Summary
Transgenic Xenopus laevis tadpoles that express a dominant negative form of the thyroid hormone receptor (TRDN) controlled by the cardiac actin muscle promoter (pCar) develop with very little limb muscle. Under the control of the tetracycline system the transgene can be induced at will by adding doxycycline to the rearing water. Pre existing limb muscle fibers begins to disintegrate within 2 days after up-regulation of the TRDN transgene. The muscle cells do not die even after weeks of transgene exposure when the myofibrils have degenerated completely and the tadpole is nearing death. A micro array analysis after 2 weeks of exposure to the transgene identified 25 muscle genes whose expression was altered in such a way that they might cause the muscle phenotype. These candidate genes are normally activated in growing limb muscle but they are repressed by the TRDN transgene. Several of these genes have been implicated in mammalian myopathies. However, the expression of only one of these genes, calsequestrin, is down regulated in 48 hrs and therefore might initiate the degeneration. Calsequestrin is one of several affected genes that encode proteins involved in calcium sequestration, transport and utilization in muscle suggesting that uncontrolled calcium influx into the growing limb muscle fibers causes rhabdomyolysis. Many of the same genes that are down regulated in the tail at the peak of metamorphic climax just before it is resorbed are suppressed in the transgenic limb muscle in effect turning the limb growth program into a tail resorption program.
Introduction
The multiple axes of a Xenopus laevis tadpole limb bud are established by Nieuwkoop-Faber (NF) stage 52 (Nieuwkoop PD, 1956) when the limb is an oblong structure with undifferentiated cells (Cameron and Fallon, 1977). The subsequent growth and differentiation of the various limb cell types is controlled by thyroid hormone (TH). The earliest effect of TH on limb development is stimulation of DNA replication in all cell types (Brown et al., 2005; Schreiber et al., 2001). Limb growth is followed by the formation of the characteristic cell types and their terminally differentiated proteins. Nerves grow into the limb as it elongates even before the appearance of muscle fibers. The limb is fully formed before it is functionally innervated. Tadpoles convert from tail to leg swimming at the climax of metamorphosis (NF60) before the tail is resorbed (Marsh-Armstrong et al., 2004).
A series of transgenic experiments, in which different cell type specific promoters control the expression of a dominant negative thyroid hormone receptor alpha (TRDN) revealed that the multiple programs of limb development are controlled independently by TH (Brown et al., 2005). A ubiquitously expressed promoter driving the TRDN reporter represses TH-induced DNA synthesis in the early limb bud (Schreiber et al., 2001) while cell-type specific promoters inhibit cell autonomously the differentiation of muscle, innervation of the limb muscle, and the growth of the limb skeleton (Brown et al., 2005). The death of muscle in the tail at metamorphic climax is also a cell autonomous TH-induced program (Das et al., 2002; Nakajima and Yaoita, 2003; Yaoita and Nakajima, 1997). Over expression of the same TRDN transgene that causes muscle degeneration in limbs protects tail muscle from TH-induced resorption (Das et al., 2002). A transgenic tadpole with the X. laevis cardiac actin promoter (pCar) driving the TRDN transgene dies at the climax of metamorphosis. The animal’s limbs never function because they have almost no muscle (Das et al., 2002).
We began a series of micro array experiments designed to compare the TH-induced gene expression profiles in the growing limb and brain with the tail (Das et al., 2006). Not only do the growth and resorption programs differ greatly but also in some cases the same genes had opposite responses to the hormone in tail compared to limb.
In this paper we have investigated the limb muscle wasting induced by the TRDN transgene. We prepared transgenic animals in which the pCar promoter drives the expression of the TRDN reporter under the control of the tetracycline inducible system. Induction of the transgene causes the degeneration of limb muscle specifically without affecting the other cell types that differentiate in the limb (Brown et al., 2005). A micro array analysis of the gene expression profile reveals a small set of muscle genes whose expression pattern is inhibited from the up-regulation that occurs normally at metamorphosis in the muscle growth program. These same genes are down regulated in the muscle death program that occurs normally during tail resorption. Several of these candidate genes have been implicated in human myopathies.
Material and method
The plasmids used for transgenesis
Control of the expression of a transgene by the tetracycline system (Urlinger et al., 2000) has been applied to X. laevis development (Das and Brown, 2004). One plasmid called pCS2+(tetO)TRDN/GFP has the tet operator adjacent to a dominant negative form of X. laevis thyroid receptor alpha. This mutant receptor lacks 12 amino acids from its C-terminus, and it is fused to GFP at its carboxy terminus by a small flexible linker. The TRDN part of the resulting protein has a nuclear localization signal so that the fluorescent protein can be visualized in the nucleus. The second plasmid has the X. laevis cardiac actin promoter (pCar) driving a modified tetracycline repressor (pCar/rTA2S-M2). This system is under positive control so that addition of the tetracycline derivative, doxycycline (Dox), induces expression of the TRDN/GFP transgene. The transgenesis procedure (Kroll and Amaya, 1996) used a mixture of the 2 plasmids (Das and Brown, 2004). A doubly transformed male parent was raised to sexual maturity and bred with a wild type female. The transgene can be induced in half of the progeny. Mixtures of DNAs are often integrated together presumably in tandem arrays (Marsh-Armstrong et al., 2004).
Raising tadpoles for microarray limb samples
The F1 progeny were raised to NF 55 without the inducer. Batches of 20 tadpoles were grown in 4 liters of 0.1MMR containing 50 ug/ml doxycycline hyclate (Dox; Sigma) (Das and Brown, 2004). The tadpoles were fed daily and the rearing water was changed twice each week. After 2 week and 6 week of Dox treatment the tadpoles had advanced to NF 56 and 61, respectively. The latter developmental stage (NF61) is metamorphic climax when the endogenous TH is highest. By NF62 the control tadpoles are swimming with their legs, and all of the transgenic animals at the same stage have the characteristic paralyzed phenotype. Control and transgenic tadpoles were sorted by their GFP fluorescence and their hind limbs were removed for RNA extraction. If allowed to continue development all of the paralyzed transgenic tadpoles will soon die. The fourth RNA sample was isolated from the hind limbs of control frogs (NF 66) that were raised in 50 ug/ml of Dox for 2 wks after completing metamorphosis. This concentration of the inducer has no effect on the development or growth of normal tadpoles or frogs. The tadpoles and frogs were fed throughout the experiment.
Three separate samples of hind limbs were collected from each of the 4 groups of animals. For the control and transgenic 2wk tadpoles, each sample consisted of hind limbs from 12 tadpoles. We used limbs from 6 tadpoles per replicate for the 6wk Dox treated climax transgenic tadpoles and limbs from 4 control frogs for each sample.
RNA purification, probe synthesis, and microarray hybridization
After amputation the limbs were homogenized immediately in Trizol (Invitrogen) to purify total RNA according to the manufacturers protocol. Probes for the microarray were synthesized with the Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent Kit # 5184-3523) that incorporates the fluorescent nucleotide Cy3 CTP (Perkin Elmer). The universal standard labeled with Cy5 CTP was derived from all stages of whole tadpoles and has been described (Das et al., 2006). These cRNA probes were hybridized with a second version of Agilent X. laevis micro array slides (AMADID # 013214). Each slide contains 21,654 sense oriented 60-mer oligonucleotides from the complete X. laevis Unigene list that was issued in February 2004 (Build 48). Duplicate entries and other details of these arrays have been described (Das et al., 2006). Since each of the four experimental points were performed in triplicate the statistical significance of expression values could be evaluated by the False Discovery Rate (FDR) method (Benjamini, 1995; Sharov et al., 2005). We only scored gene expression changes that differed from the control with an FDR value of less than 0.05. The microarray data is presented as the ratio of the log intensity of triplicate hybridization values (2 wk and 6 wk transgenic limbs and control frog limbs) relative to the control tadpole limb. The tail expression data is derived from our previous array analysis (Das et al., 2006). The NF62 climax tail values are compared to control tail at NF54. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE5249. It can also be accessed at our web site (http://www.ciwemb.edu/brownlab/index.html).
Histology, immunostaining and in situ hybridization
The methods for hematoxylin and eosin staining, immunocytology and in situ hybridization have been described (Cai and Brown, 2004). The antibodies used in this study were directed against skeletal muscle myosin (fast) clone My32 (Sigma, product No. M4276), goat anti-mouse Alexa –568 secondary antibody (Molecular Probes, catalogue No. A-11004), and anti-GFP polyclonal antibody (Torrey Pines Biolab). Probes for in situ hybridization were digoxygenin-labeled antisense RNA. We purchased cDNAs from ATCC to make the probes for the in situ hybridizations.
For electron microscopy frontal and cross sections of limbs were fixed with glutaraldehyde and paraformaldehyde followed by osmium tetroxide and uranyl acetate, dehydrated in ethanol and embedded in epoxy. Sections were cut at about 90nm thickness, stained with lead citrate and imaged on an FEI Techai 12 using a Gatan camera & software.
Results
Induction of the pCar/TRDN phenotype
The pCar/TRDN transgenic tadpoles form limbs that have almost no muscle (Das et al., 2002). In order to study this phenotype in greater detail we prepared transgenic X laevis with the pCar/TRDN-GFP transgene controlled by the positive tetracycline inducible system (Urlinger et al., 2000). In the presence of the inducer, doxycycline (Dox), the fused transgenic protein (TRDN-GFP) can be detected in limb muscle nuclei when specialized muscle proteins are first synthesized (NF54) (Fig. 1). The co-localization of GFP and myosin confirm that the pCar promoter drives expression specifically in differentiated muscle fibers. When the inducer was added at NF54 the muscle phenotype developed as the tadpoles advance to the climax of metamorphosis. The tadpoles die at NF62 after gill resorption but before tail resorption (Brown et al., 2005). When the TRDN transgene is induced with Dox as late as NF56 the pre-existing differentiated limb muscle fibers degenerate.
Fig. 1.
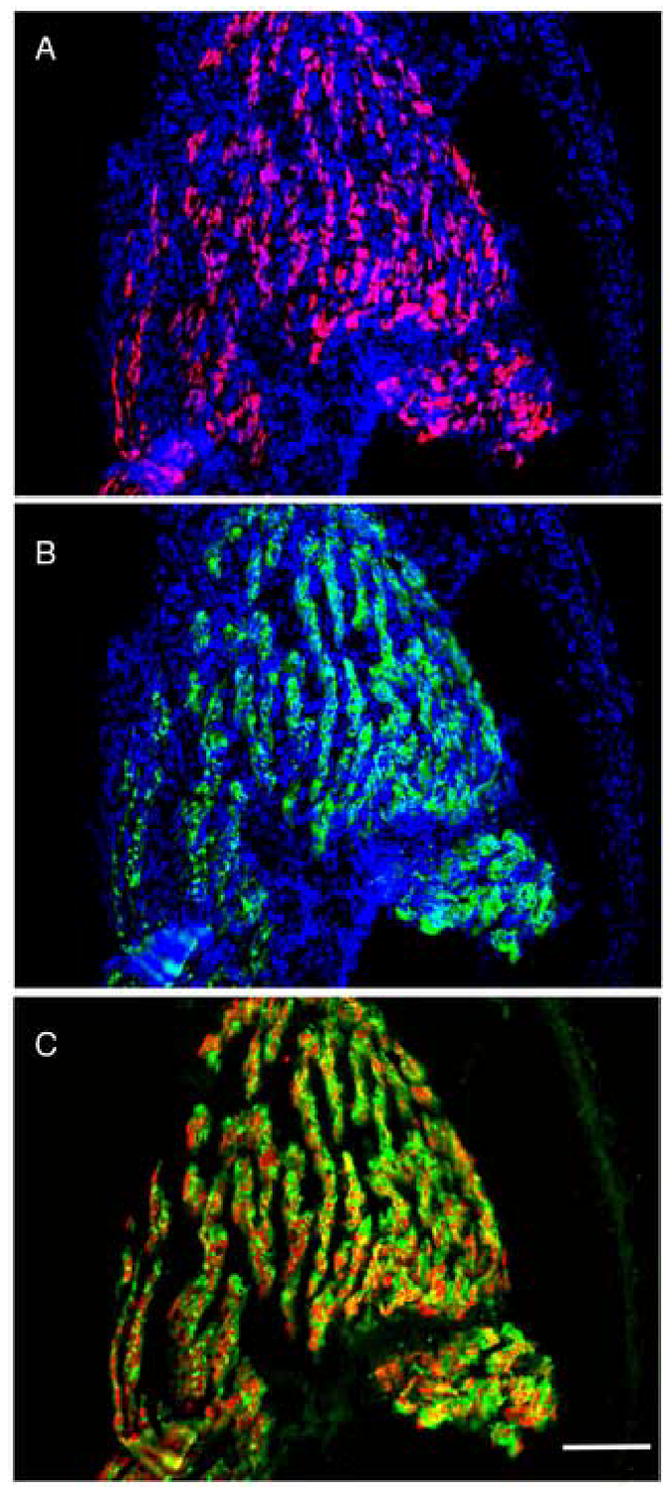
The transgene is expressed exclusively in muscle. A transgenic limb (NF55) frontal section simultaneously reacted with A) an antibody to GFP (red) and DAPI (blue); B) an antibody to myosin (My32, green) and DAPI (blue); C) merge of A and B without the DAPI. The scale bar=200μm.
The pCar/TRDN-paralyzed limb muscle was analyzed by light and electron microscopy. Muscle fibers have disintegrated, and the muscle nuclei clump together within the bundles that previously contained the myofibrils (Fig. 2). These nuclei are convoluted with prominent nucleoli. Residual patches of parallel filaments (Fig. 2B) are often visible. Mitochondria are clumped as are the sarcoplasmic reticulum. In contrast to tail muscle cells (Das et al., 2002) that are induced by TH to resorb these degraded limb muscle cells were negative for both active caspase-3 and the TUNEL reaction for apoptosis. Even at the latest stages of the phenotype the limb muscle cells are not dead. Satellite muscle cells can be identified with an antibody to Pax 7. These cells continue to incorporate Brdu (Fig. S1). As will be shown many muscle genes including the muscle cell specific transcription factors are expressed even at the late stages of the phenotype. In support of the idea that the muscle nuclei do not die we removed the inducer Dox and reversed the phenotype of one transgenic NF58 tadpole that had developed a visible limb phenotype. The animal recovered and switched to leg swimming at metamorphic climax (data not shown). Since development of these transgenic tadpoles continues inexorably to climax the time for recovery at NF58 is too narrow for efficient recovery of most tadpoles with the limb phenotype.
Fig. 2.
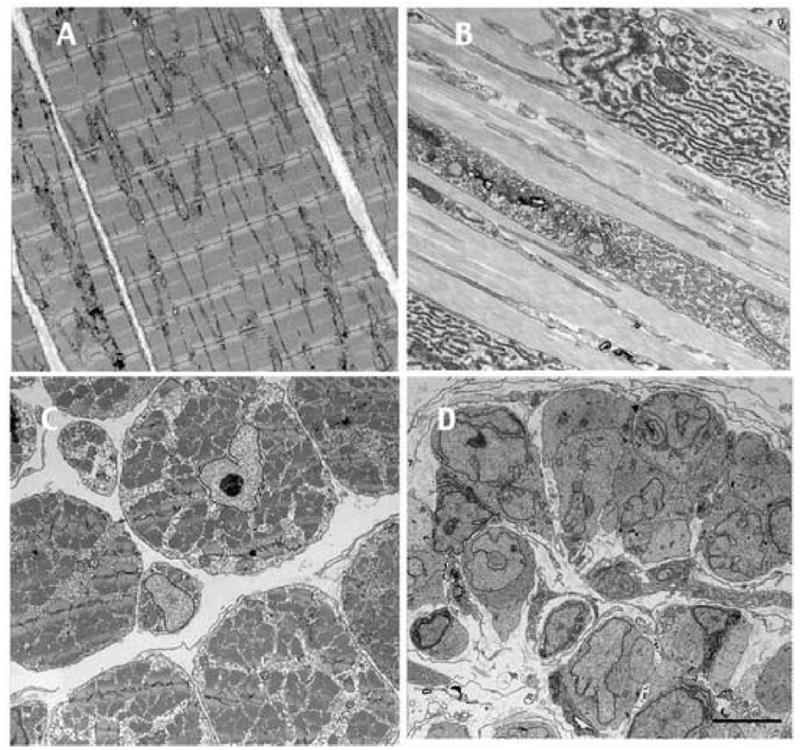
Electron micrographs of the limb muscle phenotype at NF61. A,C control, and B,D, pCar/TRDN limb. A and B are longitudinal sections and C and D are cross sections. The sibling control and pCar/TRDN tadpoles were raised for 6 wk in Dox. The scale bar = 5μm.
We have induced metamorphosed transgenic small frogs with Dox for as long as 4 months. These animals grow more slowly than controls and develop abnormal but functional limbs. They never become paralyzed but left in the inducer the frogs stop eating and die. Histology of their limbs revealed normal but smaller muscle bundles than control frogs (data not shown). The inducer was removed from the rearing water of two of these frogs, and they were raised to sexual maturity. They retained a deformed leg structure but otherwise recovered. Therefore the up-regulation of the transgene after metamorphosis has a demonstrable but much less drastic affect on the frog limb muscle than it does on tadpoles.
Gene expression profile of the developing transgenic limb
In order to probe the molecular basis of this wasting phenotype, we carried out a microarray analysis on the hind limbs of normal and Dox induced transgenic tadpoles and control frogs (Fig. 3). The gene expression profiles of the control and 2wk samples from sibling tadpoles that had been raised in the same container with Dox are remarkably similar. Out of the 21,654 entries on the micro array only 42 and 67 genes are significantly up- and down-regulated, respectively, in the transgenic 2 wk limb compared to the control limbs (Table 1). Many more genes are differentially expressed 4 weeks later (6 wk time point), when the transgenic tadpoles had progressed to the climax of metamorphosis, and in the control frog limb. The final column of Table 1 includes the number of genes that are up or down regulated in the tail at climax (NF62) compared to a growing tail (NF54). This data was obtained in our previous micro array study (Das et al., 2006). Note that there are more than twice as many up-regulated genes as down-regulated genes in the tail at climax (NF62) even though the tail has been activated to die and resorb. Most of the up regulated genes in the tail are expressed in fibroblasts while the majority of down regulated genes are expressed in muscle.
Fig. 3.
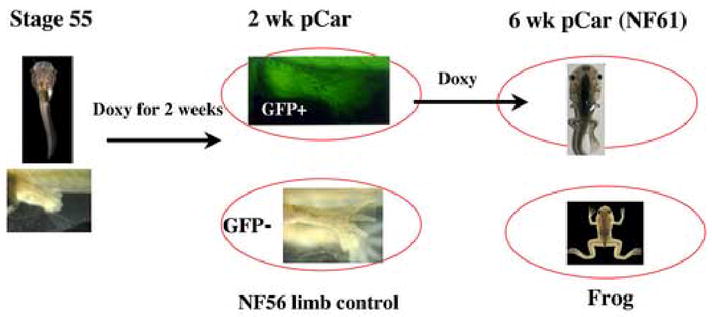
Design of the limb micro array experiment. Hind limbs were removed from the circled stages for RNA extraction. In the text the expression results always compare the transgenic limbs and the frog limbs with the GFP negative NF56 controls.
Table 1.
Summary of differentially expressed genes in the limb and tail
| 2 wk | 6 wk | Frog | Tail NF62* | |
|---|---|---|---|---|
| Up Total # | 42 | 1401 | 4242 | 1716 |
| Down Total # | 67 | 1915 | 3054 | 742 |
Tail data compares expression at climax (NF62) with tail at NF54 from Das et al., 2006. There are 21,654 oligonucleotides on the array.
The developmental changes in gene expression fall into classes (Fig. 4) that reflect the differentiation of the several cell types in the limb, the influence of TH, and the effect of the transgene. Gene expression values of transgenic limbs at the 2 wk and 6 wk time points as well as the control frog limbs are always compared with those of the GFP negative control tadpole limbs at NF56 (Fig. 3). Although we will concentrate on muscle-specific genes, the array experiments document the dramatic TH-induced transformation of the epidermis and the formation of cartilage, bone, and bone marrow that are not affected by expression of the transgene in muscle. At the 6 wk time point, which is the climax of metamorphosis (NF61), these non-muscle cell type specific genes invariably change their expression in the same direction as they do in the frog limb. We call this pattern of expression for TH-controlled genes that are unaffected by the transgene ns-up-up (Fig. 4G) or ns-down-down (Fig. 4I) referring to the three successive developmental stages compared to the control tadpole i.e. unchanged (ns) in the 2 wk transgenic, elevated (up) in both the 6 wk transgenic and the frog limb. This class of genes (ns-up-up) includes the muscle specific transcription factors that are expressed in myoblasts before the pCar promoter is activated (Table S1). There are 8 tadpole-specific muscle genes that are down regulated at metamorphosis and follow the kinetics of “ns-down-down” (Fig. 4I, Table S1). Tadpole-specific skin genes also exhibit this pattern (for example, gene 19 (U41861) (Furlow et al., 1997). The majority of genes (13,329) on the array do not change their expression during limb development at these three time points (ns-ns-ns) (Fig. 4F). There are 48 muscle genes on the array with an unchanged expression profile (Table 1). The in situ results of two of these genes titin and desmin are shown in Fig. S2.
Fig. 4.
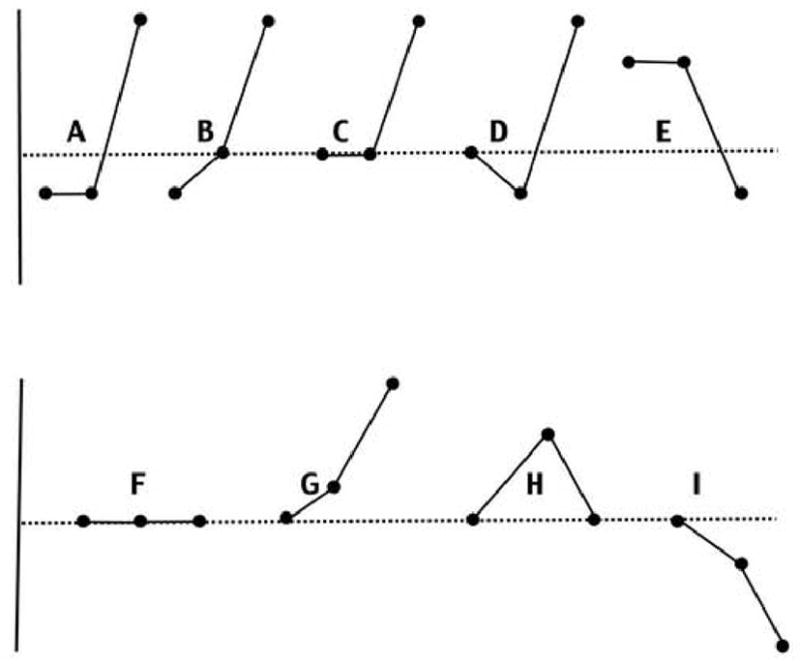
Gene expression profiles and the number of genes on the array for each profile in parenthesis. A) down-down-up (14); B) down-ns-up (18); C) ns-ns-up (3284); D) ns-down-up (28); E) up-up-down (3); F) ns-ns-ns (13329); G) ns-up-up (876); H) ns-up-ns (448); I) ns-down-down (1450). The horizontal line marks the gene expression value of the control tadpole limb that the three other limb hybridization values are compared with. Each profile has the 2 wk, 6wk and frog values from left to right. A,B, and E are primary candidate genes. C and D are secondary candidate genes; F is unchanged during development; G and I change during development but are not affected by the transgene; H follows the endogenous TH concentration. An FDR value of <0.05 was used to score a gene as up or down regulated.
Candidate genes
Several considerations narrow the search for candidate genes that might be involved in the muscle phenotype. Gene(s) responsible for the phenotype should differ in their expression levels between the control and the transgenic limbs after 2 wk of Dox induction, because the limb muscle phenotype is detectable by that time. We have established a more precise time for the onset of the muscle destruction phenotype (see below). A candidate muscle gene that normally is down regulated at metamorphosis should be up regulated in the transgenic limb. We refer to this pattern as an “up-up-down” profile (Fig. 4E). Only 3 of the 37 genes that are up regulated in the transgenic limbs after 2 wks of Dox induction have the profile of continued up regulation at 6 wks and finally down regulation in the frog limb. Their expression changes are less than twofold. These genes (uroplakin 1A, transportin 1, and an unidentified EST) are not known to be involved in muscle development or maintenance.
Genes having the opposite expression profile (down-down-up and down-ns-up) (Fig. 4A and B) could initiate the phenotype. These genes are down regulated in the transgenic limb after 2 wks of Dox, and either down regulated or unchanged in the 6 wk transgenic but always up regulated in the control frog limb. Only 62 of the 21,654 entries on the micro array are reduced in expression after 2 weeks of Dox treatment compared to the sibling control values. 32 of these 62 genes have the relevant gene expression profile (Table 2). The majority (23) of these genes are known to be expressed in muscle. The muscle localization and the change in expression caused by the transgene of two candidate genes (LIM and actin) is shown in Fig. 5. These two genes and most of the other candidate genes are up regulated in the control limb by thyroid hormone and down regulated in the control tail at the climax of metamorphosis. Candidate genes whose expression profiles have been confirmed by in situ hybridization are noted in Table 2. The remainder of the genes in Table 2 has not been studied further so they too might be expressed in muscle. 25 of these 32 genes are down regulated in the tadpole tail at climax (Table 2). The other 7 are unchanged. Many muscle genes continue being expressed in the limb even at the latest stage just before death of the transgenic tadpoles (Table S1; Fig. S2).
Table 2.
Primary candidate genes
| Accession number | Gene name | 2 wk | 6 wk | Frog | NF62 | In situ |
|---|---|---|---|---|---|---|
| BC043842 | phosphoglycerate mutase 2 | 0.13 | 0.28 | 56.91 | 0.03 | M+ |
| BC045082 | enolase 3 | 0.27 | 0.31 | 125.57 | 0.07 | M+ |
| BC043830 | phosphorylase kinase | 0.35 | 0.4 | 16.45 | 0.63 | M+ |
| AB003080 | troponin C, slow | 0.17 | 0.09 | 3.28 | NS | M+ |
| BX850052 | fast troponin I | 0.35 | NS | 8.48 | NS | M+ |
| M87307 | tropomyosin | 0.43 | NS | 14.20 | 0.36 | M+ |
| CD255184 | myosin heavy chain, E3 | 0.17 | 0.64 | 5.11 | 0.38 | M+ |
| BC046739 | actin, alpha 1 | 0.25 | 0.24 | 22.44 | 0.11 | M+ |
| BC042340 | LIM domain binding 3 | 0.48 | NS | 12.48 | 0.33 | M |
| BC041221 | LIM domain protein | 0.59 | NS | 14.71 | 0.10 | M+ |
| BC054224 | myozenin 1 | 0.34 | NS | 74.83 | 0.23 | M+ |
| BC046947 | calsequestrin 1 | 0.17 | 0.39 | 69.63 | 0.27 | M+ |
| BC044063 | ATPase, Ca++ | 0.29 | 0.57 | 7.07 | NS | M+ |
| BC042249 | creatine kinase M | 0.29 | NS | 8.61 | NS | M+ |
| CB945142 | creatine kinase B | 0.52 | NS | 24.70 | 0.33 | M |
| CF547537 | choline dehydrogenase | 0.35 | 0.35 | 9.18 | NS | |
| BX847508 | Cyt. c oxidase assembly | 0.39 | NS | 10.53 | 0.23 | M |
| BX851926 | ryanodine receptor 3 | 0.43 | NS | 4.85 | 0.37 | M |
| BC045081 | AchR alpha | 0.52 | 0.68 | 1.54 | 0.48 | M |
| U19612 | AchR epsilon | 0.53 | NS | 4.63 | 0.31 | M |
| BC043628 | AchR delta | 0.58 | NS | 2.36 | 0.21 | M |
| BJ029671 | sirtuin 1 | 0.57 | 0.62 | 25.62 | 0.23 | M |
| BC048222 | thrombospondin 3 | 0.57 | NS | 1.58 | 0.52 | |
| AY043260 | protein phosphatase 2a | 0.6 | NS | 1.62 | 0.26 | M |
| BC046852 | bridging integrator 1 | 0.49 | NS | 1.91 | 0.44 | M |
| BX851794 | Mg(2+) transporter | 0.51 | NS | 1.94 | 0.63 | |
| CB984281 | Mitoch. ADP/ATP translocase | 0.68 | NS | 5.90 | 0.24 | M |
| CD099943 | Unknown | 0.68 | 0.69 | 8.00 | NS | |
| BJ041930 | Unknown | 0.51 | NS | 1.85 | 0.41 | |
| BX854361 | ADP-ribosyltransferase 1 | 0.58 | NS | 11.60 | 0.51 | |
| AW764727 | FGF receptor substrate 3 | 0.47 | 0.63 | 9.97 | 0.23 | |
| BE026509 | Unknown | 0.38 | 0.31 | 2.48 | NS | |
| Selected secondary candidate genes: | ||||||
| BC046673 | aldolase A, fructose-bisphosphate | NS | NS | 16.21 | 0.19 | M |
| BC045015 | lactate dehydrogenase A | NS | NS | 10.66 | 0.20 | M |
| U39669 | pyruvate dehydrogenase | NS | NS | 5.35 | 0.38 | M |
| BC045100 | phosphofructokinase, muscle | NS | 0.61 | 23.60 | 0.16 | M |
| BC043781 | phosphoglycerate kinase 1 | NS | NS | 9.21 | 0.11 | M |
| BC046864 | triosephosphate isomerase 1 | NS | NS | 2.42 | 0.38 | M |
| BC054279 | EF1 alpha 2 | NS | NS | 338.77 | 4.95 | M |
M, known muscle expression; +, in situ confirmation of down regulation at the 6 wk time compared to a control at the same stage.
NS, not significant compared to 2 wk control
Fig. 5.
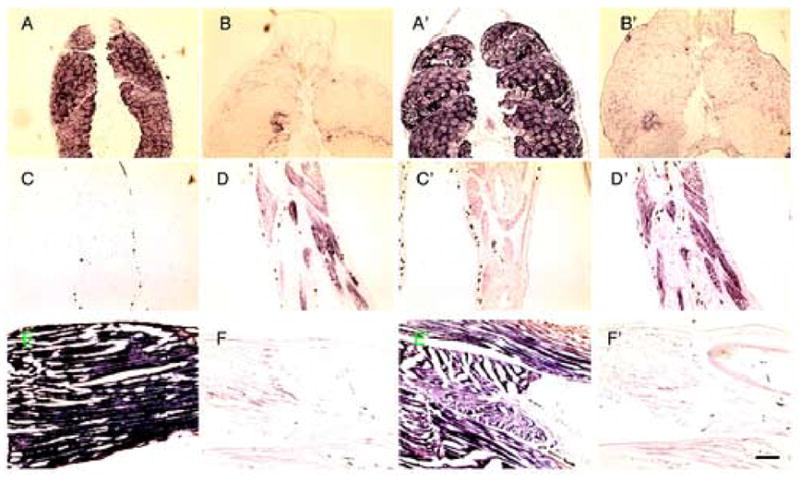
In situ hybridization with A–F, alpha actin (BC046739) and A’–F’, LIM (BC041221). A, and A’, cross sections of control NF55 tail; B and B’, cross sections of NF 62 tail; C and C’, are frontal sections of control NF56 limb. D and D’, are frontal sections of NF56 limb treated for 3days with 10 nM T3; E,E’, frontal section of control NF61 limb; F,F’, pCar transgenic limb that was induced for 6 wks. with Dox. Scale bars=200 μm
The kinetics of muscle degradation and muscle gene down regulation
The candidate genes that are down regulated in the micro array after 2 wks exposure to the inducer could include genes that respond directly or indirectly to the TRDN transgene. We analyzed the kinetics of muscle fiber degradation at varying times after induction of the transgene by whole mount immunocytology with an antibody to myosin (Fig. 6). Remarkably, patches of muscle fiber disruption were seen as early as 2 days after addition of the inducer Dox. The destruction increased at 4 days and was extensive after one week of exposure to the transgene (Fig. 6). We screened 7 of the most dramatically regulated candidate genes by in situ hybridization for the earliest time that down regulation could be detected presuming that down regulation of the gene(s) causing the phenotype must precede visible degradation of muscle fibers (Table 3). Only calsequestrin was down regulated at day 1 (Fig. 7). The other 5 genes were unchanged in expression at day 1 but down regulated partly or extensively by 4 days (Table 3; Fig. S3). The slightly later down regulation of most of these candidate genes compared to calsequestrin does not rule out their role in the degradation phenotype since the destruction begins in small patches (Fig. 6) and does not involve all of the muscle until 1 week of Dox treatment. In addition there are another 17 muscle genes that have not been tested at these early time points.
Fig. 6.
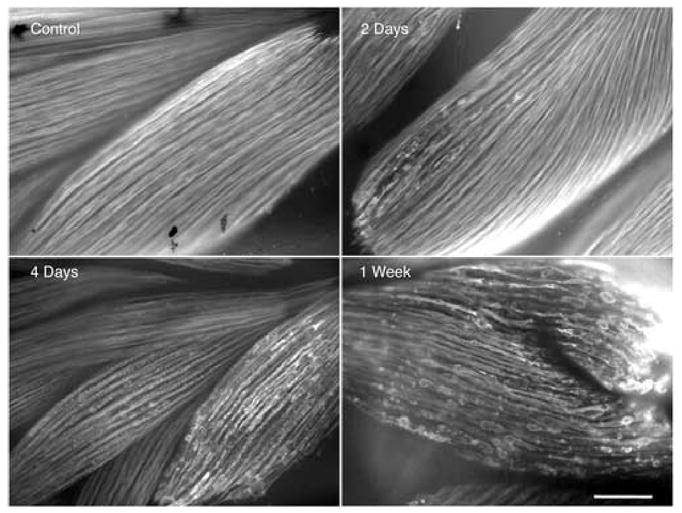
Kinetics of limb muscle fiber degradation after inducing the transgene by the addition of the inducer, Dox. Whole mount My32 (green) immunostain. Transgenic tadpoles at NF 55 were induced with Dox for 2 days, 4 days, and 1 week. Scale bar=200μm
Table 3.
Expression kinetics of candidate genes
| Gene name | 1d* | 2d | 4d | 1w |
|---|---|---|---|---|
| calsequestrin 1 | D | D | D | D |
| actin, alpha 1 | NS | NS | D | D |
| phosphoglycerate mutase 2 | NS | NS | D | D |
| enolase 3 | NS | NS | pD | D |
| phosphorylase kinase | NS | NS | pD | D |
| ATPase, Ca++ | NS | NS | NS | D |
Time after up addition of Dox. Gene expression was assessed by in situ hybridization and in each case compared to a control limb.
D, down-regulated; pD, partially down-regulated; NS, not changed
Fig. 7.
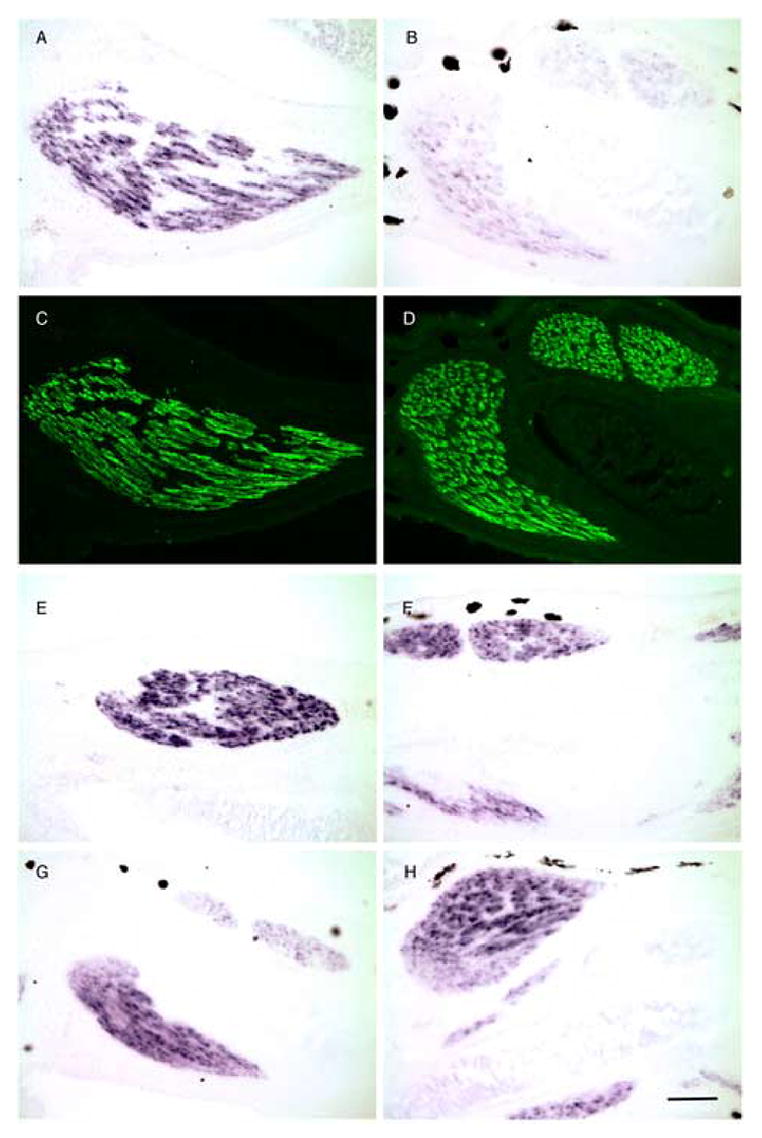
Early disappearance of calsequestrin mRNA in hind limb muscle after the up regulation of the TRDN transgene with Dox. A,B, E–H are in situ hybridization; C and D are the same sections as A and B immunostained with My32. A,C,E, and G are uninduced NF 55 transgenic tadpole limb sections; B,D,F, and H are limb sections of NF 55 transgenic tadpoles induced for 2 days with Dox. A,B) calsequestrin; E,F) actin; G,H) SERCA Ca(2+)-ATPase. Scale bar=200 μm
Muscle, glycolysis and mitochondrial energy related genes are influenced by the transgene
We have organized the genes on the array that are directly or indirectly influenced by the transgene and listed them in supplementary tables 1–4. The kinetic patterns of ns-down-up (18 genes), and ns-ns-up (2184 genes) are interpreted as indirect response genes (Table S1). 536 genes with the profile ns-ns-up are expressed at least twice the level in the frog as in the control tadpole limb. This list includes genes encoding muscle genes (Table S1), glycolytic enymes (Table S3), and many of the same genes involved in the generation of energy in muscle mitochondria (Table S4) that are down-regulated in the tail at metamorphic climax and induced by TH in growing tadpole limbs (Das et al., 2006).
Many of the candidate muscle genes have been implicated in human myopathies or shown to cause muscle degeneration in mouse knock outs. These will be discussed later. Along with the earliest responding gene, calsequestrin, a significant number of the primary candidate genes are involved in calcium delivery to or utilization by muscle.
In a previous paper (Das et al., 2006) we studied the gene expression changes in tadpole tail, limb and brain induced with TH. The sampling of X. laevis genes is large enough on these arrays to identify functional categories in which a large non-random number of genes are altered in expression by the hormone. Two such functional categories are the glycolysis pathway and the complexes that transfer electrons and synthesize ATP in mitochondria. These metabolic pathways are especially important for muscle function, and the expression of the genes encoding these functions is elevated in growing tail muscle and up regulated by TH in growing limb muscle (Das et al., 2006). There are 17 genes on the array that encode glycolytic enzymes (Suppl. Table 3). Fourteen of these genes are up regulated in the control frog limb, and either down regulated or unchanged in the transgenic limb at climax. The three remaining glycolytic genes on the array are not regulated in expression in either the frog limb or the tail at climax.
Whereas the growing limb expresses both cell cycle genes, mitochondrial energy-related genes, and genes encoding the glycolytic pathway, the wild type frog limb after metamorphosis down-regulates the cell cycle genes but continues expressing at a high level the energy-related genes required for normal functioning skeletal muscle. Many of the same set of energy-related genes that are up regulated in the limb by TH and down regulated in the tail at climax are repressed in the TRDN transgenic limb at the climax of metamorphosis (6 wk time point). A complete list of genes on the array that produce mitochondrial-localized products is presented in Table S4. To emphasize the similar gene expression response between the limbs expressing the TRDN transgene and the tail at metamorphic climax we have summarized the affected genes that are expressed in muscle, in mitochondria and in the glycolytic pathway (Table 4). Listed are the number of primary candidate genes for the limb phenotype (down-down-up, down-ns-up), secondary candidate genes (ns-ns-up) and genes that do not change (ns-ns-ns). Most of the limb muscle candidate genes are down regulated in the tail at climax while the genes that do not change in the transgenic limb also are not changed in the climax tail.
Table 4.
Many candidate genes in the limb are regulated in the tail at climax
| Tail NF62 | |||
|---|---|---|---|
| mitochondria (Suppl. Table 5) |
|
||
| ns | up | down | |
| ns-ns-ns (183) | 156 | 9 | 18 |
| ns-ns-up (141) | 72 | 1 | 68 |
| down-down-up (2) | 1 | 1 | |
| down-ns-up (2) | 2 | ||
| muscle (Suppl. Table 1) | |||
| ns-ns-ns (49) | 41 | 4 | 4 |
| ns-ns-up (47) | 24 | 2 | 21 |
| down-down-up (11) | 2 | 9 | |
| down-ns-up (13) | 2 | 11 | |
| glycolysis (Suppl. Table 4) | |||
| ns-ns-ns (3) | 3 | ||
| ns-ns-up (11) | 11 | ||
| down-down-up (2) | 2 | ||
Kinetic categories of gene expression are explained graphically in Fig. 3. Numbers in parentheses are the number of genes in the limb array in each kinetic category. The supplementary tables list all of the genes.
Other programs in the limb are not affected by the TRDN transgene
This micro array analysis confirms the cell autonomous nature of the muscle phenotype (Brown et al., 2005; Das et al., 2002). The most extreme expression profiles on the arrays demonstrate this independence. The 40 most up regulated genes in the frog limb are also up-regulated at the 6 wk time point as expected for TH-induced genes that are not repressed by the transgene. These genes include terminally differentiated genes expressed in adult skin, bone and other non-muscle cell types. The complete list of expressed genes in these arrays is accessible through GEO Series accession number GSE5249. or at our own web site (http://www.ciwemb.edu/brownlab/index.html).
Discussion
The development of the muscle wasting phenotype
Induction of the TRDN-GFP transgene with Dox began at NF55 when the pCar promoter first drives a detectable reporter in developing limb muscle. The transgenic progeny of the mutant male parent are remarkably uniform in their response. The 2 wk time point was chosen for the micro array because the tadpoles showed the first morphological external sign of the limb phenotype, yet they were growing and developing like controls. A careful analysis of muscle degradation (Fig. 6) showed beginning fiber damage as early as 2 days after Dox induction and pervasive muscle fiber destruction after 1 wk. The 50% that are transgenic developed an identical paralyzed limb phenotype and died shortly after their gills resorbed (NF62) but before tail resorption. Triplicate samples were collected at each time point for independent RNA purification and micro array hybridization so that the results could be subjected to statistical analysis. A meaningful change in gene expression required a false discovery rate (FDR) value <0.05. A control sample for the 6 wk time point was not taken. By this time the muscle fibers are almost completely degenerated in the transgenic limb (Fig. 2) yet development of the other limb cell types was unimpaired. We have shown previously (Brown et al., 2005; Das et al., 2002) that expression of the TRDN transgene in limb muscle is cell autonomous. The micro array confirms that many adult specific genes begin to change their expression at metamorphic climax (the 6 wk time point). This not only includes genes that change in non-muscle cell types such as skin and bone but a substantial number of muscle genes (Table S1). In each case the expression level at the 6 wk time point is elevated or suppressed in the same direction as the control frog limb (ns-up-up or ns-down-down). The complete gene expression data from these micro array experiments can be obtained from the NCBI database or our web site (http://www.ciwemb.edu/brownlab/index.html). Many muscle genes do not change their expression during development of the limb nor are they altered in expression by the transgene (ns-ns-ns)(Table S1). Replicating satellite cells that express Pax7 are abundant even when muscle degeneration caused by the transgene is extensive (Fig. S1). The traditional assays for cell death (activated caspase-3 and TUNEL) are negative in the transgenic limb muscle cells. Since the expression of the transgene occurs in all kinds of muscle including cardiac muscle the cause of death in the transgenic tadpoles is not known. We have considered that the phenotype might be caused by non-specific toxicity due to its accumulation in limb muscle nuclei. However expression of the transgene is higher in tail than in any other muscle judging from the intensity of GFP yet the tail is not only healthy in these induced transgenic tadpoles, but it is protected from TH-induced resorption by the transgene (Das et al., 2002). Furthermore, the limb muscle cells remain alive, and the expression of most of the muscle genes on the array is not altered by the transgene (Figs. S1 and S2; Table S1).
Candidate TH-regulated muscle genes
The micro array probably contains no more than half of all of the X. laevis expressed genes an obvious limitation to a comprehensive search for a candidate gene(s). Nevertheless this micro array analysis has revealed a small number of muscle genes (Table 2) that have the expression profile expected of a primary candidate to account for muscle degeneration and many more muscle genes whose activity is influenced secondarily by the phenotype (Tables S1–S4). A gene that is inhibited from being down regulated in limb muscle at metamorphosis might cause the necrosis phenotype. In that case the transgene is expected to up regulate that gene at the 2 wk induction time point (up-up-down, Fig. 4E). However there are only three genes with this expression profile on the entire array. Their expression change is small, and they have no relationship to muscle. The collection of genes with the opposite profile (down-down-up or down-ns-up) is much more relevant to the muscle phenotype. These genes are normally up regulated in limb muscle at metamorphosis but are repressed by the transgene. The majority of the 34 primary candidate genes on the array with this profile (Table 2) are expressed in muscle. The expression of many more muscle genes rise significantly in the frog limb muscle but are unchanged at 2 and 6 wks of transgene exposure (ns-ns-up) (Table S2) an expression profile that can be secondarily attributed to the transgene. The suppression of these genes at the 6 wk time point is likely to be the result rather than the cause of muscle degeneration. We have sought to determine the gene(s) directly responsible for the degradation phenotype by assaying their expression at varying times of Dox induction. Most of the candidate genes have been verified by in situ hybridization with regard to their muscle expression and down regulation at the 6 week time point (Table 2). We screened the down regulation kinetics of 7 of the most dramatically affected genes (Table 3). All seven were significantly down regulated 4 days after the addition of Dox, but only calsequestrin was down regulated before the appearance of muscle degradation at 1 day (Fig. 7).
The role of calcium in muscle degeneration
Calsequestrin is one of several of the candidate genes that are involved in calcium availability, the reuptake of Ca++ in skeletal muscle and muscle contraction. Calsequestrin is reduced in some patients with Duchenne and Becker type muscular dystrophy (Lucas-Heron et al., 1989), but no phenotype has been reported for calsequestrin knockouts. The protein is located in the terminal cisternae of the sarcoplasmic reticulum and functions as a calcium storage protein. The sarcoplasmic reticulum Ca(2+)-ATPase (SERCA) plays an important role in uptake of calcium into muscle fibers (MacLennan et al., 1997). Mutations in this gene have been associated with Brody’s disease a disorder that reduces the relaxation time of fast twitch muscle (Odermatt et al., 1996). Troponin C (Hoffmann et al., 2001) and tropomyosin beta (Donner, 2000) mediate Ca++ dependent muscle contraction. Creatine kinase influences the sequestration of Ca++ in muscle (Steeghs et al., 1997). The ryanodine receptors (RyR) are intracellular calcium release channels that regulate calcium transport in cardiac and skeletal muscle. RyR3 is the only isoform of this calcium release channel on the array. A knockout of the RyR3 isoform impairs muscle contraction in new born mice (Bertocchini et al., 1997). Myozenin, also called calsarcan, is a calcineurin binding protein. The down regulation of one or more of these proteins might cause an uncontrolled surge of calcium that activates calpains. These calcium dependent muscle specific proteases can lead to rhabdomyolysis (Gissel, 2006). The disruption of muscle fibers in the TRDN phenotype (Fig. 2) resembles this condition more than it does typical wasting dystrophies.
Other primary candidate genes are implicated in myopathies
Considering the interdependency of proteins that make up muscle fibers it is not surprising that mutations in a large number of genes cause muscle wasting. Even single mutations in a gene that encodes a contractile protein can disrupt the structure of myofibrils (Karlik et al., 1984). Several of the genes that are primary candidates to account for the muscle degeneration are known to be associated with mammalian myopathies. This phenotype has been documented either as a result of gene knock outs in the mouse or as known human mutations. Mutations in enolase (Comi et al., 2001) and phosphoglucomutase (DiMauro et al., 1981), genes that encode glycolysis enzymes, cause human “metabolic” myopathies. Both of these genes have isomers represented on the micro array that are expressed in other tissues. The non-muscle isomers are not influenced by the transgene. Most of the remaining genes that encode glycolytic enzymes behave like secondary response genes (ns-ns-up) (Table S3). The frog limb has highly elevated expression of these same glycolytic genes, and every one of these genes that has either a primary or secondary candidate profile is depressed in tail muscle at climax. The same few glycolytic genes that are unchanged in the limb array (ns-ns-ns) are unchanged in the tail at climax. A profile of gene expression changes in denervated rat muscle found many genes down regulated that encode both glycolysis and electron transport in mitochondria (Batt et al., 2006).
Deficiency of the alpha 1 form of actin is responsible for nemaline myopathy (Ilkovski et al., 2001). The expression of this gene is strongly depressed in the transgenic limbs and the tail at climax. A gene encoding a muscle-specific isomer of elongation factor E1alpha 2 is dramatically up-regulated in the frog limb but suppressed (ns-ns-up) in the transgenic limbs. Mutations in this gene have been implicated in a human wasting disorder (Chambers et al., 1998). Three subunits of the nicotinic cholinergic receptor (alpha, delta, and epsilon) are amongst the candidate genes. A fourth subunit (beta) is a secondary response gene while the fifth subunit (gamma) is down regulated in the frog limb. We have followed the development of the acetylcholine receptor in growing limbs using fluorescent alpha-bungarotoxin. The degraded muscle caused by the transgene has visible receptors that do not cluster at the nerve endings (data not shown).
Many of the same genes that are repressed by the transgene in the limb are down regulated in tail when it resorbs
TH induces the tail muscle of normal tadpoles to die and the limb muscle to grow. These programs are reversed when the TRDN is expressed in muscle. The tail muscle of a transgenic tadpole becomes resistant to TH (Das et al., 2002) while the limb muscle degenerates (Fig. 6). We observed (Das et al., 2006) that many of the same individual genes in several different functional pathways are regulated together but in opposite directions in the dying tail muscle at metamorphic climax compared to the TH-induced limb muscle. TH up regulates many genes that encode glycolytic enzymes and energy related proteins in the mitochondria of the growing limb and down regulates them in tail muscle at climax. We have extended this observation in these studies to the genes whose expression is influenced by the TRDN transgene. The majority of genes that do not change during development or in transgenic limbs are unchanged in tail muscle at climax. The candidate limb muscle genes that are down regulated by the transgene in the limb are also down regulated in the tail at metamorphic climax (Table 2). Table 4 summarizes the extent that three functional groups of muscle genes, those involved in mitochondrial energy metabolism, cytoplasmic-localized glycolysis, and muscle contraction and maintenance have a gene expression response caused by the TRDN transgene in the limb that is similar to the tail resorption program. Introduction of the TRDN transgene changes a limb growth program into a muscle resorption program by down-regulating many of the same group of genes that are normally expressed actively in functioning muscle. There are no transcription factors amongst the list of primary candidates (Table 2) that could explain this wholesale reversal of gene expression programs. However, one candidate gene, sirtuin, is involved in chromatin remodeling. Sirtuin has been reported to interact with nuclear receptor cofactors (Picard et al., 2004). The TRDN has an intact DNA binding domain and should bind to the same thyroid receptor response elements in DNA sequences as the unliganded thyroid receptor. However, the TRDN has lost an important site for coactivator binding in its C-terminus so that transcription complexes that surround the TR-response genes in muscle might be unable to release a bound corepressor thereby insuring a TH-resistant repression of a whole group of genes. The limb muscle growth and the tail muscle death program may have more in common than ever suspected. The TR may bind to the same set of genes but with opposite regulatory affects. By exchanging a dominant negative form of the receptor for the normal TH-responsive receptor a set of genes in limb muscle have changed their growth program to a resorption program
Supplementary Material
Muscle genes on the array sorted into kinetic groups.
All secondary candidate genes
Genes encoding glycolytic enzymes
All mitochondrial genes on the array
Muscle progenitor cells are present in the pCar/TRDN limb even at the climax of metamorphosis (NF61), and they incorporate Brdu. A) Brdu positive cells (green); B) satellite muscle cells labeled with an antibody to Pax 7 (red); C, a merged picture of A and B. Scale bar=200 μm
In situ hybridization shows that titin (A,B) and desmin (A’,B’) gene expression are not affected by the transgene. A, A’, NF61 limb without Dox; B, B’ pCar/TRDN NF61 limb after 6 wks of Dox treatment. ; C, C’ same sections as B,B’ but immunostained with My32 (myosin). Scale bar=200 μm
Kinetics of gene expression after addition of Dox to pCar/TRDN tadpoles at NF56. A1–D1) calsequestrin 1; A2–D2) actin alpha 1; A3–D3) phosphoglycerate mutase 2. A1–A3) control; B1–B3) 2 days of Dox; C1–C3) 4 days of Dox; D1–D3) 1 week of Dox. Sections of bone can be seen in some panels (black arrow) at background levels. Scale bar=200 μm
Acknowledgments
The authors are grateful to Rejeanne Juste for her expert technical assistance and Mike Sepanski for the electron micrographs. These micro array experiments depended upon the advice and assistance of Minoru Ko, Mark Carter, Yu-Lan Piao, and Alexei Sharov of the Developmental Genomics and Aging Section, Laboratory of Genetics Division, National Institute on Aging, Baltimore. Dr. Ralph Kuncl and Andrea Corse shared their expertise on human muscle pathology with us. Drs. Charles Emerson, Catherine Thompson, and Chen-Ming Fan critically reviewed the manuscript. This research was supported by grants to DDB from the NIH and the G. Harold and Leila Y. Mathers Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Batt J, Bain J, Goncalves J, Michalski B, Plant P, Fahnestock M, Woodgett J. Differential gene expression profiling of short and long term denervated muscle. Faseb J. 2006;20:115–7. doi: 10.1096/fj.04-3640fje. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Bertocchini F, Ovitt CE, Conti A, Barone V, Scholer HR, Bottinelli R, Reggiani C, Sorrentino V. Requirement for the ryanodine receptor type 3 for efficient contraction in neonatal skeletal muscles. Embo J. 1997;16:6956–63. doi: 10.1093/emboj/16.23.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Cai L, Das B, Marsh-Armstrong N, Schreiber AM, Juste R. Thyroid hormone controls multiple independent programs required for limb development in Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 2005;102:12455–8. doi: 10.1073/pnas.0505989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Brown DD. Expression of type II iodothyronine deiodinase marks the time that a tissue responds to thyroid hormone-induced metamorphosis in Xenopus laevis. Dev Biol. 2004;266:87–95. doi: 10.1016/j.ydbio.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Cameron JA, Fallon JF. Evidence for polarizing zone in the limb buds of Xenopus laevis. Dev Biol. 1977;55:320–30. doi: 10.1016/0012-1606(77)90175-0. [DOI] [PubMed] [Google Scholar]
- Chambers DM, Peters J, Abbott CM. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc Natl Acad Sci U S A. 1998;95:4463–8. doi: 10.1073/pnas.95.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi GP, Fortunato F, Lucchiari S, Bordoni A, Prelle A, Jann S, Keller A, Ciscato P, Galbiati S, Chiveri L, Torrente Y, Scarlato G, Bresolin N. Beta-enolase deficiency, a new metabolic myopathy of distal glycolysis. Ann Neurol. 2001;50:202–7. doi: 10.1002/ana.1095. [DOI] [PubMed] [Google Scholar]
- Das B, Brown DD. Controlling transgene expression to study Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 2004;101:4839–42. doi: 10.1073/pnas.0401011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Cai L, Carter MG, Piao YL, Sharov AA, Ko MS, Brown DD. Gene expression changes at metamorphosis induced by thyroid hormone in Xenopus laevis tadpoles. Dev Biol. 2006;291:342–55. doi: 10.1016/j.ydbio.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Das B, Schreiber AM, Huang H, Brown DD. Multiple thyroid hormone-induced muscle growth and death programs during metamorphosis in Xenopus laevis. Proc Natl Acad Sci U S A. 2002;99:12230–5. doi: 10.1073/pnas.182430599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Miranda AF, Khan S, Gitlin K, Friedman R. Human muscle phosphoglycerate mutase deficiency: newly discovered metabolic myopathy. Science. 1981;212:1277–9. doi: 10.1126/science.6262916. [DOI] [PubMed] [Google Scholar]
- Donner K, Ollikainen M, Pelin K, Gronholm M, Carpen O, Wallgren-Pettersson C, Ridanpaa M. World Muscle Society Abstract. Neuromuscul Disord. 2000:342–343. [Google Scholar]
- Furlow JD, Berry DL, Wang Z, Brown DD. A set of novel tadpole specific genes expressed only in the epidermis are down-regulated by thyroid hormone during Xenopus laevis metamorphosis. Dev Biol. 1997;182:284–98. doi: 10.1006/dbio.1996.8478. [DOI] [PubMed] [Google Scholar]
- Gissel H. The role of Ca2+ in muscle cell damage. Ann N Y Acad Sci. 2006;1066:166–80. doi: 10.1196/annals.1363.013. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17:524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- Ilkovski B, Cooper ST, Nowak K, Ryan MM, Yang N, Schnell C, Durling HJ, Roddick LG, Wilkinson I, Kornberg AJ, Collins KJ, Wallace G, Gunning P, Hardeman EC, Laing NG, North KN. Nemaline myopathy caused by mutations in the muscle alpha-skeletal-actin gene. Am J Hum Genet. 2001;68:1333–43. doi: 10.1086/320605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlik CC, Coutu MD, Fyrberg EA. A nonsense mutation within the act88F actin gene disrupts myofibril formation in Drosophila indirect flight muscles. Cell. 1984;38:711–9. doi: 10.1016/0092-8674(84)90266-6. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Lucas-Heron B, Mussini JM, Ollivier B. Is there a maturation defect related to calcium in muscle mitochondria from dystrophic mice and Duchenne and Becker muscular dystrophy patients. J Neurol Sci. 1989;90:299–306. doi: 10.1016/0022-510x(89)90116-0. [DOI] [PubMed] [Google Scholar]
- MacLennan DH, Rice WJ, Green NM. The mechanism of Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases. J Biol Chem. 1997;272:28815–8. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Cai L, Brown DD. Thyroid hormone controls the development of connections between the spinal cord and limbs during Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 2004;101:165–70. doi: 10.1073/pnas.2136755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yaoita Y. Dual mechanisms governing muscle cell death in tadpole tail during amphibian metamorphosis. Dev Dyn. 2003;227:246–55. doi: 10.1002/dvdy.10300. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, a FJ. Normal Table of Xenopus Laevis (Daudin) Elsevier/North-Holland; New York: 1956. [Google Scholar]
- Odermatt A, Taschner PE, Khanna VK, Busch HF, Karpati G, Jablecki CK, Breuning MH, MacLennan DH. Mutations in the gene-encoding SERCA1, the fast-twitch skeletal muscle sarcoplasmic reticulum Ca2+ ATPase, are associated with Brody disease. Nat Genet. 1996;14:191–4. doi: 10.1038/ng1096-191. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber AM, Das B, Huang H, Marsh-Armstrong N, Brown DD. Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc Natl Acad Sci U S A. 2001;98:10739–44. doi: 10.1073/pnas.191361698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–9. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Steeghs K, Benders A, Oerlemans F, de Haan A, Heerschap A, Ruitenbeek W, Jost C, van Deursen J, Perryman B, Pette D, Bruckwilder M, Koudijs J, Jap P, Veerkamp J, Wieringa B. Altered Ca2+ responses in muscles with combined mitochondrial and cytosolic creatine kinase deficiencies. Cell. 1997;89:93–103. doi: 10.1016/s0092-8674(00)80186-5. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963–8. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoita Y, Nakajima K. Induction of apoptosis and CPP32 expression by thyroid hormone in a myoblastic cell line derived from tadpole tail. J Biol Chem. 1997;272:5122–7. doi: 10.1074/jbc.272.8.5122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Muscle genes on the array sorted into kinetic groups.
All secondary candidate genes
Genes encoding glycolytic enzymes
All mitochondrial genes on the array
Muscle progenitor cells are present in the pCar/TRDN limb even at the climax of metamorphosis (NF61), and they incorporate Brdu. A) Brdu positive cells (green); B) satellite muscle cells labeled with an antibody to Pax 7 (red); C, a merged picture of A and B. Scale bar=200 μm
In situ hybridization shows that titin (A,B) and desmin (A’,B’) gene expression are not affected by the transgene. A, A’, NF61 limb without Dox; B, B’ pCar/TRDN NF61 limb after 6 wks of Dox treatment. ; C, C’ same sections as B,B’ but immunostained with My32 (myosin). Scale bar=200 μm
Kinetics of gene expression after addition of Dox to pCar/TRDN tadpoles at NF56. A1–D1) calsequestrin 1; A2–D2) actin alpha 1; A3–D3) phosphoglycerate mutase 2. A1–A3) control; B1–B3) 2 days of Dox; C1–C3) 4 days of Dox; D1–D3) 1 week of Dox. Sections of bone can be seen in some panels (black arrow) at background levels. Scale bar=200 μm


