectively, followed by four to seven whorls of 10 stamens, one whorl of 10 staminodia, and an innermost whorl of 5 free carpels.
(D) Sepal at anthesis.
(E) Petal at anthesis.
(F) Stamen close to anthesis.
(G) Staminodium at anthesis. The midvein of the staminodium is marked by arrows.
(H) Carpel at anthesis.
(I) and (N) Scanning electron microscopy images of the abaxial (I) and adaxial (N) surfaces of the sepal.
(J) and (O) Scanning electron microscopy images of the abaxial (J) and adaxial (O) surfaces of the petal.
(K) and (P) Scanning electron microscopy images of the anther (K) and filament (P) of the stamen.
(L) and (Q) Environmental scanning electron microscopy images of the staminodium.
(M) and (R) Scanning electron microscopy images of the ovary wall (M) and style (R) of the carpel. Small spheres in (M) are pollen (p).
Bars = 2 mm in (A), (B), (F), and (G), 2.5 mm in (D), (E), and (H), 50 μm in (I), (J), and (L), 100 μm in (K) and (M) to (P), 250 μm in (Q), and 300 μm in (R).
RESULTS
Wild-Type Floral Morphology of Aquilegia vulgaris
Flowers of Aquilegia are actinomorphic, pentamerous, and hypogynous with all floral parts unfused and arranged in whorls (Figures 1A to 1C) (Munz, 1946; Tucker and Hodges, 2005). There are five organ types, from outermost to innermost: one whorl of 5 sepals, one whorl of 5 petals, four to seven whorls of 10 stamens, one whorl of 10 sterile staminodia, and one whorl of 5 free carpels. Tucker and Hodges (2005) previously described floral development in A. olympica and their findings hold for A. vulgaris (data not shown). We performed additional studies on the mature floral organs of A. vulgaris. At maturity, the sepals are ovate in shape with a narrow base and have a number of characteristics normally associated with petaloid organs (Figure 1D). These include being brightly colored (in most cultivars) and having papillated cell types on the adaxial surface (Figure 1N). The abaxial sepal epidermis is distinguished by irregularly shaped cells and stomata (Figure 1I). The second whorl petals are positioned alternate to the petaloid sepals and are morphologically distinct, most notably due to the presence of a large nectar spur and nectary on each petal (Figure 1E). In contrast with the sepals, there are no conical cells on the epidermis of the petals (Figures 1J and 1O). Both the abaxial and adaxial surfaces contain relatively flat, featureless cells, with the addition of glandular trichomes on the abaxial surface. Internal to the perianth, there are four to seven whorls of stamens, which are composed of a long, narrow filament topped by a small tetrasporangiate anther (Figure 1F). They are arranged in 10 orthostichies (vertical straight rows; Figure 1C), and various elongated cell types are observed on the surfaces of the anther and the filament (Figures 1K and 1P). The number of stamen whorls varies between flowers within the inflorescence, which is cymose (determinate) and highly branched. Earlier developing flowers tend to have six to seven whorls of stamens, while later flowers have four to five (Tucker and Hodges, 2005). Following the stamen whorls, there is one whorl of 10 staminodia. These organs are arranged on the same orthostichies as the stamens (Figure 1C) and have been interpreted as being evolutionarily derived from fertile stamens (Munz, 1946; Tucker and Hodges, 2005). Morphologically, the staminodia are very different from the stamens. In A. vulgaris, they have a prominent central midrib with ruffled laminae extending to either side (Figure 1G). The cell types observed on the epidermis are not similar to those observed on any of the other floral organs, showing varying degrees of elongation and irregular margins (Figures 1L and 1Q). At maturity, the laminae of the staminodia form an interlocking sheath around the developing ovary. Finally, in the innermost whorl of the flower, there are five free carpels (Figure 1H). These exhibit a diversity of epidermal cell types, ranging from narrow, elongated cells on the style to many glandular trichomes and stomata on the ovary wall (Figures 1M and 1R).
In Situ Hybridization of AP3 and PI Homologs in Wild-Type Flowers of A. vulgaris
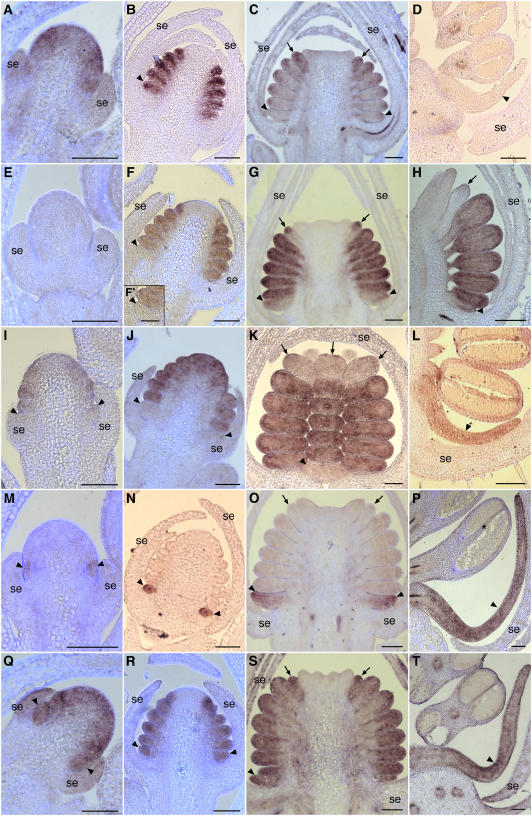
In situ hybridization was used to investigate the expression patterns of the three AP3 homologs and one PI homolog in A. vulgaris. Based on both DNA gel blot hybridization and deep EST sequencing, these appear to be the only AP3/PI homologs present in the Aquilegia genome (Kramer et al., 2003; The Institute for Genomic Research, 2005). Each of the AP3 paralogs exhibits a distinct and complex expression pattern. AqvAP3-1 is first detected in very early floral meristems as the sepal primordia initiate (Figure 2A). This expression domain includes the regions that will give rise to the petals, stamens, and staminodia. Weaker expression is occasionally observed in the sepals and presumptive carpel whorl. As the sepal primordia grow to enclose the floral meristem, AqvAP3-1 remains strongly expressed in the petal, stamen, and incipient staminodium primordia (Figure 2B). Soon after this stage, coincident with the initiation of the carpel primordia, AqvAP3-1 expression begins to decline (Figure 2C). This process starts in the petal primordia and moves acropetally until expression is only detected in the staminodia. During later stages, including the bulk of floral organ differentiation, AqvAP3-1 appears to be expressed at low levels (Figure 2D). The expression of AqvAP3-2 is delayed slightly relative to that of AqvAP3-1 and is not detected until the stage during which the stamen primordia begin to initiate (Figures 2E and 2I). AqvAP3-2 is not expressed in the petal primordia during this early phase, being restricted to the primordia that will give rise to fertile stamens and staminodia (Figures 2I, 2F, and 2J). Persistent expression is observed only in the stamens, however, as transcription appears to decline rapidly in the staminodia during the stage of carpel initiation and elaboration (Figures 2G, 2H, and 2K). Beginning at this same point, the first weak expression of AqvAP3-2 is detected in the petal primordia. This expression is maintained throughout later stages of petal development (Figure 2L). AqvAP3-3 has perhaps the most remarkable expression pattern, being petal-specific throughout the bulk of floral development (Figures 2M to 2P). AqvAP3-3 transcript is first detected after sepal initiation in the region just above the sepal primordia, which presumably gives rise to the petals (Figure 2M). As the petal primordia initiate and differentiate, AqvAP3-3 remains strongly expressed in these organs (Figures 2N to 2P). At very late stages, expression is also sometimes observed in the connective of the stamen (Figures 2P and 3). The single PI homolog, AqvPI, has an expression pattern that encompasses all of the regions where the three AP3 paralogs are observed (Figures 2Q to 2T). Early in floral development, AqvPI is detected throughout the floral meristem (Figure 2Q), but this domain quickly resolves to the petal, stamen, and staminodium primordia (Figure 2R). AqvPI appears to be strongly expressed throughout the development of all of these organs (Figures 2S and 2T).
Figure 2.
In Situ Hybridization of AqvAP3-1, AqvAP3-2, AqvAP3-3, and AqvPI in Wild-Type Floral Meristems.
(A) to (D) AqvAP3-1.
(E) to (L) AqvAP3-2.
(M) to (P) AqvAP3-3.
(Q) to (T) AqvPI.
(A), (E), and (M) show very early-stage floral meristems in which the sepals are just emerging. (I) and (Q) are slightly later, as the petal and stamen primordia are just beginning to initiate. (B), (F), (J), (N), and (R) show an early-stage floral meristem in which the sepals are growing to enclose the floral meristem and the petal, stamen, and staminodium primordia are initiating. (F′) is a magnification of the petal primordium shown in (F). (C), (G), (K), (O), and (S) correspond to the stage at which the carpel primordia are appearing and the petals, stamens, and staminodia are beginning to become morphologically distinct. (D), (H), (L), (P), and (T) show a range of later stages in which the various floral organ types are differentiating. The petals are elongating and beginning to become spurred, microsporogenesis is under way in the stamens, and the staminodia are clearly differentiated from the fertile stamens. (D), (J), (K), and (T) are tangential longitudinal sections, while the others are all radial longitudinal. se, sepals; arrowheads indicate petal primordia, and arrows indicate staminodia. The asterisk in (P) indicates weak AqvAP3-3 expression in the connective. All primordia located between the petals and staminodia are fertile stamens. Bars = 100 μm.
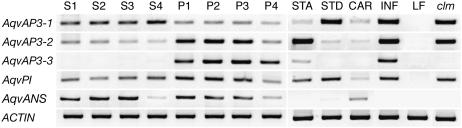
Figure 3.
Locus-Specific RT-PCR of RNA Prepared from Organs Corresponding to Stages S1 to S4 and P1 to P4 as Shown in Supplemental Figures 1 and 2 Online.
Additional reactions were performed using RNA from stamens (STA), staminodia (STD), carpels (CAR), wild-type inflorescences (INF), prevernalization vegetative leaves (LF), and clm mutant inflorescences. Six genes were analyzed: AqvAP3-1, AqvAP3-2, AqvAP3-3, AqvPI, AqvANS, and ACTIN.
Late Developmental Characteristics of the Sepals and Petals
The expression patterns observed using in situ hybridization are intriguing but also somewhat surprising in that consistent expression is not detected for any of the loci in the developing petaloid sepals. This is unexpected, in part because previous studies had found that AqvAP3-1, AqvAP3-2, and AqvPI could be detected by RT-PCR on RNA from dissected sepals (Kramer et al., 2003). In order to test the reproducibility of this result, we prepared RNA samples from four developmental stages of sepals and petals (see Supplemental Figures 1 and 2 online). It is notable that the sepals do not acquire petaloid characteristics until late in their development. Initially, they are photosynthetic, but as the flower undergoes the last phases of maturation, the sepals begin to accumulate anthocyanin and papillated cell types appear on the adaxial surface (see Supplemental Figure 1 online). Petals similarly do not accumulate anthocyanin until later developmental stages but never acquire papillated cells (Figures 1J and 1O; see Supplemental Figure 2 online). The RT-PCR experiments show that AqvAP3-1, AqvAP3-2, and AqvPI are detected in the late-stage sepals but AqvAP3-3 is not (Figure 3). These results confirm other aspects of the in situ hybridization study, particularly the differential expression of AqvAP3-1 and AqvAP3-2 between stamens and staminodia (Figure 3).
Yeast Two-Hybrid Interactions among AP3 and PI Proteins in A. vulgaris
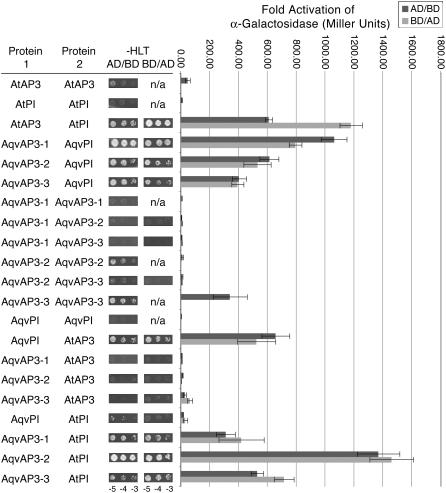
In many other taxa, the products of AP3 and PI homologs have been found to form heterodimers (Zachgo et al., 1995; Riechmann et al., 1996; Vandenbussche et al., 2004; Whipple et al., 2004). There are a few instances, however, of homodimerization, particularly of PI homologs in the monocots (Winter et al., 2002; Kanno et al., 2003). We investigated the dimerization affinities of the AP3 and PI homologs of A. vulgaris using Arabidopsis AP3 and PI as controls. This study found that all three Aquilegia AP3 paralogs could form strong heterodimers with AqvPI, particularly AqvAP3-1 (Figure 4; see Supplemental Figure 3 online). A weak homodimerization interaction was observed for AqvAP3-3 and, less consistently, for AqvAP3-2. No homodimerization interactions were observed for AqvAP3-1 or AqvPI.
Figure 4.
Protein Interactions between AP3 and PI Homologs of A. vulgaris as Determined by Growth on Selective SD Medium and Quantification of Secreted α-Galactosidase Activity (see Supplemental Methods Online).
Aquilegia and Arabidopsis proteins were cloned into both binding domain (BD) and activation domain (AD) constructs. For each protein pair listed, AD/BD indicates that protein 1 was fused with AD and protein 2 was fused with BD. BD/AD indicates the opposite situation. The colonies shown in the –HLT (−HisLeuTrp) columns represent dilution series (105, 104, and 103 colony-forming units) of each strain grown on –HLT SD medium supplemented with 20 mM 3-amino-1,2,4-triazole. n/a, not applicable. For the graph, the dark gray bars show the results from analyses of α-galactosidase activity where the first protein listed was in the AD construct and the second protein was in the BD construct. The light gray bars are for the reverse BD/AD combination. Error bars represent sd across three independent replicates.
In Situ Hybridization of AP3 and PI Homologs in clematiflora Mutant Flowers of A. vulgaris
The in situ hybridization studies in wild-type flowers suggest specific correlations between the expression of AqvAP3-2 and AqvAP3-3 and petal identity. One way to further test these associations is to investigate gene expression in a homeotic mutant. Several floral homeotic mutants are available for A. vulgaris in the horticultural trade. Of particular interest is a class of mutants that affects only the second whorl, causing the transformation of the petals into sepals (Figure 5A). Mutants in this class go by a number of different names, including stellata and clematiflora (clm), and vary in their phenotypic penetrance. We chose to work with a line of clm that exhibits complete transformation of every second whorl organ on every flower (Figure 5A). At maturity, these second whorl organs have every hallmark of wild-type sepals, including shape, color (white in this line), and epidermal morphology (Figure 5A). Histological studies indicate that the affected organs initiate in the manner of normal petal primordia (Figures 5C, 5E, and 5J) but later take on a developmental pattern characteristic of the sepals (Figures 5D, 5E, 5G, 5H, and 5L). We used in situ hybridization to study the expression of AqvAP3-1, AqvAP3-2, AqvAP3-3, and AqvPI in clm plants. At early stages, the expression pattern of AqvAP3-1 appears to be identical to that of the wild type, but the normal decline in AqvAP3-1 transcription that starts in the petal primordia initiates at an earlier developmental stage (Figure 5E). The further progression of AqvAP3-1 downregulation is similar to that of the wild type (Figure 5F). AqvAP3-2 is not normally expressed in second whorl primordia at early stages and is similarly absent in those of clm floral meristems (Figure 5G, inset). However, the late onset of AqvAP3-2 expression in the second whorl organs is never observed in the mutants (Figure 5G). Consistent with its specific expression in petals, AqvAP3-3 is never detected in this clm strain using either in situ hybridization (Figure 5H) or RT-PCR (Figure 3). The expression pattern of AqvPI is identical to that observed in the wild type until the point when the developmental trajectory of the second whorl primordia begins to diverge, at which time AqvPI expression declines to a level similar to what is normally observed in the sepals (Figures 5I to 5L).
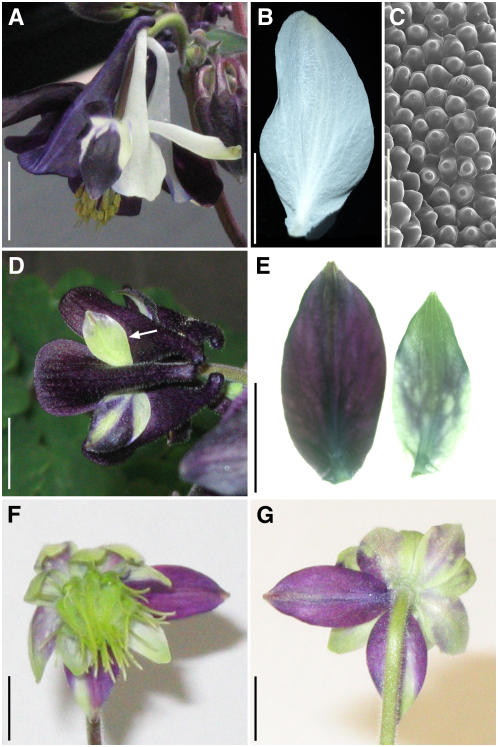
Figure 5.
Floral Morphology of A. vulgaris clm and in Situ Hybridization of AqvAP3-1, AqvAP3-2, AqvAP3-3, and AqvPI in clm Floral Meristems.
(A) Photograph of a flower from the underside. All perianth organs have the morphology of sepals (inset shows adaxial epidermis).
(B) to (D) Histological sections of developing floral meristems. Arrowheads indicate second whorl organs.
(E) and (F) AqvAP3-1.
(G) AqvAP3-2 with inset showing earlier stage.
(H) AqvAP3-3.
(I) to (L) AqvPI.
Sections in (B), (C), and (E) are tangential longitudinal, while those in (D) and (F) to (L) are radial longitudinal. Arrowheads indicate second whorl primordia, unless stated otherwise. Bars = 100 μm.
Silencing of AqvPI Using TRV-VIGS
Unfortunately, none of the commercially available floral homeotic mutants of Aquilegia exhibit morphologies that would suggest a complete loss of AqvAP3 or AqvPI function. In order to generate such a phenotype, we used TRV-based VIGS to knock down the expression of AqvPI. This approach has been used successfully in a diverse array of angiosperms, including the lower eudicot Papaver (Hileman et al., 2005). To facilitate the identification of silenced flowers, we prepared a TRV2 construct containing fragments of both AqvPI and the Aquilegia homolog of ANTHOCYANIDIN SYNTHASE (AqvANS). Loci in the anthocyanin pathway have been used as markers for floral VIGS in several other systems (Chen et al., 2004; Lee et al., 2005), and we have found that AqvANS silencing has no effect on floral morphology (B. Gould, K. Fifer, and E.M. Kramer, unpublished data). AqvANS is normally expressed at high levels in the sepals and petals, with lower levels in the carpels (Figure 3). In order to promote specific silencing, a 326-bp AqvPI fragment was derived from the 3′ end of the cDNA, including 120 bp of coding sequence and the remainder 3′ untranslated region (see Supplemental Figure 4 online). Of 12 treated plants, 4 exhibited significant levels of AqvANS silencing. Two of these displayed weak to moderate levels of silencing. In the former case, AqvANS-silenced sectors encompassed variable portions of individual perianth organs, while in the latter, silenced sectors included three to five entire perianth organs (Figures 6A and 8D below). Two other plants produced flowers with moderate to strong phenotypes, where the strong phenotypes showed silencing throughout the perianth and reproductive organs (Figures 6A to 6E). RT-PCR was used to confirm that the flowers showing AqvANS silencing were also silenced for AqvPI (Figure 7; see Supplemental Figure 5 online).
Figure 6.
Phenotypes of AqvPI-AqvANS–Silenced Floral Buds and Organs.
(A) Flower showing a moderate silencing phenotype. Half of the perianth shows AqvANS silencing, and the petal spur on that side of the flower is missing (arrow indicates the expected position).
(B) Flower showing a strong silencing phenotype. The perianth organs show only patchy AqvANS expression, all of the second whorl organs have the appearance of sepals, and the majority of internal organs are transformed into carpeloid organs. One stamen-like organ is indicated by an arrow.
(C) Flower showing a strong silencing phenotype. Two sepals were removed to expose the inner organs. No AqvANS expression is apparent, the second whorl organs are completely transformed into sepals, and all internal organs are carpels.
(D) A silenced second whorl organ from the flower shown in (A).
(E) Organs from the first (w1) and second (w2) whorls of the flower shown in (C).
(F) Scanning electron microscopy of the adaxial epidermal surface of a silenced second whorl organ.
(G) Scanning electron microscopy of the adaxial epidermal surface of a silenced first whorl organ.
(H) The carpeloid organs of a strongly silenced flower. The perianth as well as one vertical orthostichy of carpeloid organs has been removed. Arrows indicate a vertical row of transformed organs, which show carpel identity in every whorl through to the innermost whorl (asterisk), representing the position of the wild-type carpels.
(I) to (K) Individual carpeloid organs from strongly silenced flowers exhibiting a range of partial fusion.
(L) Scanning electron microscopy image of the organ shown in (I).
(M) Scanning electron microscopy image of a staminoid organ from a strongly silenced flower.
Bars = 1 mm in (A) to (E) and (H) to (M) and 100 μm in (F) and (G).
Figure 8.
Sepal Phenotypes in Flowers Treated with TRV2-AqvANS or TRV2-AqvPI-AqvANS.
(A) A flower showing silencing in response to treatment with TRV2-AqvANS.
(B) A single AqvANS-silenced sepal.
(C) Scanning electron microscopy image of the adaxial epidermis of an AqvANS-silenced sepal.
(D) A flower showing patchy silencing in response to treatment with TRV2-AqvPI-AqvANS. Note the small, green sepal (arrow).
(E) Sepals from the same whorl of a single flower with partial AqvPI-AqvANS silencing. Note the size difference between the unsilenced (left) and silenced (right) organs.
(F) and (G) Front (F) and back (G) views of a flower showing strong AqvPI-AqvANS silencing. Two first whorl organs are relatively unsilenced and show distinct expansion patterns relative to the more strongly silenced organs.
Bars = 10 mm in (A), (B), and (D) to (G) and 100 μm in (C).
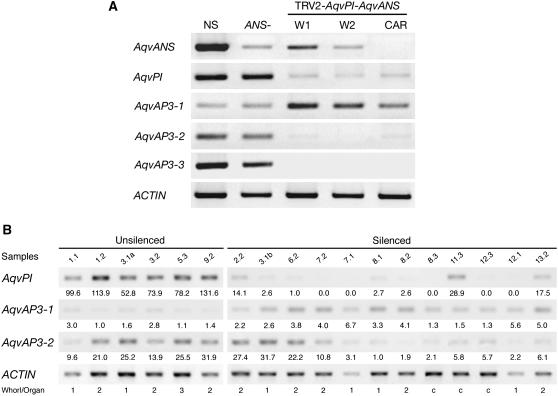
Figure 7.
Locus-Specific RT-PCR on RNA Prepared from Organs of VIGS-Treated Flowers.
(A) RNA samples from five tissue types were tested: flowers that were treated with TRV2-AqvPI-AqvANS but did not show silencing (NS), silenced flowers that were treated with TRV2-AqvANS (ANS−), first whorl organs from strongly silenced flowers treated with TRV2-AqvPI-AqvANS (w1), second whorl organs from strongly silenced flowers treated with TRV2-AqvPI-AqvANS (w2), and carpeloid organs from strongly silenced flowers treated with TRV2-AqvPI-AqvANS (car).
(B) Individual RT-PCR for 6 unsilenced and 12 silenced tissue samples. The sample numbers correspond to the bud number and whorl from which the sample was taken. For instance, sample 2.2 was a second whorl organ from the second flower to be characterized. We analyzed 33 individual flowers showing some degree of silencing as well as 12 flowers showing no silencing from the same plants. In some cases, silenced and unsilenced organs from the same whorl were analyzed separately (e.g., 3.1a and 3.1b). Unsilenced samples included sepals (whorl 1), petals (whorl 2), and stamens (whorl 3), while silenced samples included sepals (whorl 1), petaloid or sepaloid organs in whorl 2, and carpeloid organs in internal whorls (c). Normalized expression values are shown for each sample.
Dramatic homeotic phenotypes were observed in both moderately (n = 15 from four plants) and strongly (n = 18 from two plants) silenced flowers (Figures 6A to 6E). These transformations affected the staminodia, stamens, and petals. In the whorls normally occupied by the staminodia and stamens, strongly silenced flowers produced carpeloid organs (Figures 6B, 6C, and 6H). These diverged from wild-type development in that they were often incompletely fused to various degrees (Figures 6I, 6J, and 6L). The innermost carpels, representing the wild-type carpels, were properly fused, indicating that the fusion defect was specific to the transformed organs (Figures 6H and 6K). At very low frequency, 1 to 5% of organs within any given flower, stamens exhibited weak transformation, resulting in aberrant stamens or empty carpeloid organs that lacked ovules (Figures 6B and 6M). Overall, however, there were no consistent differences in the phenotypes of organs from the presumptive stamen whorl compared with those from the presumptive staminodium whorl.
In the perianth, the most striking phenotype was in the second whorl, where the petals were partially or completely transformed into sepals (Figures 6A to 6E). The second whorl organs of moderately silenced flowers retained some petaloid characteristics, such as a rounded apex, but lacked normal spur development (Figures 6A and 6D). Those from strongly silenced flowers appeared to be completely transformed into sepals, with narrow bases and lanceolate apices (Figure 6E). These second whorl organs also exhibited typical sepal cell types, most notably papillated cells on the adaxial epidermis, which are not normally observed in Aquilegia petals (cf. Figures 1O and 6F).
Determining whether silencing of AqvPI had an effect on sepal development proved difficult. Fundamentally, silenced sepals showed no differences that could be ascribed to changes in identity: their shape and epidermal cell types were identical to those of wild-type sepals (Figures 6B, 6C, 6E, and 6G). We did note, however, some subtle developmental differences in silenced versus unsilenced sepals. In flowers that showed mixed silencing in the perianth, sepals with lower levels of silencing grew to a larger size than those with strong silencing (Figures 8D to 8G). Weakly silenced sepals became fully reflexed, similar to wild-type sepals, while the strongly silenced organs remained relatively closed around the floral meristem (Figures 6B, 6C, 8F, and 8G). This is in contrast with the sepals in flowers that were only silenced for AqvANS, which were entirely wild type in their development and morphology (Figures 8A to 8C). Due to a wide range of within-inflorescence variation in flower size, it was not possible to test absolute differences in organ size between silenced and unsilenced sepals. If we focus specifically on flowers with mixed silencing in the perianth (n = 9 flowers), as shown in Figures 8F and 8G, we can compare relative length between organs within the same whorl of the same flower. In this context, we found that silenced organs were consistently 10 to 20% shorter in length from base to tip. This difference was not associated with any discernible difference in cell size between wild-type, AqvANS-silenced, or AqvANS-AqvPI–silenced organs (Figures 6F, 6G, and 8C; see Supplemental Figure 1 online). Another minor point of difference in silenced organs was their color, which was pale green (Figures 6A to 6E and 8D to 8G) rather than the bright white normally observed in AqvANS-silenced organs (Figures 8A and 8B). Examination of several nuclear and plastid molecular markers associated with photosynthesis, as well as histological analysis, did not reveal any appreciable differences between the AqvPI-AqvANS– and AqvANS-silenced organs in this regard, however.
In Arabidopsis and several other core eudicots, expression of the AP3 and PI homologs is mutually dependent due to autoregulatory and cross-regulatory interactions (Goto and Meyerowitz, 1994; Jack et al., 1994). Therefore, we used RT-PCR to examine expression levels of the three AqvAP3 paralogs in pooled tissue samples showing strong AqvPI silencing. Interestingly, while AqvAP3-1 remained expressed, the transcript levels of AqvAP3-2 and AqvAP3-3 showed a dramatic reduction (Figure 7A). These results suggest that the homeotic phenotypes observed in AqvPI-VIGS plants reflect not only the loss of AqvPI function but also of AqvAP3-2 and AqvAP3-3. We used quantitative RT-PCR to further characterize both the silencing of AqvPI and the expression levels of AqvAP3-1 and AqvAP3-2 in individual tissue samples (Figure 7B; see Supplemental Figure 5 online). AqvAP3-3 was not analyzed due to the fact that it is normally expressed only in second whorl organs, meaning that we had fewer available samples. Silencing of AqvPI ranged from 70 to 100% relative to wild-type levels, while the reduction in AqvAP3-2 expression varied from 50 to 95% of wild-type values (Figure 7B). We did not observe an absolute correlation between the reduction of AqvPI and AqvAP3-2 (Figure 7B; see Supplemental Figure 5A online), possibly due to the fact that, in some cases, VIGS-based silencing of AqvPI may have been established too late in development to significantly affect the expression of AqvAP3-2 (see Supplemental Figures 5E to 5G online). AqvAP3-1 was analyzed because initial experiments seemed to indicate that the expression levels of this locus might actually be higher in AqvPI-silenced tissue (Figure 7A). The additional RT-PCR data indicate that this inverse relationship may be real, but as with AqvAP3-2, it is not a perfect correlation (Figure 7B; see Supplemental Figure 5B online). Interestingly, the increase in AqvAP3-1 expression is more notable in first and second whorl organs than in the carpels (see Supplemental Figures 5B and 5C online), which in silenced flowers includes all of the organs internal to the perianth. The relative increase of AqvAP3-1 expression in AqvPI-silenced tissue is as much as threefold, but it remains true that in late floral tissue, AqvAP3-1 is expressed at lower levels overall than AqvAP3-2 or AqvPI.
DISCUSSION
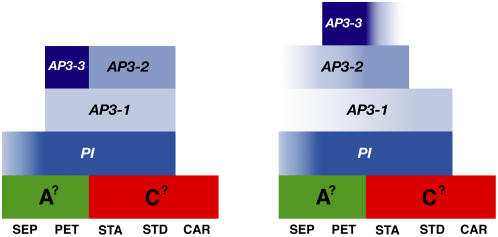
The floral morphology of Aquilegia differs significantly from that observed among core eudicot models such as Arabidopsis and Antirrhinum. The first whorl organs exhibit many of the developmental hallmarks of sepals, but by maturity, they are undeniably petaloid with floral rather than photosynthetic pigments and conical epidermal cells. Positioned between the stamens and carpels, the staminodia are morphologically unique compared with the other floral organs. They show no signs of chimerism and do not appear to represent a morphological transition zone. Thus, Aquilegia possesses five distinct organ types, and two of these are petaloid. This would seem to require two modifications of the classical ABC model: the capacity to (1) specify two types of petaloid organs and (2) determine a fifth organ identity. Detailed expression studies of the AP3 and PI homologs in Aquilegia demonstrate that each organ type expresses a specific combination of B gene homologs (Figure 9). The staminodia express both AqvAP3-1 and AqvAP3-2 with AqvPI at very early stages but quickly lose AqvAP3-2 expression. The stamens express a similar complement of loci early on but, by contrast, proceed to lose AqvAP3-1. The petals are perhaps the most complex, initially expressing AqvAP3-1 and AqvAP3-3 with AqvPI but later losing AqvAP3-1 and gaining AqvAP3-2. In the sepals, AqvAP3-1/2 and AqvPI are only detectable at very late stages. The common player across all of these organs is AqvPI, and as might be expected, downregulation of this locus affects development in all four organ types, although the sepal phenotype is subtle and not homeotic in nature.
Figure 9.
Modified ABC Models Based on the Early (Left) and Late (Right) Expression Patterns of AP3 and PI Homologs in Aquilegia.
The early-stage model corresponds to the period when sepals, stamens, and staminodia are initiating, while the late-stage model encompasses everything after carpel initiation. Color gradients indicate transient or weak expression. Each AP3 paralog has a distinct expression pattern, while the expression domain of the single PI homolog encompasses the summation of all of the AP3 expression domains. Placeholders are included for potential A and C function loci. SEP, sepals; PET, petals; STA, stamens; STD, staminodia; CAR, carpels.
Evidence for Protein and Genetic Interactions
In the core eudicot models Arabidopsis, Antirrhinum, and Petunia, AP3 and PI homologs function as obligate heterodimers, an interaction that is required for DNA binding as well as protein stability and nuclear import (Goto and Meyerowitz, 1994; Jack et al., 1994; Zachgo et al., 1995; McGonigle et al., 1996; Riechmann et al., 1996; Vandenbussche et al., 2004). Similar strict heterodimerization has been recovered in the grasses (Lee et al., 2003; Whipple et al., 2004), but the PI homologs of other monocots have some potential to homodimerize, although it remains to be determined whether these homodimers function in vivo (Winter et al., 2002; Kanno et al., 2003). Our yeast two-hybrid studies suggest that the AP3 and PI proteins of Aquilegia can function as heterodimers, with no evidence of homodimerization for AqvPI and very limited evidence for AqvAP3-3.
Studies of the clm mutant and the AqvPI-VIGS phenotype provide additional insight into the nature of AP3/PI regulation in Aquilegia and further suggest that the AP3 paralogs have distinct functions. In the case of clm, the expression studies indicate that early transcription of AqvAP3-1 and AqvPI is not sufficient to rescue petal identity. Furthermore, maintenance of AqvAP3-1 and AqvPI, as well as initiation of AqvAP3-2 and AqvAP3-3, in the second whorl is dependent on the establishment of petal identity. Although AqvAP3-3 would appear to be an obvious candidate for the clm locus, examination of the entire AqvAP3-3 region, including 1.3 kb 5′ of the transcriptional start site, reveals no sequence differences. Similarly, the other Aquilegia B gene homologs do not contain lesions in the clm mutant background.
The AqvPI-VIGS approach has built upon these findings. In strongly AqvPI-silenced flowers, the expression of AqvAP3-1 remains strong, perhaps even increasing, while that of AqvAP3-2 and AqvAP3-3 is reduced. One obvious question is whether the reduction in AqvAP3-2 and AqvAP3-3 expression reflects a genetic interaction with AqvPI or nonspecific silencing in the VIGS response. Previous experiments have shown very high specificity in TRV-based VIGS (Rhoades et al., 2002; Burch-Smith et al., 2004), including one instance in which a DNA fragment containing the highly conserved MIK domains was used to silence the Nicotiana AP3 homolog Nb DEF (Liu et al., 2004). In that case, the closely related Nb TM6 paralog was not silenced, although reduced expression of Nb GLO, the PI homolog, was observed. This finding was interpreted as evidence for a genetic interaction rather than for nonspecific silencing. In this study, we tested eight other florally expressed MADS box genes, including homologs of the A, C, and E class genes, and did not observe any instances of off-target silencing (see Supplemental Figure 6 online). Therefore, the critical point in interpreting the AqvPI-silenced phenotype is that it also encompasses the reduction of AqvAP3-2 and AqvAP3-3, creating something more analogous to an ap3 pi double mutant. Although the loss of AqvAP3-2 and AqvAP3-3 may be due to nonspecific silencing, it is also possible that the expression of AqvAP3-2 and AqvAP3-3 is somehow dependent on AqvPI function, while that of AqvAP3-1 is not. In the core eudicots, the maintained expression of AP3 and PI homologs is known to rely on functional AP3/PI heterodimers, although this autoregulation/cross-regulation can be direct or indirect (Sommer et al., 1991; Jack et al., 1992; Trobner et al., 1992; Goto and Meyerowitz, 1994; Hill et al., 1998; Honma and Goto, 2000; Vandenbussche et al., 2004; Rijpkema et al., 2006). The effect we observed in Aquilegia may be due to a similar autoregulatory loop functioning in AqvAP3-2 and AqvAP3-3. Alternatively, perhaps AqvAP3-1/AqvPI heterodimers are required for the initiation of transcription at the other two AP3 loci. The mild upregulation of AqvAP3-1 that is especially observed in perianth organs of AqvPI-silenced flowers could indicate that proper downregulation of AqvAP3-1 is dependent on AqvPI, either via negative autoregulation or perhaps through negative regulation by AqvAP3-2/AqvPI and AqvAP3-3/AqvPI heterodimers.
Together, these results emphasize the disparate expression patterns observed among the three AP3 paralogs, which are suggestive of subfunctionalization following gene duplication (Force et al., 1999). AqvAP3-1 has come to be expressed primarily at early developmental stages, whereas AqvAP3-2 is expressed primarily in stamens and late petals and AqvAP3-3 is expressed primarily in petals. Interestingly, these patterns correspond quite closely to the functions of discrete elements characterized in the Arabidopsis AP3 promoter, which has been found to contain dissociable regulatory modules controlling early-stage petal and stamen expression (Hill et al., 1998). Targeted downregulation of each of the AP3 loci is obviously the next step for this line of research.
B Class Function in Aquilegia Encompasses the Novel Staminodium as Well as Petals and Stamens
The elaborate staminodia seen in A. vulgaris are found in the majority of Aquilegia species, with the exception of A. jonesii, which has fertile stamens in place of the staminodial whorl (Munz, 1946). Similarly, in most closely related genera, fertile stamens occupy all of the whorls between the perianth and the carpels (Tucker and Hodges, 2005). The one departure is Aquilegia's sister genus Semiaquilegia, in which the innermost stamens are sometimes replaced by filamentous sterile organs. It is unclear whether these structures represent transitional organs that are homologous with the staminodia of Aquilegia or they were independently derived. Regardless, the staminodia of Aquilegia appear to have evolved quite recently, at the earliest before the last common ancestor of Aquilegia and Semiaquilegia. Due to their position and early morphological similarity to stamens, the staminodia are thought to be evolutionarily derived from stamens (Munz, 1946). Our expression studies indicate that the two organ types do share similar AP3/PI expression profiles at very early developmental stages (Figures 2 and 9), but these rapidly diverge such that AqvAP3-2 is maintained in stamens and AqvAP3-1 is maintained in staminodia. Downregulation of AqvPI and AqvAP3-2 results in the transformation of stamens and staminodia into carpeloid organs, demonstrating that B homolog function is essential to establishing identity in both organ types. From an evolutionary standpoint, this role in staminodium development represents a kind of neofunctionalization event in which a new organ identity was apparently derived from the preexisting stamen identity program. It remains to be seen whether the differential expression of AP3-1 and AP3-2 is key to the establishment of staminodium versus stamen identity. One alternative possibility is that other floral MADS box genes are differentially expressed in the staminodium and are critical to their identity.
As discussed above, the morphology of petaloid organs varies enormously across the angiosperms, leading botanists to suggest that these sterile structures have evolved many times independently (Takhtajan, 1991). In particular, the second whorl petaloid organs of the Ranunculaceae have long been considered to be recently and independently derived from outer stamens (Prantl, 1887; Worsdell, 1903). This hypothesis is based on a number of factors, including aspects of petal development, primordium position, overall appearance, and venation pattern (Tamura, 1965; Kosuge, 1994; Erbar et al., 1998). Some of these criteria result from typological thinking, such as the idea that retarded development in petals is necessarily indicative of derivation from aborted stamens (Takhtajan, 1991). Our modern understanding of the homeotic ABC program and its apparent conservation presents another scenario: that second whorl petals in the Ranunculaceae are produced by a commonly inherited genetic program. Under this model, the apparently homoplastic pattern of petal distribution across the family (Hoot, 1995) would be due to multiple independent loss events rather than independent derivations. Several recent discoveries support this alternative hypothesis. For one, the current angiosperm phylogeny is consistent with a general model whereby petaloid organs evolved very early and were subsequently lost many times (Zanis et al., 2003). In addition, homologs of the B class genes have been shown to promote the identity of petals or apparent petal derivatives across the core eudicots and in the distantly related grasses, suggesting a deeply conserved petal identity program (reviewed in Kramer and Jaramillo, 2005; Zahn et al., 2005). Likewise, comparative expression studies indicate that the vast majority of developing petaloid organs examined express AP3/PI homologs (Kim et al., 2005). What has been missing from this data set is any functional evidence from noncore eudicot taxa that possess a genuinely petaloid perianth, since the grasses produce lodicules in place of petals. Furthermore, there are conflicting results from core eudicot transgenic studies regarding whether paleo-AP3 lineage genes, which include all AP3 homologs from outside the core eudicots (Kramer et al., 1998), can function in petal identity (Lamb and Irish, 2003; Whipple et al., 2004). Thus, our data for the Aquilegia B gene homologs are particularly useful and demonstrate that AP3/PI function in this lower eudicot clearly encompasses petal identity. The critical question is whether this role reflects a commonly inherited petal identity function or was derived independently in conjunction with the evolution of unique petals in the Ranunculaceae. While we may never be able to make this distinction at a deeper level (Kramer and Irish, 2000; Kramer and Jaramillo, 2005; Jaramillo and Kramer, 2007), comparative studies of orthologs of the petal-specific AqvAP3-3 will provide insight into whether there is a conserved petal identity program across the Ranunculaceae.
Aquilegia B Gene Homologs May Not Play a Role in Sepal Identity
The sepals of Aquilegia possess several characteristics that are normally associated with petals, including the presence of a papillated cell type and the general expression of floral rather than photosynthetic pigments. Moreover, we had previously detected the expression of AqvAP3-1, AqvAP3-2, and AqvPI in the petaloid sepals. Our current analyses demonstrate, however, that Aquilegia AP3/PI homologs are not persistently expressed in the sepals during the early stages when organ identity is generally thought to be established (Bowman et al., 1989; Zachgo et al., 1995). Similar findings have been reported for the calyx-derived perianth of Aristolochia (Jaramillo and Kramer, 2004), the petaloid first whorl organs of Asparagus (Park et al., 2003, 2004), and, potentially, the petaloid sepals of Impatiens (Geuten et al., 2006). Interestingly, in Antirrhinum it has been shown that even very late expression of the B gene homologs DEFICIENS and GLOBOSA can rescue anthocyanin production and the development of papillated cell types (Carpenter and Coen, 1990), although this cannot rescue overall organ shape or size (Zachgo et al., 1995). Thus, it remained possible that the late-stage expression of AqvAP3-1/2 and AqvPI could have been involved in making these organs petaloid. Downregulation of AqvPI demonstrates that this gene, and most likely the AP3 homologs as well, may not contribute to sepal identity in general or to the production of papillated cell types in particular. This suggests that papillated cells on the first whorl organs of Aquilegia are convergent with similar cells seen on petals in the core eudicots. The cells serve the same attractive function in Aquilegia sepals, but the genetic basis for their production is different, at least in terms of being dependent on B class function. Our observations indicate that AqvPI could promote late aspects of wild-type sepal size or the complete loss of photosynthetic activity. Given that there are no consistent differences in cell size, one hypothesis is that AqvPI functions to extend the period of cell division in the sepals. This would be consistent with a role in promoting cell proliferation that has been associated with AP3 and PI in Arabidopsis and with TM6, an AP3 paralog, in tomato (Solanum lycopersicum) (Sakai et al., 1995; de Martino et al., 2006). However, investigating this function has proven difficult, largely due to the technical limitations of the VIGS approach. Silencing is not 100% complete, and plants are likely to retain some residual gene function. In addition, it is difficult to recognize silenced flowers at early developmental stages, and in some cases, silencing appears to result in mericlinal or periclinal sectors. This last point is particularly relevant because while many aspects of AP3/PI function are cell-autonomous, their effect on organ size is non-cell-autonomous (Jenik and Irish, 2001). It would be more accurate, therefore, to say that while our silencing of AqvPI was sufficient to cause homeotic transformation of staminodia, stamens, and petals, it is conceivable that this gene functions in sepal identity at a very low threshold of expression. A more detailed understanding of any potential role for AqvPI or the AP3 homologs in sepal development will depend on the development of additional functional techniques in Aquilegia.
Revisiting the question posed in the introduction of how the petaloid sepals and second whorl petals of Aquilegia are genetically differentiated, we conclude that this is most likely achieved via distinct identity pathways in which B genes primarily contribute to identity in the second whorl. This study demonstrates that although a conserved petal identity program may exist, that does not necessarily mean that all petaloid organs are dependent on B gene homologs for their identity. Also, these results stand as a caution against the assumption that late-stage expression of such genes is necessarily indicative of persistent expression or a function in organ identity.
METHODS
Scanning Electron and Light Microscopy
For scanning electron microscopy studies, dissected floral organs were fixed under vacuum in FAA (50% ethanol, 4% formalin, and 5% glacial acetic acid) and then transferred to 70% ethanol for storage. Before imaging, organs were dehydrated through an ethyl alcohol series and critical point–dried with CO2 in a Tousimis/Autosamdri-815 dryer. Material was mounted on aluminum stubs with carbon-conductive adhesive tabs (Electron Microscopy Sciences), sputter-coated with gold palladium in the Denton/Desk II sputter coater, and studied in a model Quanta 200 scanning electron microscope (FEI Company) at 5 to 20 kV. In the case of the staminodia, fixation and dehydration resulted in significant distortion of the material, so these organs were imaged using environmental scanning electron microscopy settings on the same machine described above. Light photographs of dissected organs were prepared using a Kontron Elektronik ProgRes 3012 digital camera mounted on a Leica WILD M10 dissecting microscope (Harvard Imaging Center).
In Situ Hybridization and Histology
Developing inflorescences from wild-type and clm mutant plants were collected and fixed under vacuum in freshly prepared, ice-cold FAA. After an incubation of 16 to 18 h, they were then completely dehydrated in an ethyl alcohol series, embedded in Paraplast, and stored at 4°C until use. Samples were sectioned to 8 μm with a disposable steel blade on a Reichert–Jung microtome. For in situ hybridization, DNA templates for RNA probe synthesis were obtained by PCR amplification of 300- to 400-bp fragments from cDNA clones of each locus (AqvAP3-1, AqvAP3-2, AqvAP3-3, and AqvPI). For specificity, the probe templates included 200 to 250 bp of the 3′ untranslated region in addition to 100 to 150 bp from the 3′ coding region. Fragments were cloned using the TOPO-TA plasmid vector (Invitrogen). Digoxigenin-labeled RNA probes were prepared from linearized template plasmids and alkaline hydrolyzed to 250 bp. RNA in situ hybridization was performed according to previously described methods (Kramer, 2005b). In situ hybridized sections were counterstained with calcofluor and imaged using a combination of white and fluorescent light. For histology, slides were stained with 0.025% alcian blue 8GX and 0.01% safranin O in 0.1 M acetate buffer, pH 5.0. All sections were digitally photographed using a Leica Leitz DMRD microscope equipped with a Retiga EXi imaging system (Harvard Imaging Center).
RT-PCR
Differential expression of AP3 and PI homologs was assayed using RT-PCR on RNA extracted from four late stages of developing sepals and petals (see Supplemental Figures 1 and 2 online) as well as pooled stages of stamens, staminodia, carpels, wild-type inflorescences, prevernalized leaf tissue, and clm mutant inflorescences. Total RNA was prepared using Plant RNA Reagent (Invitrogen), DNase-treated to remove genomic DNA contamination, and then 5 μg was used as template for cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen). The resulting cDNA template was diluted 1:10 and PCR-amplified using locus-specific primers (see Supplemental Table 1 online) for 25 cycles at an annealing temperature of 55°C. PCR was run on a 1% agarose gel, which was stained with ethidium bromide and digitally photographed using a Bio-Rad Gel Doc XR system.
Yeast Two-Hybrid Analysis
The IKC domains of AqvAP3-1, AqvAP3-2, AqvAP3-3, and AqvPI were used to test protein interactions in yeast assays. Corresponding domains of Arabidopsis thaliana AP3 and PI were used as controls. Each protein was fused with the GAL4 binding domain in the pGBKT7 vector (Clontech) and the GAL4 activation domain in the pGADT7 vector (Clontech). Background levels of autoactivation were suppressed by the addition of 20 mM 3-amino-1,2,4-aminotriazole to the growth medium (see Supplemental Figure 3 online). Every pairwise protein combination was tested for both growth on selective medium and α-galactosidase activity. It is important to note that interactions identified in yeast analyses do not necessarily have linear correlations to Kd values in in vitro binding studies. See Supplemental Methods online for a complete description of the experimental approach.
TRV-VIGS
TRV1 and TRV2 vectors were kindly provided by S.P. Dinesh-Kumar of Yale University (Ratcliff et al., 2001; Dinesh-Kumar et al., 2003). A 738-bp fragment of the Aquilegia vulgaris ANS locus was amplified from floral cDNA using primers designed to the EST sequence TC2856 (GenBank accession number DR946276; 5′-AGTTCATTCCCAAGGAGTATGTGC, 3′-TGGCAGTCACCCATTTTTCATC) and cloned using the TOPO-TA plasmid vector (Invitrogen). The resulting AqvANS plasmid was used as a PCR template with primers that amplified a 230-bp fragment and added XbaI and BamHI restriction sites to the respective 5′ and 3′ ends of the PCR product (5′-GGTCTAGATTGGGATTGGAAGAAGAAAGGC, 3′-AAGGATCCATGTTGAGCAAATGTGCGA). This PCR product was then digested with XbaI/BamHI and ligated into a similarly digested TRV2 vector, yielding the TRV2-AqvANS construct. To make the TRV2-AqvPI-AqvANS construct, we PCR-amplified a 326-bp fragment of AqvPI using primers that added EcoRI and XbaI sites to the respective 5′ and 3′ ends of the PCR product (5′-CGGAATTCGCTTGGCGGGAATGATAGAGAAATGGAAAATG, 3′-CGCTCTAGAGCGCCATAATCAAGAGAAACTTTAAAATCATGGATA). This PCR product was digested with EcoRI/XbaI and ligated into a similarly digested TRV2-AqvANS construct. After sequencing to confirm that the construct was correct, TRV1 and TRV2-AqvPI-AqvANS were separately transformed into Agrobacterium tumefaciens strain GV3101. Separate 50-mL liquid cultures for each construct were grown, cells were collected by centrifugation at 4000g for 12 min, and each pellet was resuspended at OD600 = 2.0 in an infiltration buffer containing 10 mM MES, 200 μM acetosyringone (3′5′-dimethoxy-4′-hydroxyacetophenone), and 10 mM MgCl2. Following a 4-h incubation at room temperature, the TRV1 and TRV2-AqvPI-AqvANS cultures were mixed in equal volume. Twelve A. vulgaris plants that had been vernalized at 4°C for 8 weeks were treated with the mixed cultures starting at 2 weeks after the plants had been removed from vernalization. Incisions were made in the basal rosette of each plant, and ∼1 to 2 mL of mixed culture was injected using a needleless 1-mL syringe. This treatment was repeated once per week for 3 weeks.
Inflorescences were examined for signs of AqvANS silencing. Flowers showing any silencing were photodocumented during development and upon maturation, and the flowers were dissected. All individual perianth organs were photographed using a Kontron Elektronik ProgRes 3012 digital camera mounted on a Leica WILD M10 dissecting microscope (Harvard Imaging Center). For every silenced flower, a selection of organs from each whorl was either frozen at −80°C for subsequent RNA analysis or fixed in freshly prepared, ice-cold FAA for scanning electron microscopy analysis (as described above). This process was also repeated for several unsilenced flowers on every treated plant. DNase-treated total RNA was prepared as described above. For Figure 6 and Supplemental Figure 6 online, total RNA from strongly silenced flowers was pooled by whorl of origin or organ type (first whorl, second whorl, carpel-like). cDNA preparation and RT-PCR were performed as described above (see Supplemental Table 1 online for primer sequences). To more accurately assess relative levels of AqvAP3-1, AqvAP3-2, AqvPI, and AqvANS expression (see Supplemental Figure 5 online), we first established the linear range of amplification for these loci, taking great care to avoid amplification levels that saturated the ethidium bromide staining (see Supplemental Table 1 online for primer sequences; 28 to 32 cycles was optimal for AqvPI, AqvANS, and ACTIN, and 30 to 37 cycles was optimal for AqvAP3-1 and AqvAP3-2). We then used quantitative RT-PCR analyses on separate cDNA pools prepared from 1.5 μg of DNase-treated total RNA from 18 individual tissue samples: 6 from TRV2-AqvPI-AqvANS–treated but unsilenced floral organs and 12 from TRV2-AqvPI-AqvANS–silenced floral organs. PCR products were conducted using 1:100 dilutions of template cDNA with 32 cycles of amplification. The products were separated by electrophoresis on a 1% agarose gel containing 0.5 mg/L ethidium bromide. DNA band intensities were imaged with UV light on an Alpha Innotech ChemiImager at levels below saturation and calibrated against a low DNA mass ladder (Invitrogen) using AlphaEase FC imaging software (Alpha Innotech), and values were normalized based on relative ACTIN values. Reactions were repeated two to four times each in order to test reproducibility, with the results from one round being shown.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF489475 to EF489476 (see also Supplemental Figure 6 online).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. All PCR Primer Sequences.
Supplemental Methods. Expanded Yeast Two-Hybrid Methods.
Supplemental Figure 1. Late Developmental Stages of the Sepals.
Supplemental Figure 2. Late Developmental Stages of the Petals.
Supplemental Figure 3. Control Data for the Yeast Two-Hybrid Experiment.
Supplemental Figure 4. Respective Nucleotide Alignments between AqvAP3-1, AqvAP3-2, or AqvAP3-3 and the AqvPI Fragment Used in the TRV2-AqvPI-AqvANS Construct.
Supplemental Figure 5. Relationships between Quantified Expression Levels of AqvPI, AqvAP3-1, and AqvAP3-2 in Plants Treated with TRV2-AqvPI-AqvANS, with Associated Organ Photographs.
Supplemental Figure 6. Locus-Specific RT-PCR for Eight Additional Floral Organ Identity Gene Homologs.
Supplementary Material
Acknowledgments
We thank members of the Kramer and Mathews laboratories, as well as three anonymous reviewers, for comments on the manuscript. We also express our gratitude to Renate Hellmiss-Peralta of the Molecular and Cellular Biology Imaging Center for help with the figures. This work was supported by National Science Foundation Grant IBN-0319103 to E.M.K. Scanning electron microscopy analysis was conducted at Harvard's Center for Nanoscale Systems, supported by National Science Foundation Infrastructure Grant 0099916.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Elena M. Kramer (ekramer@oeb.harvard.edu).
Online version contains Web-only data.
References
- Aagaard, J.E., Olmstead, R.G., Willis, J.H., and Phillips, P.C. (2005). Duplication of floral regulatory genes in the Lamiales. Am. J. Bot. 92 1284–1293. [DOI] [PubMed] [Google Scholar]
- Albert, V.A., Gustafsson, M.H.G., and Di Laurenzio, L. (1998). Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal. In Molecular Systematics of Plants, Vol. II, D. Soltis, P. Soltis, and J.J. Doyle, eds (New York: Chapman and Hall), pp. 349–374.
- Ambrose, B.A., Lerner, D.R., Ciceri, P., Padilla, C.M., Yanofsky, M.F., and Schmidt, R.J. (2000). Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. [DOI] [PubMed] [Google Scholar]
- Aoki, S., Uehara, K., Imafuku, M., Hasebe, M., and Ito, M. (2004). Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. J. Plant Res. 117 229–244. [DOI] [PubMed] [Google Scholar]
- Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith, T.M., Anderson, J.C., Martin, G.B., and Dinesh-Kumar, S.P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39 734–746. [DOI] [PubMed] [Google Scholar]
- Buzgo, M., Soltis, P.S., and Soltis, D.E. (2004). Floral developmental morphology of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 165 925–947. [Google Scholar]
- Carpenter, R., and Coen, E.S. (1990). Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 4 1483–1493. [DOI] [PubMed] [Google Scholar]
- Chen, J.C., Jiang, C.Z., Gookin, T.E., Hunter, D.A., Clark, D.G., and Reid, M.S. (2004). Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol. Biol. 55 521–530. [DOI] [PubMed] [Google Scholar]
- Christensen, K.I., and Hansen, H.V. (1998). SEM studies of epidermal patterns of petals in the angiosperms. Opera Bot. 135 5–91. [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353 31–37. [DOI] [PubMed] [Google Scholar]
- Cubas, P. (2002). Role of TCP genes in the evolution of morphological characters in angiosperms. In Developmental Genetics and Plant Evolution, Q.C.B. Cronk, R.M. Bateman, and J.A. Hawkins, eds (London: Taylor and Hawkins), pp. 247–266.
- de Martino, G., Pan, I., Emmanuel, E., Levy, A., and Irish, V. (2006). Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., Anandalakshmi, R., Marathe, R., Schiff, M., and Liu, Y. (2003). Virus-induced gene silencing. In Plant Functional Genomics, E. Grotewold, ed (Totowa, NJ: Humana Press), pp. 287–293. [DOI] [PubMed]
- Endress, P.K. (1994). Diversity and Evolutionary Biology of Tropical Flowers. (Cambridge, UK: Cambridge University Press).
- Erbar, C., Kusma, S., and Leins, P. (1998). Development and interpretation of nectary organs in Ranunculaceae. Flora 194 317–332. [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.-L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten, K., Becker, A., Kaufmann, K., Caris, P., Janssens, S., Viaene, T., Theissen, G., and Smets, E. (2006). Petaloidy and petal identity MADS-box genes in the balsaminoid genera Impatiens and Marcgravia. Plant J. 47 501–518 [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hileman, L.C., Drea, S., de Martino, G., Litt, A., and Irish, V.F. (2005). Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J. 44 334–341. [DOI] [PubMed] [Google Scholar]
- Hill, T., Day, C.D., Zondlo, S.C., Thackeray, A.G., and Irish, V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125 1711–1721. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127 2021–2030. [DOI] [PubMed] [Google Scholar]
- Hoot, S.B. (1995). Phylogeny of the Ranunculaceae based on preliminary atpB, rbcL and 18S nuclear ribosomal DNA sequence data. Plant Syst. Evol. Suppl. 9 241–251. [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76 703–716. [DOI] [PubMed] [Google Scholar]
- Jaramillo, M.A., and Kramer, E.M. (2004). APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth in Aristolochia (Aristolochiaceae). Evol. Dev. 6 449–458. [DOI] [PubMed] [Google Scholar]
- Jaramillo, M.A., and Kramer, E.M. (2007). The role of developmental genetics in understanding homology and morphological evolution in plants. Int. J. Plant Sci. 168 61–72. [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2001). The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128 13–23. [DOI] [PubMed] [Google Scholar]
- Kanno, A., Saeki, H., Kameya, T., Saedler, H., and Theissen, G. (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52 831–841. [DOI] [PubMed] [Google Scholar]
- Kim, S., Koh, J., Yoo, M.J., Kong, H.Z., Hu, Y., Ma, H., Soltis, P.S., and Soltis, D.E. (2005). Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 43 724–744. [DOI] [PubMed] [Google Scholar]
- Kim, S., Yoo, M., Albert, V.A., Farris, J.S., Soltis, P.S., and Soltis, D.E. (2004). Phylogeny and diversification of B-function genes in angiosperms: Evolutionary and functional implications of a 260-million year old duplication. Am. J. Bot. 91 2102–2118. [DOI] [PubMed] [Google Scholar]
- Kosuge, K. (1994). Petal evolution in Ranunculaceae. Plant Syst. Evol. Suppl. 8 185–191. [Google Scholar]
- Kramer, E.M. (2005. a). Floral architecture: Regulation and diversity of floral shape and pattern. In Plant Architecture and Its Manipulation, C.G.N. Turnbull, ed (Oxford, UK: Blackwell Publishing), pp. 120–147.
- Kramer, E.M. (2005. b). Methods for studying the evolution of plant reproductive structures: Comparative gene expression techniques. In Molecular Evolution: Producing the Biochemical Data, E.A. Zimmer and E.H. Roalson, eds (San Diego, CA: Elsevier Academic Press), pp. 617–635. [DOI] [PubMed]
- Kramer, E.M., Di Stilio, V.S., and Schluter, P. (2003). Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164 1–11. [Google Scholar]
- Kramer, E.M., Dorit, R.L., and Irish, V.F. (1998). Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M., and Irish, V.F. (2000). Evolution of the petal and stamen developmental programs: Evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161 S29–S40. [Google Scholar]
- Kramer, E.M., and Jaramillo, M.A. (2005). The genetic basis for innovations in floral organ identity. J. Exp. Zool. 304B 526–535. [DOI] [PubMed] [Google Scholar]
- Lamb, R.S., and Irish, V.F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 100 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Baum, S.F., Oh, S.H., Jiang, C.Z., Chen, J.C., and Bowman, J.L. (2005). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132 5021–5032. [DOI] [PubMed] [Google Scholar]
- Lee, S., Jeon, J.-S., An, K., Moon, Y.-H., Lee, S., Chung, Y.-Y., and An, G. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217 904–911. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Nakayama, N., Schiff, M., Litt, A., Irish, V.F., and Dinesh-Kumar, S.P. (2004). Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol. Biol. 54 701–711. [DOI] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., and Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99 367–376. [DOI] [PubMed] [Google Scholar]
- Martin, C., Bhatt, K., Baumann, E., Jin, H., Zachgo, S., Roberts, K., Schwarz-Sommer, Z., Glover, B., and Perez-Rodrigues, M. (2002). The mechanics of cell fate determination in petals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle, B., Bouhidel, K., and Irish, V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10 1812–1821. [DOI] [PubMed] [Google Scholar]
- Munz, P.A. (1946). Aquilegia: The cultivated and wild columbines. Gentes Herb. 7 1–150. [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Fukuda, T., Nakano, M., Hasebe, M., Kameya, T., and Kanno, A. (2005). The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. orientalis (Agapanthaceae) flowers. Plant Mol. Biol. 58 435–445. [DOI] [PubMed] [Google Scholar]
- Park, J.-H., Ishikawa, Y., Ochiai, H., Kanno, A., and Kameya, T. (2004). Two GLOBOSA-like genes are expressed in second and third whorls of homochlamydeous flowers in Asparagus officinalis L. Plant Cell Physiol. 45 325–332. [DOI] [PubMed] [Google Scholar]
- Park, J.-H., Ishikawa, Y., Yoshida, R., Kanno, A., and Kameya, T. (2003). Expression of AODEF, a B-functional MADS-box gene, in stamens and inner petals of the dioecious species Asparagus officinalis L. Plant Mol. Biol. 51 867–875. [DOI] [PubMed] [Google Scholar]
- Prantl, K. (1887). Beitrage zur Morphologie und Systematik der Ranunculacean. Bot. Jahrb. Syst. 9 225–273. [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25 237–245. [DOI] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema, A.S., Royaert, S., Zethof, J., van der Weerden, G., Gerats, T., and Vandenbussche, M. (2006). Functional divergence within the DEF/AP3 lineage: An analysis of PhTM6 in Petunia hybrida. Plant Cell 18 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Medrano, L.J., and Meyerowitz, E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378 199–201. [DOI] [PubMed] [Google Scholar]
- Sommer, H., Nacken, W., Beltran, P., Huijser, R., Pape, H., Hansen, R., Flor, P., Saedler, H., and Schwarz-Sommer, Z. (1991). Properties of deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus. Dev. Suppl. 1 169–175. [Google Scholar]
- Stellari, G.M., Jaramillo, M.A., and Kramer, E.M. (2004). Evolution of the APETALA3 and PISTILLATA lineages of MADS-box containing genes in basal angiosperms. Mol. Biol. Evol. 21 506–519. [DOI] [PubMed] [Google Scholar]
- Takhtajan, A. (1991). Evolutionary Trends in Flowering Plants. (New York: Columbia University Press).
- Tamura, M. (1965). Morphology, ecology, and phylogeny of the Ranunculaceae. IV. Sci. Rep. Osaka Univ. 14 53–71. [Google Scholar]
- Trobner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lonnig, W.E., Saedler, H., Sommer, H., and Schwarz-Sommer, Z. (1992). Globosa—A homeotic gene which interacts with deficiens in the control of Antirrhinum floral organogenesis. EMBO J. 11 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, W.C., Kuoh, C.S., Chuang, M.H., Chen, W.H., and Chen, H.H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 45 831–844. [DOI] [PubMed] [Google Scholar]
- Tucker, S.C., and Hodges, S.A. (2005). Floral ontogeny of Aquilegia, Semiaquilegia, and Isopyrum (Ranunculaceae). Int. J. Plant Sci. 166 557–574. [Google Scholar]
- Tzeng, T.Y., and Yang, C.H. (2001). A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol. 42 1156–1168. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, M., Zethof, J., Royaert, S., Weterings, K., and Gerats, T. (2004). The duplicated B-class heterodimer model: Whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Larsen, J., and Harder, L.D. (2000). The evolution of staminodes in angiosperms: Patterns of stamen reduction, loss, and functional re-invention. Am. J. Bot. 87 1367–1384. [PubMed] [Google Scholar]
- Whipple, C.J., Ciceri, P., Padilla, C.M., Ambrose, B.A., Bandong, S.L., and Schmidt, R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131 6083–6091. [DOI] [PubMed] [Google Scholar]
- Winter, K.U., Weiser, C., Kaufmann, K., Bohne, A., Kirchner, C., Kanno, A., Saedler, H., and Theissen, G. (2002). Evolution of class B floral homeotic proteins: Obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19 587–596. [DOI] [PubMed] [Google Scholar]
- Worsdell, W.C. (1903). The origin of the perianth of flowers, with special reference to the Ranunculaceae. New Phytol. 2 42–48. [Google Scholar]
- Zachgo, S., de Andrade Silva, E., Motte, P., Trobner, W., Saedler, H., and Schwarz-Sommer, Z. (1995). Functional analysis of the Antirrhinum floral homeotic Deficiens gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121 2861–2875. [DOI] [PubMed] [Google Scholar]
- Zahn, L.M., Leebens-Mack, J., dePamphilis, C.W., Ma, H., and Theissen, G. (2005). To B or not to B a flower: The role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered. 96 225–240. [DOI] [PubMed] [Google Scholar]
- Zanis, M., Soltis, P.S., Qiu, Y.-L., Zimmer, E.A., and Soltis, D.E. (2003). Phylogenetic analyses and perianth evolution in basal angiosperms. Ann. Mo. Bot. Gard. 90 129–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.