Abstract
A prominent feature of the hepatic response to injury is production of a fetal isoform of fibronectin, a splice variant containing the EIIIA region, which appears very early after injury and derives from sinusoidal endothelial cells. Previous studies have shown that it is instrumental in initiating the cellular response to injury, specifically the conversion of resting stellate cells to myofibroblast-like cells. The present work describes the regulation of this change in fibronectin expression. Using sinusoidal endothelial cells from normal or injured liver in primary culture, we show that exogenous transforming growth factor β (TGFβ) stimulates [EIIIA]Fn expression. To assess the role of TGFβ in vivo, we used a chimeric IgG containing the extracellular portion of the TGFβ type II receptor as a competitive inhibitor of the cytokine. Administered to animals at the time of injury, the inhibitor reduced expression of [EIIIA]Fn mRNA by 50% as compared to controls (P < 0.01). There was a corresponding decrease in [EIIIA]Fn protein production as judged by immunohistochemistry. Cell fractionation experiments indicated that the changes observed in whole-liver extracts were localized to sinusoidal endothelial cells. We conclude that TGFβ initiates wound repair in part by stimulating endothelial expression of [EIIIA]Fn. Results with the soluble inhibitor of the TGFβ type II receptor suggest a novel strategy for modulating wound repair in vivo.
Transforming growth factor β (TGFβ), a 25-kd homodimeric polypeptide, has been implicated in regulating repair and regeneration after tissue injury. 1,2 Prominent among its effects in an injury milieu are the stimulation of production of other cytokines and inflammatory mediators, 3,4 induction of extracellular matrix (ECM) synthesis, 5-7 and inhibition of matrix degradation. 8,9 The cellular source of TGFβ in liver varies with the type of injury and may involve resident parenchymal cells, mesenchymal cells, or recruited inflammatory cells; its effects appear to be largely autocrine or paracrine. 10,11
An important aspect of the injury response is the elaboration of ECM, including collagens I, III, IV, VI as well as fibronectin and other glycoproteins. ECM is elaborated presumably to contain the agent of injury, close the wound, and complete healing. When ECM deposition is excessive, however, it may disrupt the organization of the tissue and lead to impaired function. From numerous recent studies, it is clear that ECM in injury derives principally from stellate cells. 12-19 A pivotal event in the response of the liver to injury is the activation of these cells, which is characterized by the de novo appearance of cytokine receptors, ECM production, proliferation, and contraction. 18,20,21 Because of its importance to fibrosis and chronic liver injury, the regulation of stellate activation has been a subject of intensive study. Both soluble mediators and the ECM itself play a role. 18
Fibronectin is one of the earliest of the ECM components to be expressed after injury. Its primary transcript has three regions, termed EIIIA, EIIIB, and V, that are variably spliced. 22-24 The EIIIA and EIIIB domains are either completely included or excluded within the mature molecule. Splice variants containing EIIIA are expressed during development but are minimally present in normal adult tissue. They reappear, however, in the setting of epithelial repair; this has been studied extensively in cutaneous repair. 25 It has been demonstrated that the EIIIA isoform is also expressed in various forms of liver injury. 26,27 We have shown that this derives from hepatic sinusoidal endothelial cells (SEC) and that it critically modifies the local ECM so as to activate stellate cells. 26 Moreover, increased expression of this fibronectin variant is detectable within 12 hours of an injury, preceding any increase in expression of collagen and other structural ECM proteins. 26
The present study concerns the regulation of [EIIIA]Fn production, focusing on cytokines that are part of the injury milieu. Although TGFβ is particularly prominent in the early repair process, its effects in vivo are not well understood with respect to cellular targets and quantitative changes in Fn isoform expression. We show that it modulates production of [EIIIA]Fn in primary cultures of hepatic sinusoidal endothelial cells and in vivo. The in vivo experiments take advantage of a novel recombinant TGFβ-soluble receptor to block the action of TGFβ during wound repair. The findings point to the central role of endothelial cells in wound repair and suggest that manipulation of [EIIIA]Fn expression in injury may be beneficial in reducing neomatrix formation.
Materials and Methods
Materials
Pronase and DNase were purchased from Boehringer Mannheim (Indianapolis, IN), collagenase from Serva (Heidelburg, Germany), and Ham’s F-12, Medium 199, DME, and fetal calf and donor horse sera from Flow Laboratories (McLean, VA). Eagle’s MEM without calcium was prepared using amino acids purchased from Sigma Chemical Co. (St Louis, Mo). Accudenz was obtained from Accurate Chemicals (Westbury, NY). Collagen I from rat tail tendon was prepared in the laboratory.
TRI reagent was from Molecular Research Center, Inc., (Cincinnati, OH). Acrylamide, bis-acrylamide and agarose were from Bio-Rad (Richmond, CA). Ultrapure urea, Trypsin-EDTA, and RNaseT2 were from GIBCO BRL (Gaithersburg, MD). T7 RNA polymerase, RQ1 DNase, and RNasin were from Promega (Madison, WI). Radiolabeled cytidine-5′-triphosphate ([α-32P]CTP, >800 Ci/mmol) and 3 H-thymidine (>50 Ci/mmol) were from Amersham Corp. (Arlington Heights, IL); Trans35S-label was from ICN (Irvine, CA). Protein A sepharose CL-4B was purchased from Pharmacia (Kalamazoo, MI). Mink lung epithelial cells (CCL64) were obtained from the University of California San Francisco Cell Culture Facility (San Francisco, CA).
The monoclonal antibody IST-9, which is specific for the EIIIA domain of fibronectin, 28 was kindly provided by L. Zardi (Genoa, Italy). Biotinylated sheep anti-mouse IgG and streptavidin-linked Texas red were purchased from Amersham. Avidin-biotin complex (Vectastatin) was from Vector (Burlingame, CA). Purified TGFβ and TGFβ antibodies were purchased from R&D Systems (Minneapolis, MN). Other chemicals were from Sigma.
Soluble TGFβ Type II Receptor (sTGFβR)
The TGFβ receptor fusion protein was synthesized using the extracellular domain of the rabbit TGFβ type II receptor, which was amplified by PCR from plasmid 3F11. 29 The forward primer consisted of nucleotides 1–21 of the rabbit TGFβ type II receptor, and the reverse primer was the complement of nucleotides 466–486. To prepare a plasmid encoding the desired fusion protein, DNA fragments encoding amino acids 1–160 of the rabbit TGFβ type II receptor were ligated to a DNA fragment encoding the Fc domain of human IgG1 in the Biogen transient expression vector SAB132. DNA sequencing revealed a glycine-to-arginine mutation in the fourth amino acid of the signal peptide. The rabbit TGFβ type II receptor-IgG fusion DNA fragment was cloned into the Biogen stable expression plasmid pMDR901 to generate plasmid pMSN001, which was used to stably transfect Chinese hamster ovary cells. Fusion protein secreted by the cells was purified on a protein A sepharose affinity column as previously described. 30 The extracellular domain of the rabbit TGFβ type II receptor is 83% homologous to that of the rat. The inhibitory activity of the protein was verified and titered in a standard bioassay for TGFβ 31 as described below.
Animal Model of Liver Injury
Injury was induced in male Sprague-Dawley rats (∼400 g body weight) by laparotomy and high ligation of the bile duct. 21,32 Controls underwent laparotomy and manipulation of the bile duct but no ligation. The effects of this injury are well documented. 20 After 3 days, the liver exhibits cholestasis with periductular inflammation, and collagen I mRNA is markedly increased in stellate cells; after 5 to 7 days, periportal fibrosis is seen histologically. At the time of laparotomy, some animals received sTGFβR (5 mg/kg) or normal human IgG by slow infusion into the inferior vena cava. In preliminary experiments, this amount was found to be sufficient for maximal inhibition of TGFβ. Unless otherwise stated, animals were sacrificed 24 hours later for cell isolation. In preliminary studies with varying doses of sTGFβR, 5 mg/kg was found to maximally reduce [EIIIA]Fn expression; in the experiments presented here, this dose was used.
Endothelial Cell Isolation and Culture
SECs were isolated by in situ perfusion with pronase and collagenase and fractionation by centrifugation on a discontinuous gradient of Accudenz (15.6 and 8.2%), 33 then further purified by centrifugal elutriation. 34 Purity was monitored by labeling the cells in vivo with fluorescent acetoacetylated low density lipoprotein, administered before cell isolation, 34 and was >90%. SECs were plated at confluent density in 60 mm culture dishes precoated with a thin layer of type I collagen (∼ 40 μg/60 mm dish) to facilitate cell attachment. The culture medium was a modified Medium 199 35 with 20% serum (10% calf, 10% horse), insulin (4 mU/ml), and gentamicin (40 μg/ml). For comparison of treated and control cultures, cells from a single animal were used, initiated in culture under identical conditions.
Quantitation of mRNA
Total RNA was isolated from whole liver tissue or from purified SECs using TRI reagent. Its integrity was assessed routinely by visualization of 18S and 28S ribosomal bands on agarose/formaldehyde gel electrophoresis. Radiolabeled probe for [EIIIA]Fn was prepared as described 26 and mRNA quantified by RNase protection. Fibronectin mRNA containing the EIIIA domain protected a 280-bp fragment of the labeled cRNA probe, and that lacking EIIIA protected 109 bp. Samples were assayed also for S-14 mRNA, which encodes a ribosomal protein. 36 S-14 exhibits minimal variation after bile duct ligation (BDL) 26 and was used to control for mRNA loading.
Probes were hybridized with either 20 μg (EIIIA) or 5 μg (S14) of total RNA at 55°C for 16 to 18 hours. Unhybridized RNA was digested with ribonuclease T2. 20 The RNA-cRNA hybrids were extracted in phenol-chloroform, precipitated with ammonium acetate and ethanol, and then dissociated by boiling for 3 minutes in electrophoresis buffer containing 80% formamide. Samples were run on a 6% (EIIIA) or 5% (S-14) polyacrylamide/urea gel, dried, and visualized by autoradiography (Kodak X-OMat AR-5). Specific bands were quantitated by scanning densitometry (Hoefer Scientific Instruments, San Francisco, CA) with correction of the raw data for the size difference of the [EIIIA]-positive and [EIIIA]-negative fragments.
Nuclear Run-on Assay for Fibronectin Gene Transcription
Fibronectin transcription was measured by a modification of the nuclear run-on assay described by Kavanaugh et al. 37 SECs were isolated and cultured for 24 hours in 20% serum growth medium. After a further overnight incubation in serum-free medium, the cells were incubated for 2 hours with either TGFβ (7.5 ng/ml) or vehicle, then lysed in ice cold 10 mmol/L Tris-HCl, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2, and 0.5% NP-40. The lysate was centrifuged to pellet nuclei, which were washed and resuspended in storage buffer (20 mmol/L Tris-HCl, pH 8.0, 50% glycerol, 60 mmol/L NaCl, 100 μmol/L EDTA, 1 mmol/L DTT) to a concentration of 1 to 2 × 10 7 nuclei/100 μl. An aliquot was inspected by light microscopy and the remainder snap-frozen in liquid nitrogen until use.
Run-on analysis was performed in a total volume of 250 μl comprising 125 μl of nuclei brought to a final concentration of 50 mmol/L Tris-HCl, pH 8.0, 5 mmol/L MgCl2, 0.5 mmol/L MnCl2, 100 mmol/L KCl, 0.25 mg/ml BSA, 250 U/ml RNasin, 50 μmol/L EDTA, 1 mmol/L DTT, 0.5 mmol/L each of rCTP, rGTP, and rATP, and 125 μCi of α-32P-labeled rUTP (3000 mCi/mol, Amersham). Following incubation on a rotary shaker for 30 minutes at room temperature, the reaction was terminated with the addition of 12.5 units of DNase I for 20 minutes. Proteinase K (200 μg/ml), SDS (0.2%), and EDTA (5 mmol/L) were added, and the mixture was incubated at 37°C for 30 minutes. Nuclei were extracted using TRI reagent, and the final RNA pellet was resuspended in RNase-free water. An aliquot was counted, and an equal amount of radioactivity in each sample was used for hybridization to denatured plasmid DNA (5 μg) slot blotted on nitrocellulose filters. Hybridization was carried out at 65°C for 40 hours in a buffer consisting of 50 mmol/L PIPES, pH 6.8, 10 mmol/L EDTA, 0.2% SDS, 2.5× Denhardt’s, 100 μg/ml herring DNA, 100 μg/ml poly(A), and 600 mmol/L NaCl. The filters were then washed serially in 2× SSC, 2× SSC containing RNase A (10 μg/ml, 37°C), and finally in 2× SSC. After air drying, they were examined by autoradiography.
Immunohistochemistry of [EIIIA]Fn
After low pressure perfusion with buffered saline via the portal vein, the liver was cut into 0.5-cm 2 pieces, which were snap-frozen in liquid nitrogen and stored at −70°C until use. Cryostat sections were stained for [EIIIA]Fn with IST-9 mouse anti-EIIIA antibody; the secondary antibody was sheep anti-mouse. 26 Negative controls consisted of sections incubated with nonimmune mouse IgG and processed in parallel.
Metabolic Labeling and Immunoprecipitation of [EIIIA]Fn
SECs in culture (∼80% confluent) were placed in serum free medium containing 0.5% bovine serum albumin for 16 hours before metabolic labeling. Cells were labeled for 6 hours with Trans35S-label (200 μCi/ml) in serum free medium without methionine or cysteine. Newly synthesized [EIIIA]Fn protein in conditioned medium and in cell lysates was immunoprecipitated as described, 38 with modifications. Cells were lysed on ice for 30 minutes in buffer (0.1 mol/L Tris-HCl, pH 8.3, 0.1% SDS, 1 mmol/L N-ethylmaleimide, 1 mmol/L iodoacetic acid, 2 mmol/L phenylmethylsulfonyl fluoride, 2 mmol/L EDTA, 0.5% NP-40, 0.5% deoxycholate). The lysate DNA was then sheared by repeated passage through a 23 G needle, and the mixture was spun at 10,000 × g at 4°C for 10 minutes. The extent of radiolabel incorporation into cellular proteins was determined by trichloroacetic acid precipitation.
Before immunoprecipitation with specific antibody the supernatant was precleared by shaking for 45 minutes at 4°C with protein A sepharose beads previously equilibrated in lysis buffer containing 5 mg/ml ovalbumin. The beads were pelleted (14,000 rpm for 10 seconds), and the supernatant was incubated overnight at 4°C with the primary antibody (IST-9) or nonimmune mouse IgG. One hour before the end of the incubation, polyclonal rabbit anti-mouse IgG was added, and the mixture was incubated for 45 minutes at 4°C in a rotary shaker with 50 μl of protein A sepharose. The beads were washed four times in buffer, resuspended in an equal volume of 2× buffer (6% SDS, 125 mmol/L Tris-HCl, pH 6.8, 20% glycerol) containing 6% β-mercaptoethanol, and boiled for 5 minutes to dissociate antigen-antibody complexes. After centrifugation at 14,000 rpm for 1 minute to sediment sepharose beads, the proteins in the supernatant were resolved on a 5% SDS-PAGE gel. 39 The gel was dried and exposed to X-ray film for 2 to 7 days.
Quantitation of TGFβ Secretion
Bioactive TGFβ in conditioned medium was measured by its ability to inhibit the proliferation of mink lung epithelial cells. 31 Mink lung cells (CCL64) were maintained in DMEM/10% FCS in 75-cm 2 flasks. For the bioassay, subconfluent cultures between passages 3 and 10 were used. Cells were washed and trypsinized, and 1 × 10 4 cells were seeded onto 96-well microtiter plates in 190 μl of DMEM/0.2% FCS/10 mmol/L HEPES (pH 7.4). SECs from normal or BDL rats were isolated and plated directly in serum-free Medium 199. The conditioned medium was harvested 24 hours later and assayed directly for active TGFβ. For determining total TGFβ, samples were acidified for 10 minutes. (0.05 N HCl final concentration), followed by neutralization with 0.05 N NaOH and HEPES, pH 7.4 (0.025 mol/L final concentration); this treatment converts latent TGFβ to the active form. 31 Ten microliters of sample were then added to each well and incubated at 37°C for 24 hours. The cells were subsequently incubated with 3 H-thymidine (0.1 μCi) for 4 hours, lysed at −70°C, and the incorporated radioactivity trapped onto glass fiber filters (Cambridge Technology Inc., Watertown, MA) using a cell harvester (Cambridge Technology). The radioactivity was measured in a liquid scintillation counter. Each assay was carried out in triplicate; the concentration of TGFβ was determined by comparison with a standard curve generated using purified TGFβ1.
Statistical Analysis
Data are presented as means ± SE of at least three independent experiments. Differences between groups were analyzed using the unpaired Student’s t-test (2 groups) or the one-factor analysis of variance (multiple comparisons). The significance level for all tests was P < 0.05.
Results
[EIIIA]Fn mRNA Quantitation in Endothelial Cells after BDL
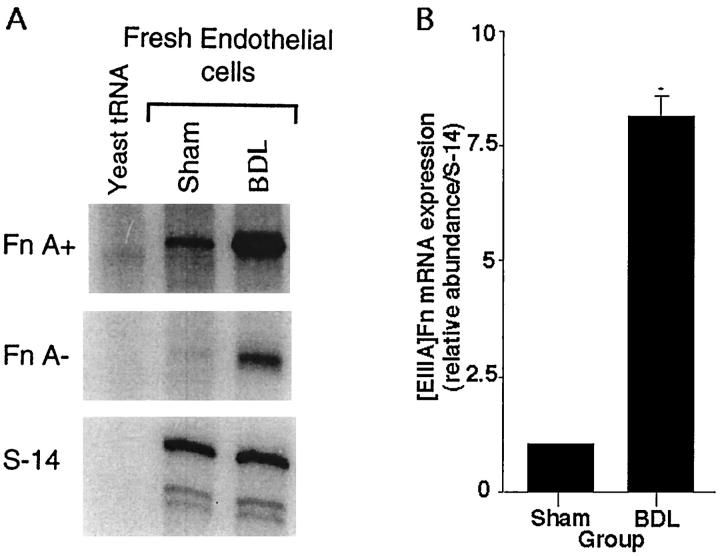
One day after BDL, [EIIIA]Fn mRNA in fresh isolates of SECs was increased eightfold compared to sham-operated controls (Figure 1 ▶ ; P < 0.0001). Although expression of [EIIIA]-negative Fn also was increased (Figure 1) ▶ , [EIIIA]-positive Fn mRNA as a percent of total Fn mRNA rose from 32% in controls to 42% in SECs from BDL rats; this difference was significant (P < 0.001). The stability of the observed changes in primary culture was examined by placing freshly isolated SECs from normal or BDL rats in culture for 24 hours under serum-free conditions. Over this period of culture, [EIIIA]Fn mRNA remained elevated about sixfold (Figure 2, A and B) ▶ in SECs from BDL animals compared to controls (P < 0.001). However, the previously noted spontaneous change in splicing 26 was confirmed: after 24 hours in culture the proportion of [EIIIA]Fn was similar in all cultures, whether derived from control or BDL rats (57% [EIIIA]-positive in cells from controls, 61% in cells from BDL animals).
Figure 1.
Expression of [EIIIA]Fn and S-14 mRNA in fresh isolates of sinusoidal endothelial cells from sham-operated or 24-hour BDL rat liver. A: A representative RNase protection assay using 20 μg of total RNA for the fibronectin probe and 5 μg for S-14. Yeast tRNA is a negative control. B: Relative abundance of [EIIIA]Fn mRNA in endothelial cells derived from sham-operated (Con) and 24-hour BDL rat liver. Values represent the mean ± SE of at least four separate experiments. The signal for [EIIIA]Fn has been corrected for the level of S-14 mRNA in the same samples. * P < 0.0001.
Figure 2.
Stability of [EIIIA]Fn expression in cultured sinusoidal endothelial cells isolated from sham-operated and 24-hour BDL rat liver. The cells from 4 different animals (2 sham-operated and 2 subjected to BDL) were placed in culture for 24 hours under serum-free conditions. Total RNA was isolated and the level of [EIIIA]Fn mRNA determined using an RNase protection assay. A: A representative result for two livers is shown, with RNA loading as for Figure 1 ▶ . B: Relative abundance of [EIIIA]Fn mRNA in endothelial cells in culture. Values represent the mean ± SE of at least four separate experiments. The signal for [EIIIA]Fn has been corrected for the level of S-14 mRNA in the same samples. * P < 0.001.
Exogenous TGFβ Up-Regulates [EIIIA]Fn Expression; Blocking Effect of sTGFβR
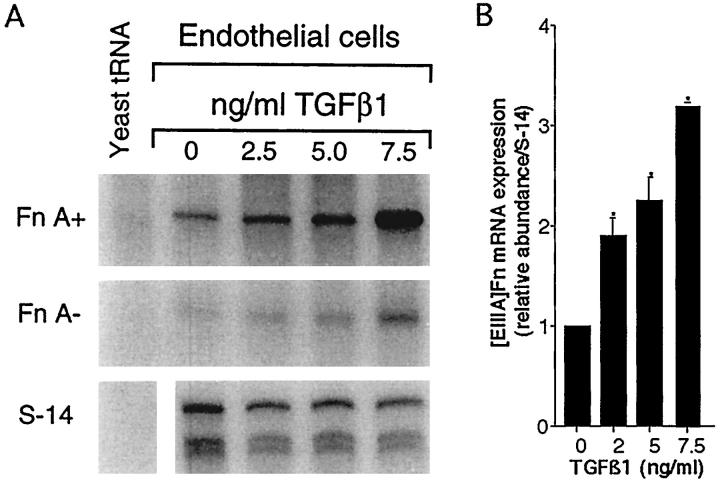
We isolated SECs and plated them for 24 hours in medium with 20% serum to allow for recovery from the isolation procedures. After two washes the medium was replaced with one without serum, containing TGFβ1 (from a stock in 4 mmol/L HCl and 1 mg/ml BSA) or vehicle. TGFβ1 produced a dose-dependent increase in [EIIIA]Fn mRNA up to threefold (Figures 3, A and B ▶ ; P < 0.05); expression of [EIIIB]Fn in the same samples was minimal (data not shown). Concentrations greater than 7.5 ng/ml appeared to be cytotoxic as judged by cellular morphology. TGFβ1 also produced a dose-dependent increase in [EIIIA]Fn mRNA splicing; at a concentration of 7.5 ng/ml TGFβ1, [EIIIA]Fn, as a proportion of total Fn, increased from 57% at baseline to an average of 70% after incubation with TGFβ1 (7.5 ng/ml). By nuclear run-on analysis, the change in fibronectin expression was at least in part transcriptional (Figure 4) ▶ . Exogenous TGFβ1 also induced [EIIIA]Fn protein synthesis, as shown by metabolic labeling and immunoprecipitation (Figure 5) ▶ .
Figure 3.
Effect of TGFβ1 on SEC expression of [EIIIA]Fn. Endothelial cells were placed in 20% serum medium for 24 hours, then washed and incubated in serum-free medium for 24 hours with the indicated concentrations of TGFβ1. A: A representative RNase protection assay, with RNA loading as for Figure 1 ▶ . B: Relative abundance of [EIIIA]Fn mRNA in endothelial cells treated with varying doses of TGFβ1. Values represent the mean ± SE of at least three separate experiments. The signal for [EIIIA]Fn has been corrected for the level of S-14 mRNA in the same samples. * P < 0.05 versus cultures without TGFβ1.
Figure 4.
Transcriptional regulation by TGFβ. SECs were incubated for 2 hours with either TGFβ1 (7.5 ng/ml) or vehicle, then lysed for harvesting of nuclei (see Methods). Isolated nuclei were probed for fibronectin transcription using a 280-bp rat cDNA as described in Methods. S-14 served as an internal control. The pGEM4Z plasmid served as a negative control for the specificity of hybridization conditions.
Figure 5.
[EIIIA]Fn protein synthesis in endothelial cell cultures treated with TGFβ1. Cells were obtained as for Figure 4 ▶ and incubated with either vehicle (4 mmol/L HCl and 1 mg/ml BSA, lanes 1 and 2) or 7.5 ng/ml TGFβ1 (lane 3). Following metabolic labeling [EIIIA]Fn protein was immunoprecipitated using the IST-9 antibody (lanes 2 and 3). The negative control (lane 1) was handled identically with non-immune mouse IgG in place of IST-9. Cell lysates containing equal amounts of radiolabel were used for immunoprecipitation.
This system was used to test the inhibitory effect of sTGFβR on TGFβ-mediated events. SEC isolates from normal rats were initiated in culture in 20% serum, then changed to serum-free medium with or without TGFβ1 (7.5 ng/ml). Plates treated with TGFβ1 were simultaneously incubated with either sTGFβR (50 μg/ml) or control IgG at the same dose. As shown in Figure 6 ▶ , the inhibitor reduced [EIIIA]Fn mRNA to the level in control cultures; there was relatively little effect on [EIIIA]−Fn.
Figure 6.

Effect of sTGFβR on TGFβ-mediated stimulation of fibronectin mRNA. Endothelial cells in serum-free culture were incubated with either TGFβ1 or vehicle. TGFβ1-treated plates were incubated simultaneously with sTGFβR or control peptide (IgG). Yeast tRNA served as the negative control.
Regulation of [EIIIA]Fn Expression in SEC by Autocrine TGFβ
We have postulated that effects of TGFβ on hepatic stellate cells involve predominantly autocrine stimulation. 12 To investigate this, we first measured release of the cytokine by SECs from injured or normal rat liver. Freshly isolated SECs from BDL rats were placed in serum-free medium, and TGFβ protein secretion was quantitated after 24 hours using the mink lung cell growth inhibition assay. Endothelial cells from bile duct ligated animals produced 2.7-fold greater active (P < 0.05) and 3.2-fold greater total (P < 0.01) TGFβ protein than did SECs isolated from sham-operated controls (Table 1) ▶ .
Table 1.
Secretion of Total and Active TGFβ Protein by Endothelial Cells Isolated from Normal and Bile Duct-Ligated (BDL) Rats
| Source of cells | TGFβ production (pg/mg protein/24 h) | |
|---|---|---|
| Total TGFβ | Active TGFβ | |
| Control rats | 135 ± 14 | 59 ± 2 |
| BDL rats | 426 ± 50* | 157 ± 29† |
Endothelial cells were isolated placed in serum-free conditions in primary culture and incubated for 24 hours before assay. Total (acid-activated) and active TGFβ were determined using the mink lung cell assay (see Methods).
*P < 0.01.
†P < 0.05.
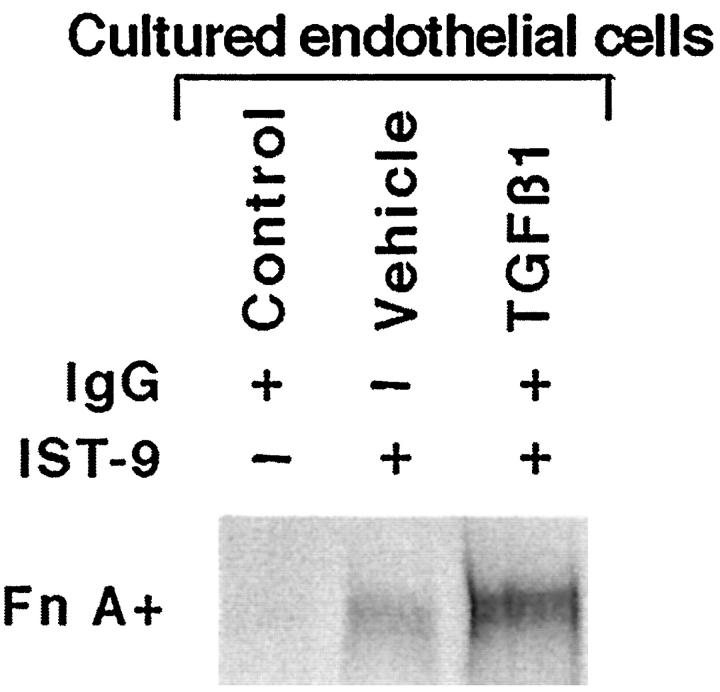
To study the role of autocrine TGFβ in regulating expression of [EIIIA]Fn, we placed SEC isolated from BDL rats in serum-free medium for 24 hours with sTGFβR (50 μg/ml) or control IgG. Incubation with the inhibitor, compared to control IgG, significantly reduced [EIIIA]Fn expression (Figure 7, A and B) ▶ ; the fraction of total Fn as [EIIIA]Fn also decreased (sTGFβR 32% versus IgG 39%; P < 0.001). The soluble receptor was similarly effective in preventing the spontaneous increase in [EIIIA]Fn expression exhibited by normal SEC placed in culture. 26 [EIIIA]Fn protein paralleled mRNA in these experiments, as documented by studies in which SEC were metabolically labeled with radiomethionine and [EIIIA]Fn isolated by immunoprecipitation (data not shown).
Figure 7.
Inhibition of autocrine TGFβ by sTGFβR. Endothelial cells from BDL rats were placed in serum-free culture for 24 hours with sTGFβR or control IgG. A: Representative RNase-protection assay of mRNA. B: Relative abundance of [EIIIA]Fn mRNA in endothelial cell cultures treated with sTGFβR. Data are mean ± SE of four experiments.
Administration of sTGFβR in Vivo Reduces EIIIA Fibronectin Expression
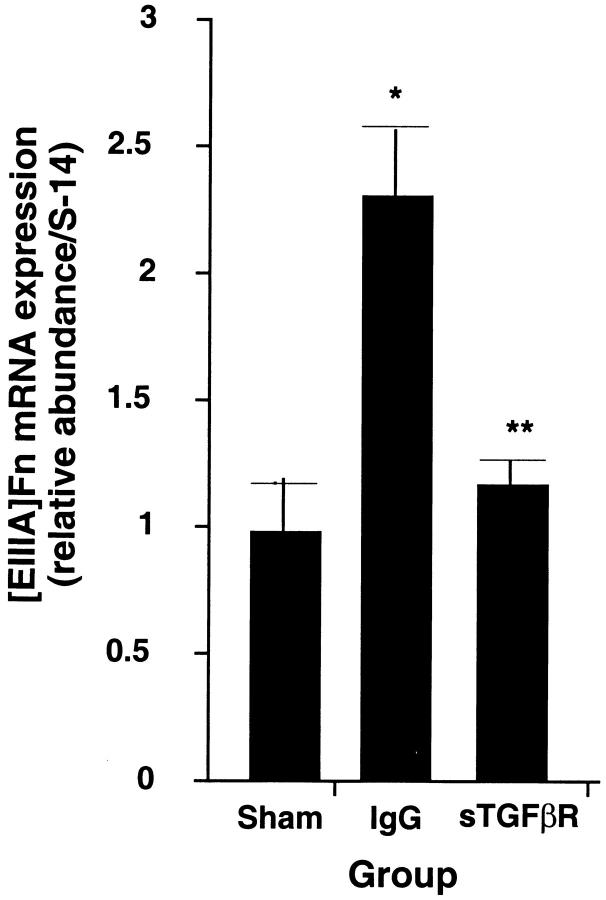
Five rats in each treatment group were subjected to either sham operation, BDL with infusion of the sTGFβR, or BDL with infusion of control human IgG. Whole liver tissue was extracted 24 hours later for assay of fibronectin mRNA. [EIIIA]Fn mRNA in whole liver extracts rose 2.3 ± 0.2-fold in bile duct-ligated rats receiving control IgG compared to sham-operated controls (P < 0.02). This was similar to the increase observed in untreated BDL rats. By contrast, the level in rats infused with the recombinant peptide increased only 1.2 ± 0.1-fold. The difference between this and the value from animals infused with control IgG was significant (Figure 8 ▶ ; P < 0.01). Infusion of sTGFβR also reduced [EIIIA]Fn mRNA splicing compared to rats receiving control peptide (Table 2 ▶ ; 6.2 ± 1.3% versus 10.4 ± 1.5%; P < 0.01). The percent of [EIIIA]Fn in rats receiving IgG was not different from that in sham-operated animals. We next examined [EIIIA]Fn mRNA expression and splicing in SECs isolated from from BDL rats that had been treated with sTGFβR or IgG (Figure 9A) ▶ . In the animals receiving sTGFβR, [EIIIA]Fn mRNA expression was less than one-half that in controls (Figure 9B ▶ ; P < 0.0003); [EIIIA]Fn as a percent of total fibronectin was also reduced by 16% (P < 0.05).
Figure 8.
Effect in vivo of sTGFβR on [EIIIA]Fn mRNA from whole liver. Rats underwent sham operation or BDL and infusion with either sTGFβR or control IgG. The liver was harvested 24 hours later and RNA extraction performed (see Methods). Values represent the mean ± SE of five separate experiments. The signal for [EIIIA]Fn has been corrected for the level of S-14 mRNA in the same samples. * P < 0.02 relative to sham-operated; ** P < 0.01 compared to animals infused with the control peptide.
Table 2.
Effect of sTGFβR In Vivo on Fibronectin Splicing
| Procedure | [EIIIA]Fn mRNA (percent of total Fn) |
|---|---|
| Sham operation | 5.3 ± 0.9 |
| Infusion with control IgG | 10.4 ± 1.5* |
| Infusion with sTGFβR | 6.2 ± 1.3† |
Animals were treated as described for Fig 8 ▶ . The value for total fibronectin was calculated by adding the densitometry values for [EIIIA+]Fn and [EIIIA−]Fn mRNAs after correcting for the difference in probe length. In total liver RNA [EIIIA]Fn comprises only ∼5% of fibronectin RNA reflecting the large amount of [EIIIA−]Fn from hepatocytes. Mean ± SE; n = 5.
*P < 0.004 relative to sham-operated.
†P < 0.002 compared to animals infused the control peptide.
Figure 9.
Effect of TGFβ blockage in vivo on [EIIIA]Fn mRNA in isolated SEC. Rats were subjected to BDL and given either sTGFβR or control peptide. After 24 hours SEC were prepared, and the fresh isolates were extracted for RNA. A: Representative RNase protection assay is shown with RNA loading as for Figure 1 ▶ . B: Relative abundance of [EIIIA]Fn mRNA in fresh endothelial cells derived from 24-hour BDL rats infused at the time of surgery with either sTGFβR or control peptide. Each group consisted of 3 animals. The signal for [EIIIA]Fn has been corrected for the corresponding level of S-14 mRNA.
The influence of sTGFβR on [EIIIA]Fn protein expression in vivo was examined by immunohistology. At 2 days after BDL, liver was harvested, fixed, and stained using the monoclonal antibody IST-9, which is specific for the EIIIA segment of fibronectin. In sections from normal or sham-operated rat liver, specific staining was limited to vessels within portal triads, as shown previously. 26 By contrast, 2 days after BDL, extensive perisinusoidal staining was noted (Figure 10A) ▶ . In animals receiving sTGFβR after BDL, the intensity of perisinusoidal staining was similar to that seen in control liver (Figure 10B) ▶ .
Figure 10.

Immunohistochemical detection of [EIIIA]Fn in rat liver after BDL. Photomicrographs of whole liver sections stained with the IST-9 monoclonal antibody are shown. A: Two days after BDL. At the time of surgery rats were infused with a control peptide (human IgG). There is extensive staining for [EIIIA]Fn in a perisinusoidal distribution. B: Two days after BDL. At the time of surgery rats were infused with sTGFβR. Positive staining is sharply reduced (arrow), resembling that in control (sham-operated) liver, as shown previously. 26
Discussion
The initial stages of the epithelial repair response to injury involve the coordinated action of several cell types under the influence of an inflammatory milieu that includes cytokines, other small molecules (eg, oxyradicals, NO), and a neo-ECM. Our previous work documented that the neo-ECM contains a novel fibronectin isoform, [EIIIA]Fn, and that its production precedes detectable up-regulation of collagen and other ECM proteins by stellate cells. 26 Moreover, [EIIIA]Fn has an important regulatory role with respect to the activation of stellate cells. 26 This has led us to examine the regulation of [EIIIA]Fn synthesis, with the finding that TGFβ plays a central role.
This cytokine has been widely implicated in the wound repair response as a direct stimulus to ECM production by fibroblasts and myofibroblasts. 7,40-44 Although it acts similarly on stellate cells in culture, 15,19 its effects are modest relative to the change in ECM expression that occurs in liver injury in vivo. 20 This suggests either that it acts indirectly or that other factors are at play in stellate activation. The present results provide both cell-culture and in vivo evidence that much of the effect of TGFβ is indirect, through stimulation of [EIIIA]Fn production by sinusoidal endothelial cells. Previous studies of fibrosing injury to the kidney 45 or lung 46 have suggested that [EIIIA]Fn expression is modulated by TGFβ but have provided no information as to the cellular source(s) or target(s) of this fibronectin variant.
The source of TGFβ in liver injury is an interesting and important question. In the current paradigm, injury elicits inflammatory cells, which release TGFβ among other cytokines and may locally stimulate SEC and stellate cells. This is a model of paracrine signaling. Although platelets are considered the richest source of this cytokine, 26 mononuclear cells express it also. 47 When TGFβ is elevated in liver (for example, in liver infected with the hepatitis C virus 48 ), it may be due in part to the mononuclear cell infiltrate that is characteristic of this injury. However, it is reasonable to consider also the possibility that TGFβ in injury derives from resident liver cells. We have shown previously that TGFβ is produced constitutively by SEC and Kupffer cells. 11 Moreover, these same cells express type I and type II TGFβ receptors. 49 Thus, a potential pathway for autocrine signaling is present. As discussed elsewhere, 11 autocrine signaling may be a mechanism for locally restricting the effects of TGFβ within the intact tissue. The present results are consistent with autocrine effects of this cytokine on SEC production of [EIIIA]Fn, at least in the context of wound repair.
The findings do not preclude a role for other factors in the initiation of stellate activation. The direct effect of TGFβ on sinusoidal endothelial cells involves a threefold increase in [EIIIA]Fn at the mRNA level. Although this is significant, it is less than the approximately eightfold increase exhibited by SECs isolated directly from the injured liver. This suggests that TGFβ is responsible for only a portion of the change in [EIIIA]Fn expression. Consistent with this, the effect of inhibitory sTGFβR on the culture-induced increase, although significant, was incomplete. Though some TGFβ may be inaccessible to the soluble inhibitor, the findings more likely reflect the presence of other regulators of stellate activation. From the perspective of using this kind of inhibitor as anti-fibrotic therapy, incomplete inhibition is desirable: total inhibition of the repair process would raise significant safety concerns.
Information from culture is valuable in that it is direct. Its confirmation in vivo, however, is required, given that culture can induce rapid change in the cell phenotype. Moreover, in autocrine or paracrine pathways, the actual (physiological) concentration of an agonist at the receptor is unknowable. Thus, the relevance of culture experiments in which agonist is added at an arbitrary concentration is necessarily uncertain. Some in vivo models have the same shortcoming, eg, infusion or transgenic overexpression of TGFβ. In a transgenic model involving constitutive overproduction and secretion of TGFβ by hepatocytes under the albumin promoter, liver fibrosis was surprisingly limited, suggesting barriers to paracrine stimulation of stellate cells. Rather, the principal site of damage was the kidneys, reflecting the high circulating levels of active TGFβ. 43
Modulation of the endogenous cytokine avoids some of the issues inherent in transgenic animals. This was accomplished by introducing a competitive inhibitor of the type II receptor, consisting of human IgG into which a peptide representing the ligand-binding extracellular domain of the receptor was inserted. The chimeric IgG competes for endogenous TGFβ, preventing its binding to receptors on endothelial (and other) cells. As an IgG, it circulates with a half-life of several days. A single injection blocked the induction of [EIIIA]Fn by about 50%, which is entirely consistent with the culture data and confirms in vivo the role of TGFβ in the early phase of the hepatic injury response. Because the [EIIIA]Fn elaborated by SEC is capable of directly stimulating stellate cells, 15 the possibility exists that blockade of TGFβ at the level of the SEC would have significant downstream effects on fibrogenesis. In ongoing work, we have found that administration of sTGFβR reduces markers of stellate cell activation including collagen I expression. 50
The apparently transcriptional effect of TGFβ and the fact that this cytokine causes both increased expression of [EIIIA]Fn and an altered pattern of splicing also are of interest. Although the change in splicing in response to TGFβ appears small, it is statistically highly significant; moreover, it is similar to previously published data. 7 The mechanisms for alternate splicing and its relationship to transcription and posttranscriptional processing are not clearly defined. Recent studies demonstrated that the extent of EIIIA splicing is dependent on promoter architecture. 51 Our data support the postulate that regulation of transcription and splicing are intimately associated. A comparison of promoter architecture in SECs and hepatocytes, respectively, could prove informative, given the complete absence of a response to injury in hepatocytes. Such studies are anticipated.
Finally, it is worth noting that these data may be broadly applicable to epithelial fibrosis. Mechanisms of fibrogenesis in kidney appear to be very similar to those in liver, including an injury-related increase in [EIIIA]Fn expression. 45,52 By analogy with the liver, it seems likely that the source of [EIIIA]Fn in the kidney is the microvascular endothelium, although this remains to be shown. Moreover, the demonstrated antifibrogenic effect of administered anti-TGFβ in this tissue 53 may well reflect inhibition of [EIIIA]Fn expression. In short, the regulatory effect of TGFβ on [EIIIA]Fn production likely is of general relevance to epithelial injury.
Acknowledgments
We gratefully acknowledge the assistance of the Culture and Microscopy Core Facilities of the UCSF Liver Core Center.
Footnotes
Address reprint requests to D. Montgomery Bissell, Box 0358, Division of Gastroenterology, University of California, San Francisco, CA 94143. E-mail: dmbiss@itsa.ucsf.edu.
Supported by grants DK31198 and DK26743 from the U.S. National Institutes of Health and by RO3 TW00717 from the Fogarty International Center, NIH. J.G. was the recipient of a Neil-Hamilton-Fairley Fellowship (Australia) and a postdoctoral fellowship from the American Liver Foundation.
References
- 1.Sporn MB, Roberts AB: TGF-β: problems and prospects. Cell Reg 1990, 1:875-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assoian RK, Sporn MB: Type β transforming growth factor in human platelets: release during platelet degranulation and action on vascular smooth muscle cells. J Cell Biol 1986, 102:1217-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn MB, Fauci AS: Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med 1986, 163:1037-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsunawaki S, Sporn M, Ding A, Nathan C: Deactivation of macrophages by transforming growth factor-β. Nature 1988, 334:260-262 [DOI] [PubMed] [Google Scholar]
- 5.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga JH, Kehrl JH, Fauci AS: Transforming growth factor type β: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci USA 1986, 83:4167-4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ignotz RA, Massague J: Transforming growth factor-β stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem 1986, 261:4337-4345 [PubMed] [Google Scholar]
- 7.Balza E, Borsi L, Allemanni G, Zardi L: Transforming growth factor β regulates the levels of different fibronectin isoforms in normal human cultured fibroblasts. FEBS Lett 1988, 228:42-44 [DOI] [PubMed] [Google Scholar]
- 8.Edwards DR, Murphy G, Reynolds JJ, Whitham SE, Docherty AJ, Angel P, Heath JK: Transforming growth factor β modulates the expression of collagenase and metalloproteinase inhibitor. EMBO J 1987, 1904, 6:1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laiho M, Saksela O, Keski-Oja J: Transforming growth factor-β induction of type-1 plasminogen activator inhibitor. Pericellular deposition and sensitivity to exogenous urokinase. J Biol Chem 1987, 262:17467-17474 [PubMed] [Google Scholar]
- 10.Kim SJ, Jeang KT, Glick AB, Sporn MB, Roberts AB: Promoter sequences of the human transforming growth factor-β 1 gene responsive to transforming growth factor-β 1 autoinduction. J Biol Chem 1989, 264:7041-7045 [PubMed] [Google Scholar]
- 11.Bissell DM, Wang SS, Jarnagin WR, Roll FJ: Cell-specific expression of transforming growth factor-β in rat liver: evidence for autocrine regulation of hepatocyte proliferation. J Clin Invest 1995, 96:447-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minato Y, Hasumura Y, Takeuchi J: The role of fat-storing cells in Disse space fibrogenesis in alcoholic liver disease. Hepatology 1983, 3:559-566 [DOI] [PubMed] [Google Scholar]
- 13.Friedman SL, Roll FJ, Boyles J, Bissell DM: Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci USA 1985, 82:8681-8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czaja MJ, Weiner FR, Flanders KC, Giambrone M-A, Wind R, Biempica L, Zern MA: In vitro and in vivo association of transforming growth factor-β 1 with hepatic fibrosis. J Cell Biol 1989, 108:2477-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuoka M, Tsukamoto H: Stimulation of hepatic lipocyte collagen production by Kupffer cell-derived transforming growth factor β: implication for a pathogenetic role in alcoholic liver fibrogenesis. Hepatology 1990, 11:599-605 [DOI] [PubMed] [Google Scholar]
- 16.Nakatsukasa H, Nagy P, Evarts RP, Hsia C-C, Marsden E, Thorgeirsson SS: Cellular distribution of transforming growth factor-β1 and procollagen types I, III, and IV transcripts in carbon tetrachloride-induced rat liver fibrosis. J Clin Invest 1990, 85:1833-1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner FR, Giambrone M-A, Czaja MJ, Shah A, Annoni G, Takahashi S, Eghbali M, Zern MA: Ito-cell gene expression and collagen regulation. Hepatology 1990, 11:111-117 [DOI] [PubMed] [Google Scholar]
- 18.Friedman SL: The cellular basis of hepatic fibrosis: mechanisms and treatment strategies. N Engl J Med 1993, 328:1828-1835 [DOI] [PubMed] [Google Scholar]
- 19.Knittel T, Janneck T, Muller L, Fellmer P, Ramadori G: Transforming growth factor β1-regulated gene expression of Ito cells. Hepatology 1996, 24:352-360 [DOI] [PubMed] [Google Scholar]
- 20.Maher JJ, McGuire RF: Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J Clin Invest 1990, 86:1641-1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Racine-Samson L, Rockey DC, Bissell DM: The role of α1β1 integrin in wound contraction A quantitative analysis of liver myofibroblasts in vivo and in primary culture. J Biol Chem 1997, 272:30911-30917 [DOI] [PubMed] [Google Scholar]
- 22.Kornblihtt AR, Vibe-Pedersen K, Baralle FE: Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J 1984, 3:221-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarzbauer JE, Tamkun JW, Lemischka IR, Hynes RO: Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell 1983, 35:421-431 [DOI] [PubMed] [Google Scholar]
- 24.Odermatt E, Tamkun JW, Hynes RO: Repeating modular structure of the fibronectin gene: relationship to protein structure and subunit variation. Proc Natl Acad Sci USA 1985, 82:6571-6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French-Constant C: Alternative splicing of fibronectin: many different proteins but few different functions. Exp Cell Res 1995, 221:261-271 [DOI] [PubMed] [Google Scholar]
- 26.Jarnagin WR, Rockey DC, Koteliansky VE, Wang SS, Bissell DM: Expression of variant fibronectins in wound healing: cellular source and biological activity of the EIIIA segment in rat hepatic fibrogenesis. J Cell Biol 1994, 127:2037-2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odenthal M, Neubauer K, Meyer zum Buschenfelde KH, Ramadori G: Localization and mRNA steady-state level of cellular fibronectin in rat liver undergoing a CCl4-induced acute damage or fibrosis. Biochim Biophys Acta 1993, 1181:266–272 [DOI] [PubMed]
- 28.Borsi L, Carnemolla B, Castellani P, Rosellini C, Vecchio D, Allemanni G, Chang SE, Taylor-Papadimitriou J, Pande H, Zardi L: Monoclonal antibodies in the analysis of fibronectin isoforms generated by alternative splicing of mRNA precursors in normal and transformed human cells. J Cell Biol 1987, 104:595-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.di Clemente N, Wilson C, Faure E, Boussin L, Carmillo P, Tizard R, Picard JY, Vigier B, Josso N, Cate R: Cloning expression and alternative splicing of the receptor for anti-Mullerian hormone. Mol Endocrinol 1994, 8:1006-1020 [DOI] [PubMed] [Google Scholar]
- 30.Sanicola M, Hession C, Worley D, Carmillo P, Ehrenfels C, Walus L, Robinson S, Jaworski G, Wei H, Tizard R, Whitty A, Pepinsky RB, Cate R: Glial cell line-derived neurotrophic factor-dependent RET activation can be mediated by two different cell-surface accessory proteins. Proc Natl Acad Sci USA 1997, 94:6238-6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danielpour D, Dart LL, Flanders KC, Roberts AB, Sporn MB: Immunodetection and quantitation of the two forms of transforming growth factor-β (TGF-β1 and TGF-β2) secreted by cells in culture. J Cell Physiol 1989, 138:79-86 [DOI] [PubMed] [Google Scholar]
- 32.Bienkowski RS, Cowan MJ, MacDonald JA, Crystal RG: Degradation of newly synthesized collagen. J Biol Chem 1978, 253:4356-4363 [PubMed] [Google Scholar]
- 33.Friedman SL, Roll FJ: Isolation and culture of hepatic lipocytes Kupffer cells and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal Biochem 1987, 161:207-218 [DOI] [PubMed] [Google Scholar]
- 34.Irving MG, Roll FJ, Huang S, Bissell DM: Characterization and culture of sinusoidal endothelium from normal rat liver: lipoprotein uptake and collagen phenotype. Gastroenterology 1984, 87:1233-1247 [PubMed] [Google Scholar]
- 35.Bissell DM, Guzelian PS: Phenotypic stability of adult rat hepatocytes in primary monolayer culture. Ann NY Acad Sci 1980, 349:85-98 [DOI] [PubMed] [Google Scholar]
- 36.Rhoads DD, Dixit A, Roufa DJ: Primary structure of human ribosomal protein S14 and the gene that encodes it. Mol Cell Biol 1986, 6:2774-2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavanaugh WM, Harsh GR, Starksen NF, Rocco CM, Williams LT: Transcriptional regulation of the A and B chain genes of platelet-derived growth factor in microvascular endothelial cells. J Biol Chem 1988, 263:8470-8472 [PubMed] [Google Scholar]
- 38.Peters JH, Trevithick JE, Johnson P, Hynes RO: Expression of the alternatively spliced EIIIB segment of fibronectin. Cell Adh Commun 1995, 3:67-89 [DOI] [PubMed] [Google Scholar]
- 39.Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227:680-685 [DOI] [PubMed] [Google Scholar]
- 40.Cui W, Fowlis DJ, Cousins FM, Duffie E, Bryson S, Balmain A, Akhurst RJ: Concerted action of TGF-β 1 and its type II receptor in control of epidermal homeostasis in transgenic mice. Genes Dev 1995, 9:945-955 [DOI] [PubMed] [Google Scholar]
- 41.Lee M-S, Gu D, Feng L, Curriden S, Arnush M, Krahl T, Gurushanthaiah D, Wilson C, Loskutoff DL, Fox H, Sarvetnick N: Accumulation of extracellular matrix and developmental dysregulation in the pancreas by transgenic production of transforming growth factor-β1. Am J Pathol 1995, 147:42-52 [PMC free article] [PubMed] [Google Scholar]
- 42.O’Kane S, Ferguson MW: Transforming growth factor βs and wound healing. Int J Biochem Cell Biol 1997, 29:63-78 [DOI] [PubMed] [Google Scholar]
- 43.Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS: Hepatic expression of mature transforming growth factor β 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA 1995, 92:2572-2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milani S, Herbst H, Schuppan D, Stein H, Surrenti C: Transforming growth factors β1 and β2 are differentially expressed in fibrotic liver disease. Am J Pathol 1991, 139:1221-1229 [PMC free article] [PubMed] [Google Scholar]
- 45.Isaka Y, Brees DK, Ikegaya K, Kaneda Y, Imai E, Nobel NA, Border WA: Gene therapy by skeletal muscle expression of decorin prevents fibrotic disease in rat kidney. Nat Med 1996, 2:418-423 [DOI] [PubMed] [Google Scholar]
- 46.Giri SN, Hyde DM, Braun RK, Gaarde W, Harper JR, Pierschbacher MD: Antifibrotic effect of decorin in a bleomycin hamster model of lung fibrosis. Biochem Pharmacol 1997, 54:1205-1216 [DOI] [PubMed] [Google Scholar]
- 47.Brandes ME, Wakefield LM, Wahl SM: Modulation of monocyte type I transforming growth factor-β receptors by inflammatory stimuli. J Biol Chem 1991, 266:19697-19703 [PubMed] [Google Scholar]
- 48.Castilla A, Prieto J, Fausto N: Transforming growth factors β1 and α in chronic liver disease: effects of interferon alfa therapy. New Engl J Med 1991, 324:933-940 [DOI] [PubMed] [Google Scholar]
- 49.Roulot D, Sevcsik AM, Coste T, Strosberg AD, Marullo S: Role of transforming growth factor β type II receptor in hepatic fibrosis: studies of human chronic hepatitis C and experimental fibrosis in rats. Hepatology 1999, 29:1730-1738 [DOI] [PubMed] [Google Scholar]
- 50.George J, Roulot D, Koteliansky VE, Bissell DM: In vivo inhibition of rat stellate cell activation by soluble transforming growth factor β type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci USA 1999, 96:12719-12724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR: Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA 1997, 94:11456-11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gould VE, Martinez-Lacabe V, Virtanen I, Sahlin KM, Schwartz MM: Differential distribution of tenascin and cellular fibronectins in acute and chronic renal allograft rejection. Lab Invest 1992, 67:71-79 [PubMed] [Google Scholar]
- 53.Border WA, Okuda S, Languino LR, Sporn MB, Ruoslahti E: Suppression of experimental glomerulonephritis by antiserum against transforming growth factor β1. Nature 1990, 346:371-374 [DOI] [PubMed] [Google Scholar]