Abstract
Biliary tract carcinoma carries a poor prognosis, and difficulties with clinical management in patients with advanced disease are often due to frequent late-stage diagnosis, lack of serum markers, and limited information regarding biliary tumor pathogenesis. RNA-based global analyses of gene expression have led to the identification of a large number of up-regulated genes in several cancer types. We have used the recently developed Affymetrix U133A gene expression microarrays containing nearly 22,000 unique transcripts to obtain global gene expression profiles from normal biliary epithelial scrapings (n = 5), surgically resected biliary carcinomas (n = 11), and biliary cancer cell lines (n = 9). Microarray hybridization data were normalized using dCHIP (http://www.dCHIP.org) to identify differentially up-regulated genes in primary biliary cancers and biliary cancer cell lines and their expression profiles was compared to that of normal epithelial scrapings using the dCHIP software as well as Significance Analysis of Microarrays or SAM (http://www-stat.stanford.edu/∼tibs/SAM/). Comparison of the dCHIP and SAM datasets revealed an overlapping list of 282 genes expressed at greater than threefold levels in the cancers compared to normal epithelium (t-test P <0.1 in dCHIP, and median false discovery rate <10 in SAM). Several pathways integral to tumorigenesis were up-regulated in the biliary cancers, including proliferation and cell cycle antigens (eg, cyclins D2 and E2, cdc2/p34, and geminin), transcription factors (eg, homeobox B7 and islet-1), growth factors and growth factor receptors (eg, hepatocyte growth factor, amphiregulin, and insulin-like growth factor 1 receptor), and enzymes modulating sensitivity to chemotherapeutic agents (eg, cystathionine β synthase, dCMP deaminase, and CTP synthase). In addition, we identified several “pathway” genes that are rapidly emerging as novel therapeutic targets in cancer (eg, cytosolic phospholipase A2, an upstream target of the cyclooxygenase pathway, and ribosomal protein S6 kinase and eukaryotic translation initiation factor 4E, two important downstream mediators of the mitogenic Akt/mTOR signaling pathway). Overexpression of selected up-regulated genes was confirmed in tissue microarrays of biliary cancers by immunohistochemical analysis (n = 4) or in situ hybridization (n = 1), and in biliary cancer cell lines by reverse transcriptase PCR (n = 2). The majority of genes identified in the present study has not been previously reported in biliary cancers, and represent novel potential screening and therapeutic targets of this cancer type.
Biliary tract carcinomas, which include cancers of the gallbladder and intra- and extrahepatic biliary tree, affect 7500 individuals in the United States each year, and nearly 3500 patients die as a direct consequence of this lethal disease. 1 Once established, biliary tract cancers are notoriously challenging to diagnose and treat. At present, only surgical excision of detectable malignancy is associated with improvement in 5-year survival. Distant metastases, extensive regional lymph node metastasis, and vascular encasement or invasion preclude resection. 2 In general, the outcome for patients with advanced biliary tract cancer at any site is dismal, and neither radiation nor conventional chemotherapy significantly improves survival or quality of life. Early diagnosis has a significant impact on prognosis of biliary cancers. For example, patients with lesions confined to the gallbladder mucosa have a 32% 5-year survival rate, while patients with more advanced lesions have only a 10% 5-year survival rate. 3 Thus, urgent efforts are needed for identification of reliable tumor markers that will facilitate the early detection of biliary cancer in at-risk individuals. In addition, we need to identify cancer-specific cellular targets that would form the basis for novel therapeutic approaches for established biliary tract malignancies.
Global expression platforms such as oligonucleotide microarrays are a robust technique for identification of differentially up-regulated cancer-specific genes in tumors. This technology has recently been successfully applied for the identification of novel tumor markers in a large variety of human neoplasms. 4-7 To identify differentially expressed genes in biliary cancers versus non-neoplastic biliary epithelium, we applied RNA-based global gene expression profiling using the recently developed Affymetrix U133A expression microarrays to a series of surgically resected biliary cancers, biliary cancer cell lines, and biliary epithelial scrapings. We report the identification of 282 genes, conforming to a variety of potential tumorigenic pathways, expressed threefold or greater in biliary cancers compared to normal biliary epithelium. The majority of these genes has not been previously described in biliary tract carcinoma and may serve as potential therapeutic targets and novel tumor markers for this lethal disease.
Materials and Methods
Non-Neoplastic Epithelial Scrapings, Primary Biliary Cancers, and Cell Lines
Permission for this study was obtained through The Johns Hopkins Joint Committee on Clinical Investigation. Fresh scrapings from five non-neoplastic biliary epithelial samples were collected as previously described 8 from patients undergoing Whipple resection or incidental cholecystectomies for non-biliary disorders at The Johns Hopkins Hospital; this method has been shown to yield highly enriched sheets of epithelial cells without contaminating stromal elements. The epithelial scrapings were collected within 10 minutes of surgical resection and stored at −80°C, and included 3 extrahepatic biliary 2 gallbladder epithelial samples. Eleven cancer samples were collected from patients undergoing surgery for biliary tract cancer, and included 7 gallbladder carcinomas, 2 intrahepatic cholangiocarcinomas, and 2 distal bile duct carcinomas. All tumor samples were collected within 10 minutes of surgical resection, snap-frozen in liquid nitrogen, and stored at −80°C. Hematoxylin and eosin-stained sections from adjacent frozen tissue were prepared before sample harvest to confirm the diagnosis and assess neoplastic cellularity. RNA was extracted from tumor samples containing >50% neoplastic cells on frozen section examination.
The nine human biliary cancer cell lines used for this study included EGI-1 and TFK-1 9,10 (obtained from the German Collection of Microorganisms and Cell Cultures Department, Braunschweig, Germany), HUH28 11 and HUCCT-1 12 (obtained from the Health Science Research Resources Bank, Osaka, Japan), SNU 245, SNU 308, and SNU 1079 13 (obtained from the Korean Cell Line Bank, Seoul, Korea), GB-H3, and GB-D1. 14 Of these, SNU308, GB-H3, and GB-D1 cell lines were derived from gallbladder carcinomas, HuH28, HuCCT-1, and SNU 1079 were derived from intrahepatic cholangiocarcinomas, and EGI-1, TFK-1, and SNU 245 were derived from extrahepatic biliary cancers. All cell lines except EGI-1 were grown in RPMI (Life Technologies Inc., Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Life Technologies Inc.); EGI-I was grown in Dulbecco’s MEM (Life Technologies Inc.) supplemented with 10% FBS. Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air.
RNA Extraction and Hybridization
Sample preparation and processing procedure was performed at the Roswell Park Cancer Institute Microarray Core Facility, as described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix Inc., Santa Clara, CA). Briefly, frozen tumor tissues were crushed in TRIzol (Invitrogen Inc., Carlsbad, CA) by using a Polytron homogenizer (Brinkman Instruments, Westbury, NY). Total RNA was then extracted from the crushed tissue and cleaned using RNeasy columns according to manufacturer’s protocol (Qiagen Inc., Valencia, CA). For biliary cancer cell lines and biliary epithelial scrapings, the RNeasy protocol for human cell lines was directly used for extraction of total RNA. The integrity of total RNA was confirmed in each case using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Using 5 to 40 μg of total RNA, double-stranded cDNA was synthesized following SuperScript Choice system (Invitrogen Inc.). T7-(dT24) oligomer was used for priming the first-strand cDNA synthesis. The resultant cDNA was purified using Phase Lock Gel, phenol/chloroform extraction, and precipitated with ethanol. The cDNA pellet was collected and dissolved in appropriate volume. Using cDNA as template, cRNA was synthesized using a T7 MegaScript In-Vitro Transcription (IVT) kit (Ambion, Austin, TX). Biotinylated-11-CTP and 16-UTP ribonucleotides (Enzo Diagnostics Inc., Farmingdale, NY) were added to the reaction as labeling reagents. IVT reactions were carried out at 37°C for 6 hours and, the labeled cRNA obtained was purified using RNeasy columns (Qiagen Inc.). The cRNA was fragmented in a fragmentation buffer (40 mmol/L Tris-acetate, pH 8.1, 100 mmol/L KOAc, 30 mmol/L MgOAc) for 35 minutes at 94°C. Fragmented cRNA (10 to 11 μg/probe array) was used to hybridize to human U133A GeneChip array at 45°C for 24 hours in a hybridization oven with constant rotation (60 rpm). The chips were washed and stained using Affymetrix fluidics stations. Staining was performed using streptavidin phycoerythrin conjugate (SAPE; Molecular Probes, Eugene, OR), followed by the addition of biotinylated antibody to streptavidin (Vector Laboratories, Burlingame, CA), and finally with streptavidin phycoerythrin conjugate. Probe arrays were scanned using fluorometric scanners (Hewlett Packard Gene Array Scanner; Hewlett Packard Corporation, Palo Alto, CA). The scanned images were inspected and analyzed using established quality control measures.
Data Filtering and Analysis
The 25 .CEL files generated by the Affymetrix Microarray Suite (MAS) version 5.0 were converted into .DCP files using dCHIP (www.dCHIP.org), as described previously by Li and Wong. 15 The .DCP files were normalized, and raw gene expression data generated using the dCHIP system of model-based analysis. To evaluate how the 25 samples grouped together according to the similarity of their gene expression profiles, we used hierarchical clustering with the average linkage method, with a subset of 2308 genes demonstrating the largest variation across samples (SD/mean ≥1). For hierarchical cluster analysis, data were log-transformed, median-centered, and visualized using the CLUSTER and TREEVIEW programs. 16 For comparison of global gene expression profiles between normal and cancer samples, a two-pronged strategy was used. The first comparison was performed using the dCHIP software itself, wherein the five biliary epithelial scrapings were designated as “baseline” (B), and the 20 biliary cancer specimens designated as “experiment” (E). Genes expressed threefold or higher in the cancers versus normal samples were then identified by defining the appropriate filtering criteria in the dCHIP software (mean E/mean B >3; mean E − mean B = 100, P < 0.1, t-test). The second comparison was performed using significance analysis of microarrays or SAM v1.13 (http://www-stat.stanford.edu/∼tibs/SAM/), 17,18 which contains a sliding scale for false discovery rate (FDR) of significantly up-regulated genes. The output criteria selected for SAM included threefold or greater expression in the biliary cancers as compared to normal tissues, and a significance threshold expected to produce a median FDR of less than 10 genes.
Immunohistochemistry
A biliary cancer tissue microarray was generated from 40 biliary tract cancers (15 gallbladder carcinomas, 10 intrahepatic cholangiocarcinomas, and 15 extrahepatic cholangiocarcinomas), as previously described. 19 Each cancer specimen was represented by four 1.4-mm cores on the tissue microarrays, to obtain adequate representation of neoplastic cells. In addition, non-neoplastic biliary tissues, and additional control tissues from extra-biliary sites were also included on the tissue array. Slides were deparaffinized in fresh xylenes and rehydrated through sequential graded ethanol steps. Antigen retrieval was performed by citrate buffer incubation (18 mmol/L citric acid, 8.2 mmol/L sodium citrate, pH 6.0) using a household vegetable steamer (Black and Decker) for 60 minutes. Slides were incubated for 5 minutes with 3% hydrogen peroxide, washed in TBS/T (20 mmol/L Tris, 140 mmol/L NaCl, 0.1% Tween-20, pH 7.6), and incubated in appropriate antibody dilutions for cdc2(p34) (Zymed, South San Francisco, CA, 1:100), topoisomerase II α [topoIIα] (Neomarkers, Fremont, CA, 1:3200), bone morphogenetic protein receptor 1A [BMPR1A/Activin A receptor/ALK3] (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, 1:500), and geminin (1:500, a generous gift from Dr. Anindya Dutta, Brigham and Woman’s Hospital, Boston, MA) for 60 minutes at room temperature. Normal saline was substituted for the primary antibody in control sections. The avidin-biotin-peroxidase complex method from DAKO (Glostrup, Denmark) was used and slides were subsequently counterstained with hematoxylin. Assessment of immunohistochemical labeling in the tissue microarrays was performed by two of the authors (D.E.H. and A.M.). For the three nuclear markers (cdc2, geminin, and topo IIα), scoring was performed as follows: negative, <5% nuclear labeling; focal, 5 to 25% nuclear labeling; and diffuse, >25% nuclear labeling; for BMPR1A, labeling was scored as negative (<5% cytoplasmic and membranous staining), focal (5 to 25% cytoplasmic and membranous staining), and diffuse (>25% cytoplasmic and membranous staining).
In Situ Hybridization
Non-radioisotopic in situ hybridization was performed for the transcription factor islet-1 (ISL-1). Sense and antisense riboprobes were prepared from the corresponding sequence-verified I.M.A.G.E. clone (Invitrogen, Inc.), as previously described, 20 and labeled using Digoxin RNA Labeling mix (Boehringer Mannheim, Germany). Biliary cancer tissue microarrays were rehydrated through ethanol gradients (as described above), incubated for 10 minutes in 1% hydrogen peroxide (Super G Brand), washed in TBS, treated with proteinase K for 30 minutes at 37°C, and incubated overnight in 300 μl of diluted sense or antisense probe at 50°C. The next day, slides were washed in 2X SSC, treated with 250 μl Rnase A/T1 cocktail (Ambion, Austin, TX) for 30 minutes at 37°C, washed in 2X SSC with 50% formamide (American Bioanalytical, Natick, MA), washed in 0.8X SSC, blocked, incubated with rabbit HRP-anti-DIG (DAKO), and developed in the dark with biotinyl-tyramide (DAKO). Slides were then counterstained as previously described. 20 The specificity of hybridization was assessed by absence of signal in the sense riboprobe slide; the antisense riboprobe slide was then used for analysis of transcript expression pattern in neoplastic versus non-neoplastic biliary cells.
Semi-Quantitative RT-PCR
The differential expression of two genes, homeobox B7 (HoxB7) and dickkopf 1 homolog (Dkk-1), was validated by reverse transcriptase PCR. Total RNA was prepared from five biliary cancer cell lines (GB-H3, SNU1079, SNU308, SNU245, and HuCCT1), and three normal biliary epithelial scrapings using the RNeasy Mini kit (Qiagen, Inc.). For cDNA preparation, a mix of 4 μg total RNA and Oligo (dT) primers (Invitrogen, Inc.) was incubated at 70°C for 10 minutes to denature RNA and briefly placed on ice. A final buffer mixture of 1X First Strand Buffer, 20 mmol/L DTT, and 2 mmol/L dNTP (Invitrogen) was added, samples were incubated at 42°C for 2 minutes, 200 units of Super Script II RNase H-reverse transcriptase (Invitrogen, Inc.) were added, and samples were again incubated at 42°C for 50 minutes. A final incubation at 70°C for 15 minutes was performed. Polymerase chain reaction was performed in a final mixture of 1X Platinum PCR Supermix (Invitrogen, Inc.), 400 nmol/L forward and reverse primer, and 2 μl cDNA. PCR reactions were performed in a Thermo Hybaid MBS 0.2S cycler as follows: 96°C for 5 minutes, 35 cycles of 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 3 minutes, followed by 72°C for 5 minutes, then 4°C. PCR products were visualized by 1% agarose gel electrophoresis, followed by eithidium bromide staining. The primer sequences were as follows: HoxB7 foward primer 5′-AGCCTCAAGTTCGGTTTTCG-3′, Hox B7 reverse primer 5′-GCGTCAGGTA-GCGATTGTAG-3′, Dkk-1 forward primer 5′-ACCAGACCATTGACAACTAC-3′, Dkk-1 reverse primer 5′-GTGTCTAGCACAACACAATC-3′. Standardization was performed using GAPDH; forward primer and reverse primers were used as previously described. 4
Results
Identification of Differentially Up-Regulated Genes in Biliary Cancers
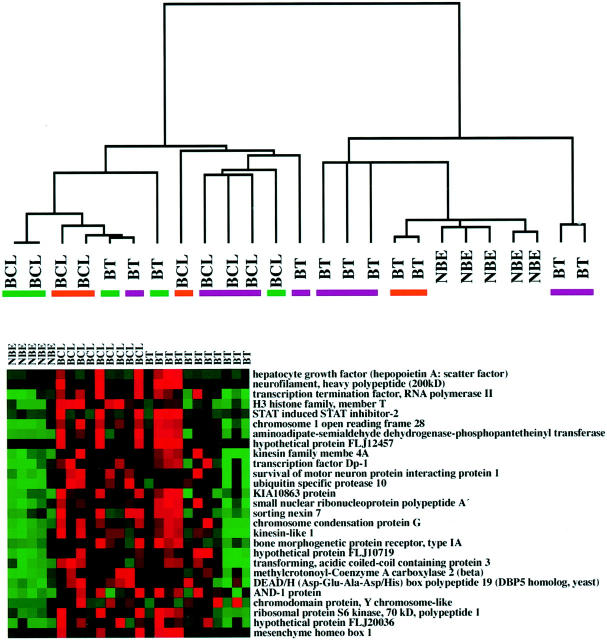
RNA samples extracted from neoplastic and non-neoplastic biliary tissues and cell lines were hybridized to the Affymetrix U133A expression microarray containing ∼22,000 unique transcripts, and normalized gene expression data were generated using the dCHIP software. Hierarchical clustering was performed using a subset of 2308 genes with the greatest variation between 25 samples. Based on gene expression profiling, two major clusters were identified, the first containing 9 of 9 cell lines and 4 of 11 resected tumors, and the second containing 5 of 5 normal biliary epithelial scrapings and 7 of 11 primary tumors (Figure 1A) ▶ . As expected, all five normal biliary scrapings clustered with remarkable identity and separately from the nine biliary cancer cell lines on the dendrogram; the clustering of a subset of primary biliary cancers near the biliary epithelial samples was not unexpected given the presence of residual stroma and non-neoplastic epithelium in many of the resected tumor samples. Although there were examples of site-of-origin-specific clustering (for example, three gallbladder carcinoma lines on the same branch of the dendrogram), in general, we found intermingling on the dendrogram between cancers from different sites in the biliary tree (Figure 1A) ▶ . Thus, while most cancer specimens, especially cell lines, clustered separately from normal epithelium, there was no unequivocal site-of-origin clustering. A pictorial representation of the principal component analysis of biliary cancers and normal biliary epithelium is illustrated in Figure 1B ▶ , with red representing relative overexpression and green representing relative underexpression (TREEVIEW software).
Figure 1.
Cluster analysis of biliary cancers. A: Cluster analysis of 9 biliary cancer cell lines (BCL), 11 primary biliary cancers (BT), and 5 normal biliary epithelial samples (NBE). The colored bars under the tumor samples indicate their location in the biliary tree: purple, gallbladder; green, extrahepatic bile duct; orange, intrahepatic bile duct. B: Representative panel demonstrating results of principal component analysis (PCA) of biliary cancers and normal biliary epithelium. Red indicated relative overexpression, green indicates relative underexpression (TREEVIEW software).
To achieve a high level of stringency for identification of differentially up-regulated genes in biliary cancers, a two-pronged strategy was used. First, the dCHIP normalized hybridization data from biliary cancers and normal epithelium was compared using the dCHIP software itself, which yielded 512 Affymetrix fragments expressed at threefold or greater intensity in the cancers versus normal samples (P <0.1, t-test). Second, the dCHIP normalized hybrdidization data were exported into, and analyzed by SAM, which yielded 373 Affymetrix fragments expressed at threefold or greater intensity in the cancers, using a threshold median discovery rate (FDR) of <10 genes. The merging of the two datasets from dCHIP and SAM analysis yielded 347 overlapping Affymetrix fragments. After purging this set of 347 Affymetrix fragments for duplicate fragments from the same gene, unnamed hypothetical proteins, and ESTs, we identified 282 unique known genes up-regulated at least threefold or higher in primary resected cancers and biliary cancer cell lines versus normal epithelial scrapings. Table 1 ▶ lists a representative subset of 50 annotated genes identified in our analysis; the complete list of 282 up-regulated genes is available publicly on the Johns Hopkins gallbladder and bile duct cancer website (http://pathology2.jhu.edu/gbbd/microarray). We then performed a similar series of analyses comparing only the nine biliary cancer cell lines with the five biliary epithelial scrapings. This direct comparison of neoplastic versus non-neoplastic epithelium yielded 700 Affymetrix fragments by dCHIP analysis, 717 fragments by SAM analysis, and 638 fragments that were common to both analyses. The 638 fragments were then parsed for duplicates, unnamed hypothetical proteins, and ESTs, identifying 514 known genes that were overexpressed threefold or greater in the cell lines versus normal epithelium (data available publicly on our website). The larger number of differentially expressed genes when only cell lines were compared with normal epithelium possibly reflects either the “dilution” effect caused by non-neoplastic epithelial and stromal elements within primary cancers, or the effects of in vitro culture. In passing, it should be mentioned that we also identified 513 genes that were down-regulated threefold or greater in the cancers versus normals; however, since the objective of the current study was to identify novel tumor markers, the ensuing discussion will focus on up-regulated genes only.
Table 1.
Representative Subset of Differentially Overexpressed Genes in Biliary Cancers
| Affymetrix tag number | Gene name | Fold change | p Value | Chromosome | Function |
|---|---|---|---|---|---|
| 205239_at | Amphiregulin (schwanomma-derived growth factor) | 5.64793 | 0.011387 | 4q13-q21 | Autocrine growth factor, mitogen |
| 204832_s_at | Bone morphogenetic protein receptor, type IA | 3.92410 | 0.000314 | 10q22.3 | Integral membrane protein, TGF-β mediator |
| 209642_at | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | 3.37973 | 0.000275 | 2q14 | Spindle assembly checkpoint regulator |
| 216602_s_at | Calreticulin | 3.08473 | 0.000315 | 19p13.3-p13.2 | Protein folding, calcium storage |
| 206075_s_at | Casein kinase 2, alpha 1 polypeptide | 3.41682 | 0.000074 | 20p13 | Phosphorylation of proteins (eg, p53) |
| 203968_s_at | CDC6 cell division cycle 6 homolog | 5.16898 | 0.000004 | 17q21.3 | DNA replication checkpoint control |
| 203213_at | Cell division cycle 2, G1 to S and G2 to M | 3.51015 | 0.000139 | 10q21.1 | Cell cycle regulator |
| 205394_at | CHK1 checkpoint homolog (S. pombe) | 3.64472 | 0.000261 | 11q24 | Cell cycle regulator |
| 202613_at | CTP synthase | 4.20409 | 0.000019 | 1p34.1 | Phospholipid and nucleic acid biosynthesis; multidrug resistance associated |
| 203418_at | Cyclin A2 | 5.02650 | 0.000076 | 4q27 | Cell cycle regulator |
| 200953_s_at | Cyclin D2 | 3.61139 | 0.005114 | 12p13 | Cell cycle regulator |
| 205034_at | Cyclin E2 | 4.01328 | 0.000196 | 8q21.3 | Cell cycle regulator |
| 212816_s_at | Cystathione beta synthase | 7.39167 | 0.00009 | 21q22.3 | Homocysteine metabolism; induces susceptibility to cytosine arabinoside |
| 201571_s_at | dCMP deaminase | 3.87652 | 0.000056 | 4q35.1 | Target enzyme for gemcitabine |
| 202576_s_at | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) | 3.29770 | 0.000058 | 16q22.3 | mRNA nuclear export |
| 204602_at | Dickkopf homolog 1 (Xenopus laevis) | 12.2781 | 0.000046 | 10q11.2 | Wnt signaling antagonist |
| 213789_at | Emopamil binding protein (sterol isomerase) | 3.50690 | 0.024251 | Xp11.23-p11.22 | Binds tamoxifen, SR31747A |
| 203499_at | EphA2 | 3.75649 | 0.001234 | 1p36 | Receptor tyrosine kinase |
| 207981_s_at | Estrogen-related receptor gamma | 5.67041 | 0.006618 | 1q41 | Binds estrogen response elements |
| 201436_at | Eukaryotic translation initiation factor 4E | 4.03826 | 0.000002 | 4q21-q25 | Akt/mTOR signaling pathway |
| 218350_s_at | Geminin | 3.90463 | 0.000120 | 6p21.32 | Cell proliferation antigen |
| 210997_at | Hepatocyte growth factor (hepapoietin A; scatter factor) | 7.70259 | 0.016513 | 7q21.1 | Cell development and growth |
| 203138_at | Histone acetyltransferase 1 | 4.75385 | 0.000009 | 2q31.2-33.1 | Histone acetylation and gene expression |
| 204779_s_at | Homeo box B7 | 7.98564 | 0.000113 | 17q21-q22 | Gut and lymphoid development |
| 203627_at | Insulin-like growth factor 1 receptor | 3.57119 | 0.000542 | 15q25-q26 | Insulin receptor signaling |
| 206104_at | ISL1 transcription factor, LIM/homeodomain, (islet-1) | 13.3186 | 0.003433 | 5q11.1 | Insulin enhancer-binding protein |
| 20908_s | Jagged-1 | 5.41679 | 0.004756 | 20p12.1 | Notch ligand |
| 216815_at | Laminin receptor 1 | 3.18114 | 0.00713 | 3p21.3 | Receptor for cell adhesion molecule laminin |
| 205397_x_at | MAD, mothers against decapentaplegic homolog 5 (Drosophila) | 3.50543 | 0.002022 | 5q31 | TGF-β receptor signaling, transcription |
| 203362_s | MAD2 mitotic arrest deficient-like 1 | 3.85297 | 0.000011 | 4q27 | Mitotic checkpoint control |
| 203552_at | Mitogen-activated protein kinase kinase kinase kinase 5 | 3.08865 | 0.000236 | 14q11.2-q21 | Ser/Thr protein kinase |
| 219148_at | PDZ-binding kinase; T-cell originated protein kinase | 3.39109 | 0.000268 | 8p21.1 | Mitotic kinase |
| 204186_s_at | Peptidylprolyl isomerase D (cyclophilin D) | 7.04929 | 0.000001 | 4q31.3 | Estrogen receptor complex member |
| 210145_at | Phospholipase A2, group IVA (cytosolic) | 3.54762 | 0.000421 | 1q25 | Catalyzes arachidonic acid release |
| 213226_at | Polymyositis/scleroderma autoantigen 1 | 5.16640 | 0.000039 | 4q28.1 | Nucleolar and nucleoplasm protein |
| 205512_s_at | Programmed cell death 8 (apoptosis-inducing factor) | 3.43048 | 0.000079 | Xq25-26 | Cell death mediator |
| (Table continues) |
Multiple classes of differentially up-regulated genes were identified in this study and included proliferation and cell cycle antigens (cyclins D2 and E2, cdc2/p34, and geminin), 21-23 transcription factors (homeobox B7 and islet-1), 24,25 growth factors and growth factor receptors (hepatocyte growth factor, amphiregulin, and insulin-like growth factor 1 receptor), 26-28 and enzymes modulating sensitivity to chemotherapeutic agents (cystathionine β synthase, dCMP deaminase, and CTP synthase) 29-31 Subsets of genes involved in biologically relevant pathways were also identified, such as genes in the transforming growth factor β pathway (bone morphogenetic protein receptor 1A [BMPR1A], mothers against decapentaplegic homolog 5 [MADH5], and TGF-β receptor-associated protein 1 [TRAP-1]), 32-34 or genes involved in steroid metabolism, particularly in the estrogen/androgen pathways (estrogen-related receptor-γ, sterol isomerase, and steroid-5-α-reductase). 35-37 Interestingly, we identified several genes that are related to pathways rapidly emerging as novel therapeutic targets in many cancer types. Notable among these were cytosolic phospholipase A2, which generates arachidonic acid, the substrate for the cyclooxygenase (COX) enzyme, 38,39 and two important downstream mediators of the tumorigenic Akt/mammalian target of rapamycin (mTOR) pathway in humans, ribosomal protein S6 kinase, and eukaryotic translation initiation factor 4E. 40
A literature search of PubMed (www.ncbi.nlm.nih.gov/PubMed) revealed that several of the genes up-regulated in this study have previously been reported as overexpressed, either singly or through global microarray expression analyses in other cancer types, in principle validating our approach. A second PubMed search using the gene name and either “cholangiocarcinoma,” “biliary,” “bile duct,” or “gallbladder” in the text search revealed that only three up-regulated genes (hepatocyte growth factor, proliferating cell nuclear antigen, and cytosolic phospholipase A2) have been previously reported as specifically overexpressed in biliary cancers. 41-43 Thus, the overwhelming majority of up-regulated genes reported in this study represent novel tumor markers and cellular targets for this cancer type.
Validation of Differentially Up-Regulated Genes in Biliary Cancers
The differential overexpression of a subset of genes (n = 7) was validated using a combination immunohistochemistry and in situ hybridization in archival biliary cancers and RT-PCR on biliary cancer cell lines. A biliary cancer tissue microarray containing 40 biliary cancers was immunolabeled with antibodies against BMPR1A, geminin, topoIIα, and cdc2/p34 to confirm the overexpression of the corresponding protein product. The results of immunohistochemical analysis are summarized in Table 2 ▶ .
Table 2.
Immunohistochemical Analysis of Candidate Overexpressed Genes in Tissue Microarrays of Biliary Cancer
| Diffuse | Focal | Negative | Not evaluable | |
|---|---|---|---|---|
| BMPR1A | 23/40 (58%) | 13/40 (33%) | 3/40 (7%) | 1/40 (2%) |
| Geminin | 22/40 (55%) | 12/40 (30%) | 5/40 (13%) | 1/40 (2%) |
| Topoisomerase II | 15/40 (38%) | 13/40 (33%) | 11/40 (27%) | 1/40 (2%) |
| Cdc2 (p34) | 21/40 (53%) | 12/40 (30%) | 6/40 (15%) | 1/40 (2%) |
BMPR1A is a member of the cell surface receptor TGF-β superfamily and is expressed in human skeletal muscle, heart, and placenta under normal conditions. 32 BMPR1A immunolabeling was absent in normal biliary epithelium (<5% labeling) (Figure 2A) ▶ . In contrast, 91% of cancers demonstrate diffuse or focal cell membrane and cytoplasmic immunolabeling of BMPR1A (Figure 2B) ▶ . Similarly, 85% of biliary cancers demonstrate diffuse or focal nuclear overexpression of the novel protein geminin, 23 while normal biliary epithelium was negative (Figure 2, C and D) ▶ . The cell cycle protein cdc2/p34 serves as a promoter of mitosis and is regulated by tyrosine phosphorylation. 44 Normal biliary epithelium lacked nuclear cdc2/p34 expression (Figure 2E) ▶ , whereas 83% of biliary tract cancers demonstrated diffuse or focal nuclear expression (Figure 2F) ▶ . Similarly, labeling with topoIIα, which regulates the topology of DNA, 45 was absent in normal biliary tract epithelium (Figure 2G) ▶ , but was diffusely or focally expressed in 71% of biliary cancers (Figure 2H) ▶ (Table 2) ▶ .
Figure 2.
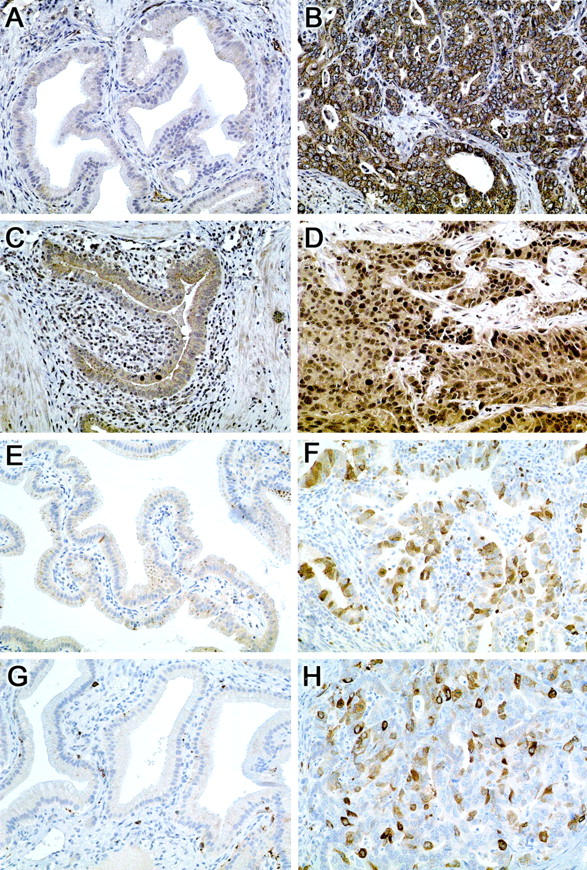
Immunohistochemical validation of genes overexpressed in biliary cancers using a biliary cancer tissue microarray. Bone morphogenetic protein receptor type 1A (BMPR1A) expression in normal biliary epithelium (A) and in adenocarcinoma of the biliary tract (B). BMPR1A is absent in normal biliary epithelium, but is diffusely expressed in a cytoplasmic and membranous distribution in the invasive adenocarcinoma. Geminin expression is absent in normal biliary epithelium (C) while biliary tract adenocarcinoma is diffusely positive (D). Cdc2 (p34) expression is absent in normal biliary epithelium (E), while it is diffusely expressed in biliary tract adenocarcinoma (F). Topoisomerase II α expression is absent in normal biliary epithelium (G) but diffuse nuclear labeling is seen in adenocarcinoma of the biliary tract (H).
In situ hybridization for the transcription factor islet-1 (ISL-1) was performed using the biliary cancer tissue microarray to confirm differential overexpression. ISL-1 is a downstream target of the Sonic hedgehog pathway, and is essential for development of the endocrine pancreas. 46,47 In situ hybridization for ISL-1 demonstrated neoplastic epithelial expression in 26 of 40 (65%) biliary tract carcinomas, while minimal expression was seen in non-neoplastic biliary epithelium (Figure 3,A and B) ▶ ; sense control yielded no signal in the cancer cells (Figure 3C) ▶ .
Figure 3.
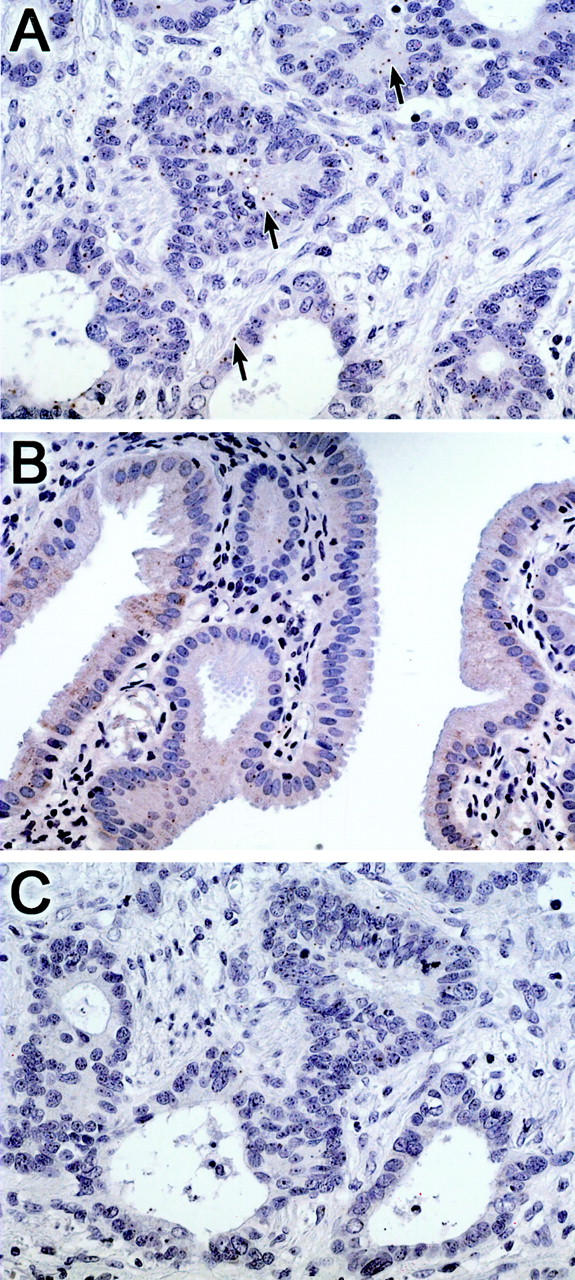
Non-radioisotopic in situ hybridization for validation of Islet-1 expression in biliary cancer using a biliary cancer tissue microarray. Islet-1 transcripts are detected in a biliary adenocarcinoma using the ISL-1 antisense riboprobe (arrows) (A), but transcripts are not detectable in normal biliary epithelium using an antisense riboprobe (B), or in the serial section of the cancer using the ISL-1 sense (control) riboprobe (C).
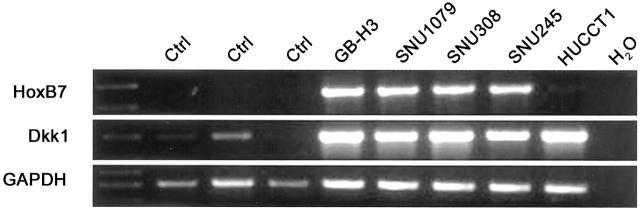
We additionally confirmed the overexpression of two genes, homeobox B7 (HoxB7) and dickkopf 1 (Dkk-1), in five biliary cancer cell lines and three non-neoplastic biliary epithelial scrapings by semi-quantitative RT-PCR. HoxB7 is a homeobox containing gene normally expressed during development and aberrantly overexpressed in several human cancers, 48 whereas Dkk-1 is a p53-responsive gene that inhibits canonical Wnt signaling pathways. 49 Both transcripts were expressed and up-regulated in the cancer cell lines we examined in comparison to normal biliary epithelium (Figure 4) ▶ .
Figure 4.
RT-PCR validation of overexpressed genes in biliary cancers. Two genes overexpressed in the Affymetrix analysis, homeobox B7 (HoxB7) and dickkopf-1 homolog (Dkk1), were selected for RT-PCR in five biliary cancer cell lines (GBH3, SNU1079, SNU308, SNU245, and HuCCT1), and three samples of normal biliary epithelium. HoxB7 expression is absent in the normal epithelium, but is present in 4 of 5 biliary cancer cell lines; water control is appropriately negative (top panel). Dkk-1 expression is present in normal epithelium, but its expression is up-regulated in 5 of 5 cell lines; water control is appropriately negative (middle panel). GAPDH is used as an external control (bottom panel).
Discussion
Biliary tract carcinoma represents the second most common primary cancer of the hepatobiliary system; each year as many as 3500 individuals succumb to this disease in the United States alone, while mortality rates are far higher in some parts of the world such as the Far East, Chile, and parts of Southeast Asia. 2 Known risk factors include primary sclerosing cholangitis, cholelithiasis, chronic parasitic infection, and chemical exposure. 3 Although early preventative steps can be taken to lower the risk of developing biliary cancers, the overall prognosis once cancer develops is poor, with only a 10% 5-year survival rate for advanced disease. 3 New methods for early detection, a better understanding of the biological mechanisms underlying cancer progression, and cancer-targeted treatment modalities are urgently needed to reduce the mortality from this lethal disease.
The availability of global expression platforms such as oligonucleotide and cDNA microarrays has greatly facilitated the identification of novel tumor markers in many cancer types. For example, oligonucleotide microarrays have enabled us to identify nearly 100 novel tumor markers with immediate diagnostic and therapeutic relevance in pancreatic adenocarcinomas. 4 In this study, we identify for the first time the up-regulation of a large number of genes in biliary cancers relative to non-neoplastic biliary epithelium using the recently developed Affymetrix U133A expression microarrays. Of the 282 genes identified as overexpressed threefold or greater in biliary cancers, only three have been reported previously in the specific context of biliary cancers. 41-43 Thus, this study has enormous potential in terms of expanding our knowledge of biliary cancer pathogenesis and identification of novel cellular targets in this cancer type. In addition to using the recently developed Affymetrix U133A microarray that probes for ∼22,000 transcripts on a single platform, other methodological strengths of our study include the use of a large number of biliary cancer cell lines representing different sites in the biliary tree, verification of neoplastic cellularity of primary resected cancers using frozen section examination, and the appropriate use of enriched biliary epithelium (as opposed to “full thickness” biliary ductal sections) as controls for comparison of gene expression. The dCHIP and SAM software used for data analysis in this study have emerged as two of the most widely used and robust programs for two-class comparison of microarray expression data. 17,18,50-52 Finally, we have validated the gene expression data for a subset of genes using a second independent technique; all seven genes were confirmed as overexpressed in biliary cancers compared to non-neoplastic biliary epithelium, irrespective of the detection technique (immunohistochemistry, in situ hybridization, or RT-PCR). While we only chose to identify, validate, and discuss overexpressed genes in this study, it should be mentioned that we have also identified over 500 genes that are underexpressed threefold or more in the tumors compared to normal samples; although not discussed in the context of this manuscript, these genes should serve as an invaluable resource in future studies for identification of tumor suppressor genes that are the target of genetic or epigenetic inactivation in biliary cancers.
Of the 282 genes identified as differentially overexpressed in biliary cancers compared to normal epithelium, many conform to cellular processes intuitive to tumor cells, such as proliferation, DNA synthesis and RNA processing, and metabolism. A subset of these genes have emerged as candidates for novel molecular therapy in several cancer models, and thus, their reported overexpression in biliary cancers provides an important basis for future in vitro and in vivo trials, targeting the corresponding proteins. For example, the cdc2/p34 protein, which we confirmed as overexpressed in our biliary cancer tissue microarrays, is the catalytic subunit of a protein kinase complex, called the M-phase promoting factor that induces entry into mitosis and is universal among eukaryotes. 22 Recently, cdc2/p34 has also been demonstrated to play a role in promoting cell survival, mediated by its site-specific phosphorylation and activation of the anti-apoptotic protein, survivin. 53 Survivin interferes with spontaneous and chemotherapy-induced cell death, and thus loss of survivin activation by pharmacological inhibition of cdc2/p34 has emerged as a novel chemotherapeutic strategy. Topoisomerase IIα, the overexpression of which was also confirmed in our tissue microarrays, is important in mediating susceptibility to drugs that specifically target this enzyme; response to topoisomerase inhibitors is enhanced in breast cancers that overexpress topo II, 54,55 providing an excellent example of how molecular events can direct choice of therapeutic regimens.
Growth factors and growth factor receptors that were up-regulated in biliary cancers included, among others, hepatocyte growth factor (HGF) (7-fold), and the insulin-like growth factor 1 (IGF-1) receptor (3.5-fold). HGF has been previously reported as up-regulated in biliary cancers, often in conjunction with its receptor, c-met. 42 Although c-met was not included in the gene list that compared all cancers versus normals, c-met was one of the additional genes that emerged in the comparison of cell lines versus normals (http://pathology2.jhu.edu/gbbd/microarray). HGF is identical to scatter factor, and it is a potent mitogen for hepatocytes. Biliary epithelial cells produce little or no HGF, while biliary cancers acquire the ability to secrete this growth factor in conjunction with up-regulation of c-met. 56 In addition to biliary cancers, dysregulation of the HGF/c-met pathway has been reported in hepatocellular, lung, prostate, and breast cancers, glioblastomas, and melanomas, wherein this pathway promotes angiogenesis, cellular motility, growth, and invasion. 26,57,58 HGF may stimulate tumor invasion in an autocrine loop through c-met, via down-regulation of cellular adhesion molecules such as E-cadherin. 59 Besides providing an avenue for therapy by signal antagonism, HGF is also useful as a potential diagnostic tool in biliary cancer patients. Elevated serum HGF levels are found in a variety of cancer types, and may correlate with prognosis. 60-62 Similarly, overexpression of the IGF-1 receptor is commonly found in many cancers, and its level of expression may also correlate with prognosis. 28,63 As a cell surface molecule upstream of many critical signaling pathways, the IGF-1 receptor represents an important target for cancer therapy. Indeed, antagonism of cellular IGF-1 receptor by overexpression of dominant-negative mutant forms or by interference using small interfering RNA (siRNA) results in decreased tumorigenicity in in vitro models. 64,65 Thus, the IGF-1 receptor represents another potential target for therapy identified in biliary cancers.
The mammalian target of rapamycin (mTOR) is a member of a recently identified family of protein kinases termed phosphoinositide 3-kinase (PI3K)-related kinases, which are involved in many critical regulatory cellular functions pertaining to cell cycle progression, DNA repair, and DNA damage. mTOR functions downstream of the proto-oncogenes PI3K and Akt, and is itself upstream of two distinct pathways that control translation of specific subsets of mRNA, one involving ribosomal protein p70 synthase kinase (p70s6k), and the other eukaryotic initiation factor 4E binding protein-1 (4EBP1). 66 In response to PI3K/Akt signaling, mTOR rapidly phosphorylates both p70s6k and 4EBP1, the latter leading to release of eIF4E, resulting in initiation of translation; these actions of mTOR can be inhibited by the administration of the macrolide rapamycin. Currently, CCI-779, a water soluble analog of rapamycin is undergoing evaluation as an anti-tumor agent in a variety of cancers. 40 It is likely that a subset of biliary cancers will be resistant to inhibition of mTOR. The identification of two pathway genes, p70s6k and eIF4E, that are differentially up-regulated in biliary cancers compared to normal epithelium provide alternative downstream targets for inhibition.
Up-regulation of the pro-inflammatory enzyme cytosolic phospholipase A2 (cPLA2) has been previously reported in biliary cancers. 41 Phospholipase A2s are a group of enzymes that catalyze the hydrolysis of the sn-2-ester bond of phospholipids, resulting in production of free fatty acid and lysophospholipids, which can then be further metabolized to produce eicosanoids and platelet-activating factor. 67 These lipid molecules are crucial for various cellular responses, such as inflammation, signal transduction, and cell proliferation. cPLA2 is a key enzyme in the liberation of arachidonic acid (AA) from membrane phospholipids for subsequent production of bioactive eicosanoids in activated cells; notably, cPLA2 is itself activated in response to signaling mediated by another differentially overexpressed gene, hepatocyte growth factor. 68 Increasing evidence has suggested a causal relationship between cancer development, including cholangiocarcinoma, and increased expression of eicosanoid-forming enzymes. For example, increased expression of cyclooxygenase (COX) enzymes has been reported in biliary cancers. 69,70 Similar to the anti-proliferative effects of COX inhibition, targeting cPLA2 also results in decreased growth of cholangiocarcinoma cells. 41 Thus, the demonstration of up-regulated cPLA2 in biliary cancers provides another important potential therapeutic target for the control of biliary cancer and, perhaps, chemoprevention in patients with precursor conditions such as primary sclerosing cholangitis.
Several genes involved in steroid metabolism, specifically the estrogen/androgen pathways (estrogen-related receptor-γ, sterol isomerase, cyclophilin D, and steroid-5-α-reductase) were also up-regulated in biliary cancers compared to normal epithelium. 35-37 Biliary cancers, especially those arising in the gallbladder, occur more frequently in women, 71 and this has been attributed to the increased incidence of gallstones in women; 2 however, direct or indirect estrogen-related effects may also function to promote carcinogenesis. The expression of sterol isomerase (emopamil-binding protein) correlates with poor prognosis in breast cancers, although the mechanistic aspects remain poorly understood. 72 Notably, SR31747A, a recently described synthetic ligand with anti-proliferative activity in numerous cell lines binds to and inhibits sterol isomerase, and overexpression of the enzyme reverts the anti-proliferative effects of this drug. 73 Sterol isomerase also binds to and is inhibited by the synthetic estrogen homolog 4-hydroxytamoxifen. 74 Additional functional studies are warranted to demonstrate whether inhibition of sterol isomerase can emerge as a therapeutic strategy in biliary cancers. We also identified an up-regulation of the estrogen-related receptor-γ gene in our study. Estrogen-related receptor-γ is a member of an orphan nuclear receptor family that shares significant homology with estrogen receptors and, although this receptor does not recognize naturally occurring estrogens, is capable of binding to the estrogen response element. 75,76 Like sterol isomerase, estrogen-related receptor-γ binds to and is inhibited by tamoxifen. 77
Cystathionine β synthase (CBS), which is mutated in patients with homocystinuria, is a cytosolic enzyme that also modulates sensitivity to the cytotoxic drug, cytosine arabinoside (ara-C). 29,78 Neoplastic cells with high levels of CBS demonstrate increased sensitivity to ara-C. Therefore, a sevenfold up-regulation of CBS transcripts in biliary cancers merits considering ara-C-containing regimens for therapy of biliary cancers. Finally, we also report up-regulation of dCMP deaminase, whose product is the target enzyme for the cytotoxic agent 2′,2′-difluorodeoxycytidine (gemcitabine). 30,79 Gemcitabine has emerged as a widely used chemotherapeutic agent for solid gastrointestinal malignancies, including biliary cancers. Fourfold up-regulation of dCMP deaminase in biliary cancers may suggest that, while gemcitabine should be considered as a viable therapeutic option, the corresponding dosages may need to be calibrated accordingly.
In summary, we report for the first time a global gene expression analysis of biliary cancers using Affymetrix oligonucleotide microarrays and a combination of primary resected tumors, biliary cell lines, and non-neoplastic biliary epithelial scrapings. We report 282 unique known genes as being differentially up-regulated threefold or greater in the cancers compared to normal epithelium, and validate a subset of genes using an alternative platform. The differentially overexpressed genes are publicly available on our website at http://pathology2.jhu.edu/gbbd/microarray, and represents an enormous resource for the scientific community to study the pathogenesis of this lethal disease and develop appropriate cancer-specific targeted therapies.
Table 1A.
(continued)
| 201202_at | Proliferating cell nuclear antigen | 4.02376 | 0.000006 | 20pter-p12 | Cell cycle regulator |
|---|---|---|---|---|---|
| 209228_x_at | Putative prostate cancer tumor suppressor | 5.91911 | 0.000976 | 8p22 | Transmembrane protein, CpG methylated |
| 209849_s_at | RAD51 homolog C (S. cerevisiae) | 4.54601 | 0.000179 | 17q23.1 | DNA repair, DNA recombination; “17q23 amplicon” |
| 204146_at | RAD51-interacting protein | 5.17207 | 0.000000 | 12p13.2-p13.1 | Genetic recombination |
| 217457_s_at | RAP1, GTP-GDP dissociation stimulator 1 | 3.21201 | 0.000089 | 4q21-q25 | GTP-GDP dissociation stimulator |
| 215098_at | Retinoid X receptor, beta | 3.13613 | 0.000124 | 6p21.3 | Transcription factor |
| 204171_at | Ribosomal protein S6 kinase, 70kD | 3.10282 | 0.001765 | 17q23.1 | Akt/mTOR pathway; “17q23 amplicon” |
| 211056_s_at | Steroid-5-alpha-reductase, alpha polypeptide 1 | 5.17492 | 0.00052 | 5p15 | Androgen metabolism |
| 202817_s_at | Synovial sarcoma translocation, chromosome 18 | 3.26773 | 0.000037 | 18q11.2 | Chromosomal translocation |
| 205210_at | TGF beta receptor associated protein 1 | 5.56609 | 0.000628 | 2q12.1 | TGF-β receptor binding protein |
| 201291_s_at | Topoisomerase (DNA) II alpha (170kD) | 3.90944 | 0.000021 | 17q21-q22 | DNA maintenance and structure |
| 204147_s_at | Transcription factor Dp-1 | 4.29968 | 0.000410 | 13q34 | Cell cycle regulator, transcription |
| 203856_at | Vaccinia related kinase 1 | 3.19922 | 0.000418 | 14q32 | Cell cycle regulator |
| 212533_at | WEE1+ homolog (S. pombe) | 8.28110 | 0.000007 | 11p15.3-p15.1 | Cell cycle regulator |
| 217821_s_at | WW domain binding protein 11 | 3.62317 | 0.000076 | 12p13.31 | SH3 domain-binding protein |
Acknowledgments
We thank Leighton Stein for his expert assistance.
Footnotes
Address reprint requests to Anirban Maitra, M.D., 720 Rutland Avenue, Ross 632 Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21212. E-mail: amaitra1@jhmi.edu.
Supported by the family of Margaret Lee. This work was also partially supported by shared resources of the Roswell Park Cancer Center Support Grant P30 CA16056.
References
- 1.Greenlee RT, Hill-Harmon MB, Murray T, Thun M: Cancer statistics, 2001. CA Cancer J Clin 2001, 51:15-36 [DOI] [PubMed] [Google Scholar]
- 2.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM: Biliary tract cancers. N Engl J Med 1999, 341:1368-1378 [DOI] [PubMed] [Google Scholar]
- 3.Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, Alonso de Ruiz P, Aristi Urista G, Nervi F: Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin 2001, 51:349-364 [DOI] [PubMed] [Google Scholar]
- 4.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH: Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 2002, 160:1239-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I: Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 2001, 98:13784-13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D: Molecular portraits of human breast tumours. Nature 2000, 406:747-752 [DOI] [PubMed] [Google Scholar]
- 7.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM: Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412:822-826 [DOI] [PubMed] [Google Scholar]
- 8.Maitra A, Wistuba II, Virmani AK, Sakaguchi M, Park I, Stucky A, Milchgrub S, Gibbons D, Minna JD, Gazdar AF: Enrichment of epithelial cells for molecular studies. Nat Med 1999, 5:459-463 [DOI] [PubMed] [Google Scholar]
- 9.Saijyo S, Kudo T, Suzuki M, Katayose Y, Shinoda M, Muto T, Fukuhara K, Suzuki T, Matsuno S: Establishment of a new extrahepatic bile duct carcinoma cell line, TFK-1. Tohoku J Exp Med 1995, 177:61-71 [DOI] [PubMed] [Google Scholar]
- 10.Steffen M, Zuehlke I, Scherdin U: Motility factors identified in supernatants of human cholangiocarcinoma cell lines. Int J Oncol 2001, 18:1107-1112 [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto H, Nishino H: Effect of recombinant human basic fibroblast growth factor (bFGF) on the growth of human tumor cell lines. Hum Cell 1996, 9:129-140 [PubMed] [Google Scholar]
- 12.Miyazaki M, Ohashi R, Tsuji T, Mihara K, Gohda E, Namba M: Transforming growth factor-β 1 stimulates or inhibits cell growth via down- or up-regulation of p21/Waf1. Biochem Biophys Res Commun 1998, 246:873-880 [DOI] [PubMed] [Google Scholar]
- 13.Ku JL, Yoon KA, Kim IJ, Kim WH, Jang JY, Suh KS, Kim SW, Park YH, Hwang JH, Yoon YB, Park JG: Establishment and characterisation of six human biliary tract cancer cell lines. Br J Cancer 2002, 87:187-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Shimura H, Aoki Y, Date K, Matsumoto K, Nakamura T, Tanaka M: Hepatocyte growth factor stimulates the invasion of gallbladder carcinoma cell lines in vitro. Clin Exp Metastasis 1998, 16:74-82 [DOI] [PubMed] [Google Scholar]
- 15.Li C, Hung Wong W: Model-based analysis of oligonucleotide arrays: model validation, design issues, and standard error application. Genome Biol 2001, 2RESEARCH0032 [DOI] [PMC free article] [PubMed]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001, 98:5116-5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Selaru FM, Yin J, Zou TT, Shustova V, Mori Y, Sato F, Liu TC, Olaru A, Wang S, Kimos MC, Perry K, Desai K, Greenwald BD, Krasna MJ, Shibata D, Abraham JM, Meltzer SJ: Artificial neural networks and gene filtering distinguish between global gene expression profiles of Barrett’s esophagus and esophageal cancer. Cancer Res 2002, 62:3493-3497 [PubMed] [Google Scholar]
- 19.van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, De Marzo AM, Hruban RH, Maitra A: Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol 2002, 161:1541-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE: Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol 2002, 160:91-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega S, Malumbres M, Barbacid M: Cyclin D-dependent kinases, INK4 inhibitors, and cancer. Biochim Biophys Acta 2002, 1602:73-87 [DOI] [PubMed] [Google Scholar]
- 22.O’Connor DS, Wall NR, Porter AC, Altieri DC: A p34(cdc2) survival checkpoint in cancer. Cancer Cell 2002, 2:43-54 [DOI] [PubMed] [Google Scholar]
- 23.Wohlschlegel JA, Kutok JL, Weng AP, Dutta A: Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am J Pathol 2002, 161:267-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Care A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, Peschle C, Colombo MP: HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res 2001, 61:6532-6539 [PubMed] [Google Scholar]
- 25.Watanabe Y, Nakamura H: Control of chick tectum territory along dorsoventral axis by Sonic hedgehog. Development 2000, 127:1131-1140 [DOI] [PubMed] [Google Scholar]
- 26.Trusolino L, Comoglio PM: Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat Rev Cancer 2002, 2:289-300 [DOI] [PubMed] [Google Scholar]
- 27.Normanno N, Bianco C, De Luca A, Salomon DS: The role of EGF-related peptides in tumor growth. Front Biosci 2001, 6:D685-707 [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Rohan T: Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst 2000, 92:1472-1489 [DOI] [PubMed] [Google Scholar]
- 29.Taub JW, Huang X, Ge Y, Dutcher JA, Stout ML, Mohammad RM, Ravindranath Y, Matherly LH: Cystathionine-β-synthase cDNA transfection alters the sensitivity and metabolism of 1-β-D-arabinofuranosylcytosine in CCRF-CEM leukemia cells in vitro and in vivo: a model of leukemia in Down syndrome. Cancer Res 2000, 60:6421-6426 [PubMed] [Google Scholar]
- 30.Rieger J, Durka S, Streffer J, Dichgans J, Weller M: Gemcitabine cytotoxicity of human malignant glioma cells: modulation by antioxidants, BCL-2, and dexamethasone. Eur J Pharmacol 1999, 365:301-308 [DOI] [PubMed] [Google Scholar]
- 31.Politi PM, Xie F, Dahut W, Ford H, Jr, Kelley JA, Bastian A, Setser A, Allegra CJ, Chen AP, Hamilton JM: Phase I clinical trial of continuous infusion cyclopentenyl cytosine. Cancer Chemother Pharmacol 1995, 36:513-523 [DOI] [PubMed] [Google Scholar]
- 32.Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, Launonen V, Virta S, Pilarski R, Salovaara R, Bodmer WF, Conrad BA, Dunlop M, Hodgson SV, Iwama T, Jarvinen H, Kellokumpu I, Kim JC, Leggett B, Markie D, Mecklin JP, Neale K, Phillips R, Piris J, Rozen P, Houlston RS, Aaltonen LA, Tomlinson IP, Eng C: Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet 2001, 69:704-711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Kinzler KW, Vogelstein B: Going mad with Smads. N Engl J Med 1999, 341:1144-1146 [DOI] [PubMed] [Google Scholar]
- 34.Wurthner JU, Frank DB, Felici A, Green HM, Cao Z, Schneider MD, McNally JG, Lechleider RJ, Roberts AB: Transforming growth factor-β receptor-associated protein 1 is a Smad4 chaperone. J Biol Chem 2001, 276:19495-19502 [DOI] [PubMed] [Google Scholar]
- 35.Ariazi EA, Clark GM, Mertz JE: Estrogen-related receptor α and estrogen-related receptor γ associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res 2002, 62:6510-6518 [PubMed] [Google Scholar]
- 36.Villalva C, Trempat P, Greenland C, Thomas C, Girard JP, Moebius F, Delsol G, Brousset P: Isolation of differentially expressed genes in NPM-ALK-positive anaplastic large cell lymphoma. Br J Haematol 2002, 118:791-798 [DOI] [PubMed] [Google Scholar]
- 37.Brawley OW, Ford LG, Thompson I, Perlman JA, Kramer BS: 5-α-reductase inhibition and prostate cancer prevention. Cancer Epidemiol Biomarkers Prev 1994, 3:177-182 [PubMed] [Google Scholar]
- 38.Osterstrom A, Dimberg J, Fransen K, Soderkvist P: Expression of cytosolic and group X secretory phospholipase A(2) genes in human colorectal adenocarcinomas. Cancer Lett 2002, 182:175-182 [DOI] [PubMed] [Google Scholar]
- 39.Graff JR, Konicek BW, Deddens JA, Chedid M, Hurst BM, Colligan B, Neubauer BL, Carter HW, Carter JH: Expression of group IIa secretory phospholipase A2 increases with prostate tumor grade. Clin Cancer Res 2001, 7:3857-3861 [PubMed] [Google Scholar]
- 40.Hidalgo M, Rowinsky EK: The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 2000, 19:6680-6686 [DOI] [PubMed] [Google Scholar]
- 41.Wu T, Han C, Lunz JG, 3rd, Michalopoulos G, Shelhamer JH, Demetris AJ: Involvement of 85-kd cytosolic phospholipase A(2) and cyclooxygenase-2 in the proliferation of human cholangiocarcinoma cells. Hepatology 2002, 36:363-373 [DOI] [PubMed] [Google Scholar]
- 42.Sirica AE, Lai GH, Zhang Z: Biliary cancer growth factor pathways, cyclo-oxygenase-2, and potential therapeutic strategies. J Gastroenterol Hepatol 2001, 16:363-372 [DOI] [PubMed] [Google Scholar]
- 43.Batheja N, Suriawinata A, Saxena R, Ionescu G, Schwartz M, Thung SN: Expression of p53 and PCNA in cholangiocarcinoma and primary sclerosing cholangitis. Mod Pathol 2000, 13:1265-1268 [DOI] [PubMed] [Google Scholar]
- 44.Lee MG, Norbury CJ, Spurr NK, Nurse P: Regulated expression and phosphorylation of a possible mammalian cell-cycle control protein. Nature 1988, 333:676-679 [DOI] [PubMed] [Google Scholar]
- 45.Tsai-Pflugfelder M, Liu LF, Liu AA, Tewey KM, Whang-Peng J, Knutsen T, Huebner K, Croce CM, Wang JC: Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21–22. Proc Natl Acad Sci USA 1988, 85:7177-7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Drucker DJ: The LIM domain homeobox gene isl-1 is a positive regulator of islet cell-specific proglucagon gene transcription. J Biol Chem 1995, 270:12646-12652 [DOI] [PubMed] [Google Scholar]
- 47.van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, Van Deventer SJ, Peppelenbosch MP: Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology 2001, 121:317-328 [DOI] [PubMed] [Google Scholar]
- 48.Naora H, Yang YQ, Montz FJ, Seidman JD, Kurman RJ, Roden RB: A serologically identified tumor antigen encoded by a homeobox gene promotes growth of ovarian epithelial cells. Proc Natl Acad Sci USA 2001, 98:4060-4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shou J, Ali-Osman F, Multani AS, Pathak S, Fedi P, Srivenugopal KS: Human Dkk-1, a gene encoding a Wnt antagonist, responds to DNA damage and its overexpression sensitizes brain tumor cells to apoptosis following alkylation damage of DNA. Oncogene 2002, 21:878-889 [DOI] [PubMed] [Google Scholar]
- 50.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA: “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 2002, 298:597-600 [DOI] [PubMed] [Google Scholar]
- 51.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ: Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417:78-83 [DOI] [PubMed] [Google Scholar]
- 52.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, Pellegata NS, de la Chapelle A: Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci USA 2001, 98:15044-15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC: Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA 2000, 97:13103-13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coon JS, Marcus E, Gupta-Burt S, Seelig S, Jacobson K, Chen S, Renta V, Fronda G, Preisler HD: Amplification and overexpression of topoisomerase IIα predict response to anthracycline-based therapy in locally advanced breast cancer. Clin Cancer Res 2002, 8:1061-1067 [PubMed] [Google Scholar]
- 55.Di Leo A, Gancberg D, Larsimont D, Tanner M, Jarvinen T, Rouas G, Dolci S, Leroy JY, Paesmans M, Isola J, Piccart MJ: HER-2 amplification and topoisomerase IIα gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res 2002, 8:1107-1116 [PubMed] [Google Scholar]
- 56.Yokomuro S, Tsuji H, Lunz JG, 3rd, Sakamoto T, Ezure T, Murase N, Demetris AJ: Growth control of human biliary epithelial cells by interleukin 6, hepatocyte growth factor, transforming growth factor β1, and activin A: comparison of a cholangiocarcinoma cell line with primary cultures of non-neoplastic biliary epithelial cells. Hepatology 2000, 32:26-35 [DOI] [PubMed] [Google Scholar]
- 57.Haddad R, Lipson KE, Webb CP: Hepatocyte growth factor expression in human cancer and therapy with specific inhibitors. Anticancer Res 2001, 21:4243-4252 [PubMed] [Google Scholar]
- 58.Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R: Role of the hepatocyte growth factor receptor, c-met, in oncogenesis and potential for therapeutic inhibition. Cytokine Growth Factor Rev 2002, 13:41-59 [DOI] [PubMed] [Google Scholar]
- 59.Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M: Down-regulation of E-cadherin and desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene 2001, 20:8125-8135 [DOI] [PubMed] [Google Scholar]
- 60.Naughton M, Picus J, Zhu X, Catalona WJ, Vollmer RT, Humphrey PA: Scatter factor-hepatocyte growth factor elevation in the serum of patients with prostate cancer. J Urol 2001, 165:1325-1328 [PubMed] [Google Scholar]
- 61.Junbo H, Li Q, Zaide W, Yunde H: Increased level of serum hepatocyte growth factor/scatter factor in liver cancer is associated with tumor metastasis. In Vivo 1999, 13:177-180 [PubMed] [Google Scholar]
- 62.Takigawa N, Segawa Y, Maeda Y, Takata I, Fujimoto N: Serum hepatocyte growth factor/scatter factor levels in small cell lung cancer patients. Lung Cancer 1997, 17:211-218 [DOI] [PubMed] [Google Scholar]
- 63.Chakravarti A, Loeffler JS, Dyson NJ: Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res 2002, 62:200-207 [PubMed] [Google Scholar]
- 64.Scotlandi K, Maini C, Manara MC, Benini S, Serra M, Cerisano V, Strammiello R, Baldini N, Lollini PL, Nanni P, Nicoletti G, Picci P: Effectiveness of insulin-like growth factor I receptor antisense strategy against Ewing’s sarcoma cells. Cancer Gene Ther 2002, 9:296-307 [DOI] [PubMed] [Google Scholar]
- 65.Nakamura K, Hongo A, Kodama J, Miyagi Y, Yoshinouchi M, Kudo T: Down-regulation of the insulin-like growth factor I receptor by antisense RNA can reverse the transformed phenotype of human cervical cancer cell lines. Cancer Res 2000, 60:760-765 [PubMed] [Google Scholar]
- 66.Aoki M, Blazek E, Vogt PK: A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc Natl Acad Sci USA 2001, 98:136-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dennis EA: Diversity of group types, regulation, and function of phospholipase A2. J Biol Chem 1994, 269:13057-13060 [PubMed] [Google Scholar]
- 68.Skouteris GG, Schroder CH: Cytosolic phospholipase A2 is activated by the hepatocyte growth factor receptor-kinase in Madin Darby canine kidney cells. J Cell Sci 1997, 110:1655-1663 [DOI] [PubMed] [Google Scholar]
- 69.Hayashi N, Yamamoto H, Hiraoka N, Dono K, Ito Y, Okami J, Kondo M, Nagano H, Umeshita K, Sakon M, Matsuura N, Nakamori S, Monden M: Differential expression of cyclooxygenase-2 (COX-2) in human bile duct epithelial cells and bile duct neoplasm. Hepatology 2001, 34:638-650 [DOI] [PubMed] [Google Scholar]
- 70.Yoon JH, Higuchi H, Werneburg NW, Kaufmann SH, Gores GJ: Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology 2002, 122:985-993 [DOI] [PubMed] [Google Scholar]
- 71.Henson DE, Albores-Saavedra J, Corle D: Carcinoma of the gallbladder: histologic types, stage of disease, grade, and survival rates. Cancer 1992, 70:1493-1497 [DOI] [PubMed] [Google Scholar]
- 72.Simony-Lafontaine J, Esslimani M, Bribes E, Gourgou S, Lequeux N, Lavail R, Grenier J, Kramar A, Casellas P: Immunocytochemical assessment of σ-1 receptor and human sterol isomerase in breast cancer and their relationship with a series of prognostic factors. Br J Cancer 2000, 82:1958-1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labit-Le Bouteiller C, Jamme MF, David M, Silve S, Lanau C, Dhers C, Picard C, Rahier A, Taton M, Loison G, Caput D, Ferrara P, Lupker J: Antiproliferative effects of SR31747A in animal cell lines are mediated by inhibition of cholesterol biosynthesis at the sterol isomerase step. Eur J Biochem 1998, 256:342-349 [DOI] [PubMed] [Google Scholar]
- 74.Cho SY, Kim JH, Paik YK: Cholesterol biosynthesis from lanosterol: differential inhibition of sterol δ 8-isomerase and other lanosterol-converting enzymes by tamoxifen. Mol Cells 1998, 8:233-239 [PubMed] [Google Scholar]
- 75.Giguere V: To ERR in the estrogen pathway. Trends Endocrinol Metab 2002, 13:220-225 [DOI] [PubMed] [Google Scholar]
- 76.Heard DJ, Norby PL, Holloway J, Vissing H: Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: tissue-specific isoforms are expressed during development and in the adult. Mol Endocrinol 2000, 14:382-392 [DOI] [PubMed] [Google Scholar]
- 77.Tremblay GB, Bergeron D, Giguere V: 4-Hydroxytamoxifen is an isoform-specific inhibitor of orphan estrogen-receptor-related (ERR) nuclear receptors β and γ. Endocrinology 2001, 142:4572-4575 [DOI] [PubMed] [Google Scholar]
- 78.Taub JW, Matherly LH, Stout ML, Buck SA, Gurney JG, Ravindranath Y: Enhanced metabolism of 1-β-D-arabinofuranosylcytosine in Down syndrome cells: a contributing factor to the superior event-free survival of Down syndrome children with acute myeloid leukemia. Blood 1996, 87:3395-3403 [PubMed] [Google Scholar]
- 79.Plunkett W, Huang P, Searcy CE, Gandhi V: Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol 1996, 23:3-15 [PubMed] [Google Scholar]