Abstract
FSAP (Factor VII-activating protease) can inhibit neointima formation and VSMC (vascular smooth-muscle cell) proliferation by cleavage of PDGF-BB (platelet-derived growth factor-BB). Negatively charged polyanions lead to autoactivation of the FSAP, but no information is available concerning the potential regulation of FSAP activity and its metabolism in the vessel wall. In the present study, we demonstrate that the enzymatic activity of FSAP can be inhibited by the serine protease inhibitor, PN-1 (protease nexin-1), that is found in the vasculature. This leads to the loss of the inhibitory effect of FSAP on PDGF-BB-mediated DNA synthesis and mitogen-activated protein kinase phosphorylation in VSMCs. The FSAP–PN-1 complexes bind to the LRP (low-density lipoprotein receptor-related protein) and are subsequently internalized. This binding is inhibited by receptor-associated protein, an antagonist of LRP, as well as heparin. While PDGFβR (PDGFβ receptor) is internalized by an LRP-dependent mechanism after stimulation of cells by PDGF-BB, the FSAP–PN-1 complex neither influenced PDGF-BB-mediated phosphorylation of PDGFβR nor its internalization via LRP. Hence, PN-1 inhibits the enzymatic activity of FSAP and neutralizes its effect on PDGF-BB-mediated VSMC proliferation. The FSAP–inhibitor complexes are internalized via LRP without influencing the PDGF-BB signal transduction pathway.
Keywords: atherosclerosis, Factor VII-activating protease (FSAP), low-density lipoprotein receptor-related protein (LRP), platelet-derived growth factor (PDGF), protease nexin-1 (PN-1), serine protease inhibitor (Serpin)
Abbreviations: BrdU, bromodeoxyuridine; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; FSAP, Factor VII-activating protease; FCS, fetal calf serum; LRP, low-density lipoprotein receptor-related protein; MAPK, mitogen-activated protein kinase; MEF, mouse embryo fibroblast; PAI, plasminogen activator inhibitor; PDGF-BB, platelet-derived growth factor-BB; PDGFβR, PDGFβ receptor; PN-1, protease nexin-1; RAP, receptor-associated protein; Serpin, serine protease inhibitor; TBS, Tris-buffered saline; VSMC, vascular smooth-muscle cell; uPA, urokinase-type plasminogen activator
INTRODUCTION
The FSAP (Factor VII-activating protease) is a serine protease containing three EGF (epidermal growth factor) domains, a kringle domain and a serine protease domain [1]. We have shown previously that FSAP is a potent inhibitor of PDGF-BB (platelet-derived growth factor-BB)-dependent proliferation and migration of VSMCs (vascular smooth-muscle cells) [2]. These are important processes in the development of atherosclerotic disease and restenosis after angioplasty or stenting. Furthermore, the level of active FSAP in the vessel wall is a key determinant of neointima formation in a mouse model of injury-induced stenosis [3].
FSAP is auto-activated in the presence of negatively charged polyanions such as heparin, hyaluronic acid or nucleic acid [4–7]. The Marburg I single nucleotide polymorphism leads to a single amino acid exchange (G534E) in the protease domain of the molecule and exhibits diminished proteolytic activity compared with wild-type FSAP [8]. The inhibitory effect of Marburg I FSAP on PDGF-BB-dependent cell proliferation and neointima formation was diminished compared with wild-type FSAP [3]. This indicates that the proteolytic activity of FSAP is a key determinant of the VSMC activation status in vivo. The enzymatic activity of FSAP is inhibited by plasma Serpins (serine protease inhibitors), such as C1-inhibitor, α2-anti-plasmin, anti-thrombin/heparin or aprotinin [9]. The Serpin PN-1 (protease nexin-1) is produced mainly by VSMCs [10], pericytes [11] and fibroblasts [12], and it is not known if it influences FSAP activity.
LRP (low-density lipoprotein receptor-related protein) is a member of a large receptor family responsible for the endocytosis of a variety of ligands [13]. LRP mediates the internalization of proteases, protease–inhibitor complexes and lipoproteins [14]. The ligands, once bound to LRP, are internalized and directed to endosomes, where they undergo degradation, whereas LRP is recycled to the cell membrane [15]. uPA (urokinase-type plasminogen activator) and tPA (tissue plasminogen activator) bind to LRP alone, as well as in complex with Serpins [13]. It is not known if FSAP can interact with LRP. It has been reported recently [14] that LRP is a signal transduction receptor that is involved in the regulation of the PDGF-BB–PDGFβR (PDGFβ receptor) signalling pathways [14].
In the present study, we demonstrate that the Serpin PN-1, found in the vasculature, can inhibit FSAP. FSAP–PN-1 complexes, but not FSAP alone, interact with the scavenger receptor LRP and are subsequently internalized. PN-1 inhibits FSAP-mediated cleavage and inhibition of PDGF-BB, but FSAP-PN-1 complexes do not influence the PDGF-BB–PDGFβR internalization by LRP or signal transduction processes.
MATERIALS AND METHODS
Cell culture
Wild-type and LRP−/− MEFs (mouse embryo fibroblasts) were cultivated in DMEM (Dulbecco's modified Eagle's medium; Invitrogen) with 10% (v/v) FCS (fetal calf serum; HyClone), 10 units/ml penicillin, 10 μg/ml streptomycin, 2 mM L-glutamine and 1 mM sodium pyruvate (Invitrogen). Mouse VSMCs were cultured in Iscov's modified medium (Invitrogen) with the same supplements as above. Cells were growth arrested in serum-free medium for 18 h prior to experiments.
Immunocytochemistry
Cells in 8-well chamber slides were incubated with the test substances for the indicated times, washed with serum-free DMEM containing 1% (w/v) BSA (Sigma) and then fixed with PBS containing 3.7% (w/v) paraformaldehyde. The cells were permeabilized with 0.2% Triton X-100 (Sigma) in TBS (Tris-buffered saline; 25 mM Tris/HCl, pH 7.4, 150mM NaCl and 2.7 mM KCl) and then blocked with 3% (w/v) BSA in TBS. After incubation with the following primary antibodies: anti-FSAP (mAb 1189 or mAb 677; ZLB Behring) and anti-LRP (rabbit polyclonal #2629, generously provided by Dr Dudley Strickland, American Red Cross, Rockville, MD, U.S.A.). Secondary antibodies labelled with FITC or Rhodamine Red-X (Dianova) were used for visualization. Finally, cells were washed and preserved in Vecta-shield (Linaris Wehrtheim–Bettingen), containing DAPI (4′,6-diamidino-2-phenylindole) to stain the nuclei. Slides were analysed using a Leica fluorescence microscope and the images were prepared with the Metamorph software (Visitron).
DNA-synthesis assays
VSMCs were stimulated in medium containing 0.2% FCS for 24 h with the test substances. For the last 4 h BrdU (bromodeoxyuridine) was added and the cells were processed with a BrdU detection kit (Roche Diagnostics) following the manufacturer's instructions.
MAPK (mitogen-activated protein kinase)-phosphorylation
Test samples were pre-incubated for 60 min in serum-free medium and the cells were stimulated for 5–15 min. Thereafter, SDS sample buffer [62.5 mM Tris/HCl, pH 6.8, 1.35 % (w/v) SDS, 10% (v/v) glyerol and 0.05% Bromphenol Blue] containing 1 mM orthovanadate was used to lyse the cells. Anti-(phospho-MAPK) antibody (p42/44MAPK) was from Cell Signaling Technology and the total anti-MAPK antibody was from Upstate (Millipore).
Western blot analysis
After SDS/PAGE (10% gels), proteins were transferred to PVDF membranes. Western blotting was performed using enhanced chemiluminescence plus reagent (GE Healthcare) as described by the manufacturer. For Western blot analysis of FSAP, a mixture of antibodies directed against the N-terminal end (mAb 1189) and against the C-terminal end (mAb 677) was used as described previously [2] in order to detect degraded forms of FSAP, if they were present.
FSAP activity assay
FSAP was isolated from human plasma as described previously [5]. To determine activation and enzymatic activity of FSAP, the hydrolysis of the chromogenic substrate H-D-isoleucyl-L-prolyl-L-arginine-p-nitroaniline dihydrochloride (S-2288) (Haemochrom) at a final concentration of 0.2 mM was measured over a time period of 60 min at 37 °C in a microplate reader (EL808, BioTek Instruments). Recombinant PN-1 was produced in Sf9 insect cells and purified in Blue Agarose and Sephacryl S-200 as described previously [16]. Concentration of active PN-1 was determined by inhibition of thrombin and determination of the residual thrombin activity as described [16]. PN-1 and aprotinin were pre-incubated with FSAP for 30 min prior to the enzymatic activity assay. The assays were performed with a final FSAP concentration of 1 μg/ml in TBS containing 10 μg/ml heparin.
FSAP binding to LRP
LRP was isolated from human placenta as described previously [17]. LRP and the respective control buffer (140 mM NaCl, 10 mM Hepes, pH 7.4, 2 mM CaCl2, 1 mM MgCl2 and 0.6% CHAPS) containing 2 μg/ml BSA were immobilized in 50 mM Tris/HCl (pH 7.4) containing 150 mM NaCl in a Maxisorp plate (Nunc). The samples were then blocked with control buffer containing 3% (w/v) BSA. Test substances or mixtures were pre-incubated for 30 min prior to their application to the coated wells for 60 min. FSAP was detected by a monoclonal anti-FSAP antibody (mAb 677), followed by detection with a horseradish-peroxidase-conjugated secondary antibody (Dako). The binding to wells coated with control buffer containing 2 μg/ml BSA was used as a blank in all experiments and was subtracted to obtain specific binding.
Expression and purification of RAP (receptor-associated protein)
The constructed plasmid containing human RAP cDNA with a His-tag and an ampicillin-resistance gene was a kind gift from Dr T. Willnow (Molecular Cardiovascular Reseach, Max Delbrück Centre, Berlin, Germany). Extracts were purified over a ProBond™ column (Promega) and fractions containing the protein were dialysed against TBS (pH 8.0) containing 5 mM GSH and 1 mM GSSG for refolding and finally dialysed against TBS (pH 8.0.)
RESULTS
Inhibition of FSAP by PN-1 and the binding of the FSAP–PN-1 complex to LRP
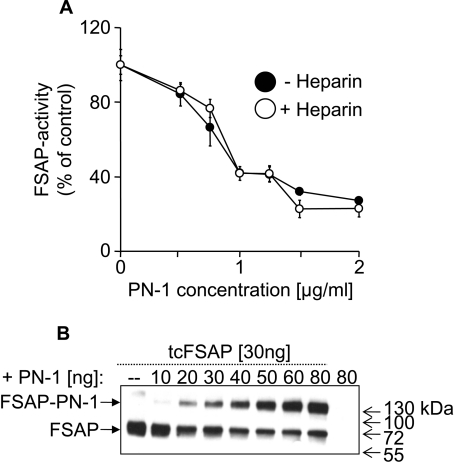
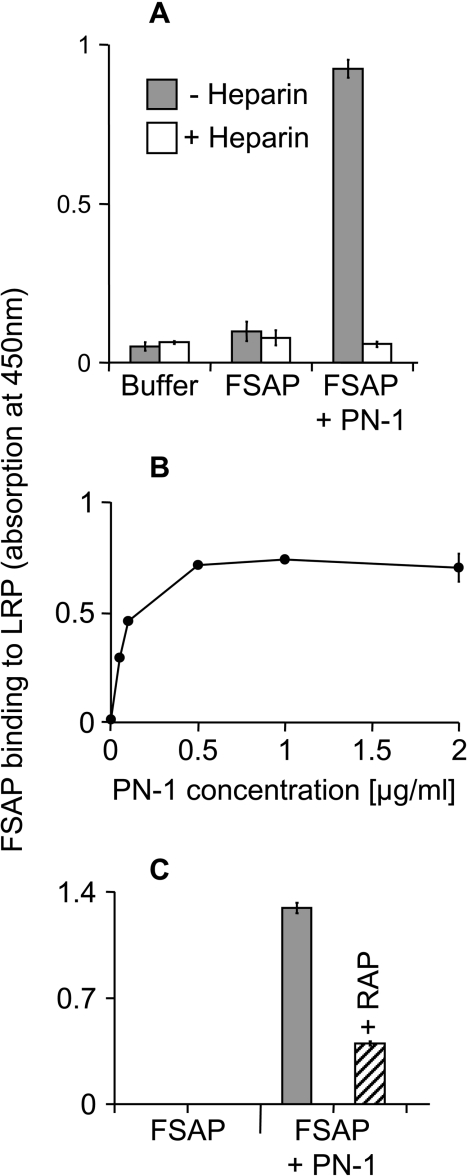
PN-1 is a Serpin produced locally in the vessel wall and it neutralized the enzymatic activity of FSAP, as measured by the direct chromogenic substrate S-2288 (Figure 1A). The presence of heparin did not influence the FSAP inhibition characteristics (Figure 1A). The formation of complexes between FSAP and PN-1 could be observed in a dose-dependent manner by Western blotting (Figure 1B). Several protease–protease-inhibitor complexes, such as uPA and tPA in complex with PN-1 or PAI (plasminogen activator inhibitor)-1, are known to bind to LRP, hence the binding of FSAP–inhibitor complexes to the immobilized LRP was investigated. The FSAP–PN-1 complex showed a strong specific binding, whereas FSAP by itself, or in complex with aprotinin (results not shown), exhibited no binding at all to LRP (Figure 2A). The binding of the FSAP–PN-1 complex was inhibited by heparin (Figure 2A). Concentration-dependent analysis confirmed that FSAP–PN-1 complex binding showed saturation (Figure 2B). The binding of FSAP–PN-1 complex could be inhibited by RAP (Figure 2C), a 39 kDa LRP antagonist, known to inhibit ligand binding to LRP [18]. Maximal inhibition (approx. 70%) was observed with 10 μg/ml RAP.
Figure 1. Inhibition of FSAP by PN-1 and complex formation.
(A) FSAP (1 μg/ml) was pre-incubated with 0.5–2.0 μg/ml PN-1 in the absence or presence of 10 μg/ml heparin for 30 min. FSAP activity was measured with the conversion of 0.2 mM specific chromogenic substrate S-2288. The results are shown as means±S.D. (n=3). (B) FSAP (30 ng/lane) was pre-incubated with 10–80 ng of PN-1, in the presence of 10 μg/ml heparin. Western blot analysis was performed using two monoclonal antibodies against FSAP. These experiments were repeated three times with similar results. Molecular-mass markers are shown to the right-hand side of the Figure in kDa.
Figure 2. FSAP–PN-1 complex binding to LRP.
(A) FSAP (1 μg/ml) was pre-incubated for 30 min with 2 μg/ml PN-1 to allow complex formation, and the binding to immobilized LRP (1 μg/ml) was measured in absence (grey bars) or presence (open bars) of 10 μg/ml heparin using a monoclonal antibody against FSAP. The results are shown as means±S.D. (n=3). (B) FSAP (1 μg/ml) was pre-incubated with 0.05–2 μg/ml PN-1 for 30min to allow complex formation. Binding of the FSAP–inhibitor complexes to immobilized LRP (1 μg/ml) and BSA was measured using a specific monoclonal antibody against FSAP. The results are shown as means±S.D. (n=3). (C) FSAP (1 μg/ml) was pre-incubated with 2 μg/ml PN-1 for 30 min to allow complex formation. Binding of the FSAP–inhibitor complexes to immobilized LRP (1 μg/ml), without (grey bars) or with (striped columns) 10 μg/ml RAP, and control buffer containing BSA (2 μg/ml) was measured using a specific monoclonal antibody against FSAP. The results are shown as means±S.D. (n=3). These experiments were repeated three times with similar results.
Binding of FSAP–PN-1 complex to LRP on cells
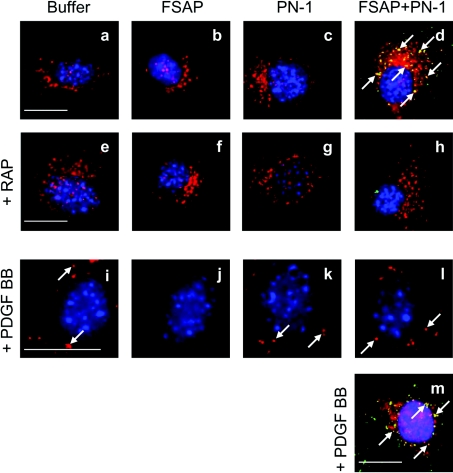
The binding of FSAP alone and FSAP–PN-1 complex to cells was analysed by immunofluorescence microscopy. The application of FSAP alone to VSMCs did not lead to an accumulation of cellular FSAP (Figure 3b), whereas FSAP–PN-1 complexes were internalized by VSMC (Figure 3d). Co-staining of FSAP (Figure 3d; green) and LRP (Figure 3d; red) in VSMCs shows the co-localization of the ligand and the receptor in the intracellular compartment. Pre-incubation of VSMCs with RAP, prior to application of FSAP–PN-1 complex, completely inhibited the internalization of the complex and the co-localization of FSAP and LRP (Figure 3h). Time course analysis showed that FSAP–PN-1 complexes were internalized within minutes (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/404/bj4040191add.htm) and that heparin inhibited this internalization (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/404/bj4040191add.htm). FSAP–PN-1 complex, but not FSAP alone, was internalized by wild-type MEFs, but not by LRP−/− MEFs (see Supplementary Figure 3 at http://www.BiochemJ.org/bj/404/bj4040191add.htm). FSAP–PN-1 complex binds to LRP and then undergoes internalization via the scavenger receptor and is directed to lysosomes.
Figure 3. Binding of FSAP–PN-1 complex to LRP and the influence of this complex on PDGFβR distribution in VSMCs.
Buffer control (a and e), FSAP (1 μg/ml) alone (b and f) and PN-1 (2μg/ml) alone (c and g) or FSAP and PN-1 together (d and h) were pre-incubated for 30 min to allow complex formation and added to VSMC without (a–d) or with RAP (10 μg/ml) (e–h) for 45 min. FSAP was detected by FITC-labelled secondary antibody (green), LRP by Rhodamine Red-X labelled secondary antibody (red) and nuclei by DAPI staining (blue) (a–h). The yellow colour indicates co-localization of FSAP and LRP and is highlighted by white arrows. PDGF-BB (20 ng/ml) was pre-incubated with buffer (i), FSAP (1 μg/ml) (j), PN-1 (2 μg/ml) (k) or FSAP–PN-1 complex (l) for 60 min at 37 °C. VSMCs were incubated with these mixtures for 30 min on 37 °C. PDGFβR was detected by Rhodamine Red-X labelled secondary antibody (red) and nuclei were stained by DAPI (blue) (i–l). The white arrows highlight examples of intracellular accumulated PDGFβR. To examine the co-localization of FSAP and PDGFβR cells were prepared as in (l), and FSAP was detected by FITC-labelled secondary antibody (green), PDGFβR with Rhodamine Red-X labelled secondary antibody (red) and nuclei were stained by DAPI (blue). Yellow colour and white arrows highlight examples of intracellular co-localization of FSAP–PN-1 complex with PDGFβR (m). Scale bar=20 μm. This staining pattern was observed in 80% of cells on a slide and this experiment was repeated twice.
Effect of FSAP–PN-1 complex on PDGF-BB-induced internalization of PDGFβR
Since FSAP is a strong inhibitor of the PDGF-BB signalling pathway and LRP is also involved in modulating the same pathway, we investigated the interactions between FSAP, LRP and PDGFβR. In unstimulated cells, PDGFβR is diffusely distributed over the whole cell membrane, and upon stimulation with PDGF-BB internalization of PDGFβR into lysosomes is observed (Figure 3i; red), which has been described previously [19,20]. This PDGF-BB-induced internalization of PDGFβR was not influenced by the presence of the FSAP–PN-1 complex (Figure 3l; red). Hence, the FSAP–PN-1 complex is internalized via LRP, but this does not influence the internalization of PDGF-BB–PDGFβR by LRP, even though there was co-localization of the FSAP–PN-1 complex (Figure 3m; green) with internalized PDGFβR (Figure 3m; red). No PDGFβR staining was observed in the presence of PDGF-BB and FSAP, owing to cleavage and inactivation of PDGF-BB, so that the diffuse distribution of PDGFβR remained unchanged (Figure 3j). Although FSAP–PN-1 complexes, as well as the PDGF-BB–PDGFβR complexes, bind to LRP, their internalization was completely independent of each other.
Effect of the FSAP–PN-1 complex on PDGF-BB-dependent cell activation
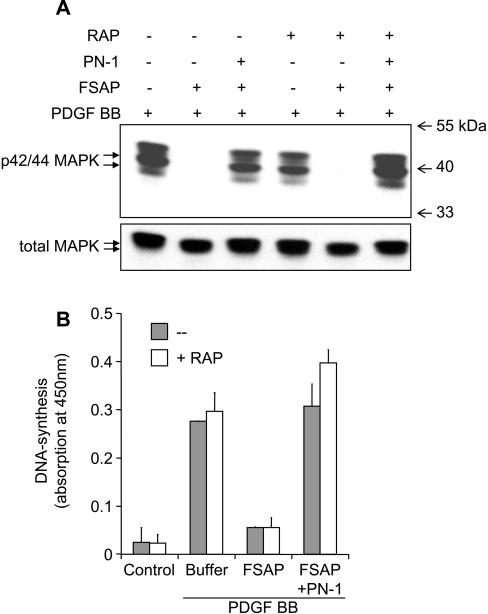
Finally, we have investigated if the FSAP–PN-1 complex can influence PDGF-BB-mediated signal transduction. Pre-incubation of PDGF-BB with FSAP diminished p42/44MAPK phosphorylation in VSMCs (Figure 4A), because of cleavage and inactivation of PDGF-BB (see Supplementary Figure 4 at http://www.BiochemJ.org/bj/404/bj4040191add.htm). Pre-incubation with FSAP–PN-1 did not alter PDGF-BB-mediated phosphorylation of p42/44MAPK in VSMCs (Figure 4A). Inhibition of the proteolytic activity of FSAP by PN-1 led to neutralization of its ability to inhibit PDGF-BB-mediated DNA synthesis (Figure 4B). Neither RAP nor the FSAP–PN-1 complex had any significant influence on PDGF-BB-mediated DNA synthesis, indicating that LRP is not regulatory under these conditions in these cells.
Figure 4. Influence of LRP on the inhibitory effect of FSAP on PDGF-BB-dependent VSMC activation.
(A) In serum-free medium containing 10 μg/ml heparin, VSMCs were stimulated with PDGF-BB (20 ng/ml), PDGF-BB pre-incubated with FSAP (1 μg/ml) or PDGF-BB pre-incubated with FSAP–PN-1 complex in the absence or presence of RAP (10 μg/ml) for 15 min. Western blot analysis was performed using a monoclonal antibody against phospho-42/44MAPK p42/44 MAPK). As a loading control, a polyclonal antibody against total MAPK was used on the stripped membrane. Molecular-mass markers are shown to the right-hand side of the Figure in kDa. (B) As in the above experiment, VSMCs were stimulated in the absence (grey bars) or presence (open bars) 10 μg/ml RAP. DNA synthesis was measured using a kit to measure BrdU incorporation into newly synthesized DNA. The results are shown as means±S.D. (n=3), and similar results were obtained in three independent experiments.
Generally, more PDGFβR was present in LRP−/− MEFs than in wild-type MEFs, and higher levels of tyrosine phosphorylation were observed in LRP−/− MEFs than in WT MEFs after PDGF-BB stimulation (see Supplementary Figure 5 at http://www.BiochemJ.org/bj/404/bj4040191add.htm). These results confirm the known role of LRP in regulating PDGF-BB activity. No effect of the FSAP–PN-1 complex was observed on this pattern of expression or phosphorylation of PDGFβR (see Supplementary Figure 5).
DISCUSSION
Inhibition of PDGF-BB-mediated VSMC proliferation by FSAP was observed in vitro [2] and in vivo in a mouse model of neointima formation [3]. The naturally occurring Marburg I form of FSAP has diminished proteolytic activity and is a weaker inhibitor of VSMC proliferation [3]. This could explain why this form of FSAP is a strong risk factor for cardiovascular diseases in general [21,22]. Hence, the proteolytic activity of FSAP is a key determinant in this scheme of atherothrombosis. PN-1 is a prominent Serpin in the vessel wall produced by VSMCs and it is capable of inhibiting the enzymatic activity of FSAP. There are numerous studies indicating that PN-1 is up-regulated in hypertension [10] and atherosclerosis [23]. Hence, through inhibition of FSAP, PN-1 can regulate the activity of FSAP towards PDGF-BB.
FSAP is homologous to factors from the coagulation and the fibrinolysis system, and LRP plays a critical role in the regulation of their activity and metabolism. Through internalization of the active Factor IXa [24] and Factor VIIIa [25], LRP also indirectly regulates the activity of the coagulation system. Complexes of PAI-1 with proteases are internalized by LRP and this process is heparin-dependent [26]. The binding of the PN-1—thrombin complex, but not the PN-1–uPA complex, was also heparin-dependent [27]. With some exceptions, such as PN-1 [28] or Factor VIII [29], the binding of individual factors to LRP is weak.
The FSAP–PN-1 complexes bound to LRP and were internalized by cells as are other protease–inhibitor complexes [30]. Internalization was not observed in LRP−/− MEFs or in the presence of the specific LRP inhibitor, RAP. Neither FSAP alone nor the FSAP–aprotinin complex binds to LRP, indicating that the recognition by LRP is only possible when FSAP is in complex with PN-1. Hence, PN-1 contributes binding elements necessary for the binding of the FSAP–PN-1 complex to LRP. The binding of the FSAP–PN-1 complex was inhibited by heparin, the probable reason for this is that PN-1 is a strong heparin-binding protein [31] analogous to FSAP. Our preliminary data show that another Serpin, PAI-1, also inhibits FSAP (results not shown) and the FSAP–PAI-1 complex binds to LRP and is internalized.
LRP also plays an important role in regulating the signal transduction pathway at many levels. LRP seems to bind to PDGF-BB directly [32], but not bFGF (basic fibroblast growth factor), thereby increasing the local concentration of PDGF-BB. LRP is phosphorylated upon stimulation of cells with PDGF-BB and there is a cross-talk between PDGFβR and LRP. Phosphorylation of LRP generates an Shc (Src homology 2 domain)-docking site propagating the signal [32–34]. In its unphosphorylated state, LRP functions as an endocytic receptor and inhibits PDGF-BB-mediated signal transduction. In the absence of LRP, PDGF-BB-dependent signal transduction is increased in vivo and leads to atherosclerosis [35,36].
Since FSAP–inhibitor complexes also bind to LRP, we have hypothesized that PDGF-BB signal transduction might be altered, since this is partially dependent on LRP. The binding of the FSAP–PN-1 complex to LRP does not influence the function of LRP with respect to the PDGF-BB–PDGFβR axis. The possible reasons are that the FSAP–PN-1 complex binds to a different region of LRP than the PDGF-BB–PDGFβR complex and no competition ensues. Alternatively, the LRP that is involved in internalization of the FSAP–inhibitor complex is localized in a different cellular compartment compared with LRP involved in PDGF-BB–PDGFβR signal transduction. Another reason could be that the LRP system is a high capacity system that is not easily saturated, which results in the lack of competition observed between the different LRP ligands.
We have shown recently [3] that a naturally occurring variant of FSAP, the Marburg I variant, has reduced proteolytic activity and cannot inhibit neointima formation in vivo in a mouse injury model. Hence, the inhibition of FSAP activity by a locally produced Serpin, such as PN-1, is a probable regulatory element in this pathway. The FSAP–inhibitor complexes are internalized via LRP without influencing the signal-transduction-related functions of LRP with respect to PDGF-BB and PDGFβR. In human atherosclerotic plaques, high intracellular FSAP staining has been observed in VSMCs and macrophages, indicating that this mechanism is likely to function in vivo [2].
Online data
Acknowledgments
The assistance of Karin Hersemeyer (Institute for Biochemistry, Justus-Liebig-University Giessen, Germany) with immunofluoresence microscopy is greatly appreciated. This study was financed from a grant from the Deutsche Forschungsgemeinschaft to S.M.K. (SFB 547:C14).
References
- 1.Choi-Miura N. H., Tobe T., Sumiya J., Nakano Y., Sano Y., Mazda T., Tomita M. Purification and characterization of a novel hyaluronan-binding protein (PHBP) from human plasma: it has three EGF, a kringle and a serine protease domain, similar to hepatocyte growth factor activator. J. Biochem. (Tokyo). 1996;119:1157–1165. doi: 10.1093/oxfordjournals.jbchem.a021362. [DOI] [PubMed] [Google Scholar]
- 2.Kannemeier C., Al-Fakhri N., Preissner K. T., Kanse S. M. Factor VII-activating protease (FSAP) inhibits growth factor-mediated cell proliferation and migration of vascular smooth muscle cells. Faseb J. 2004;18:728–730. doi: 10.1096/fj.03-0898fje. [DOI] [PubMed] [Google Scholar]
- 3.Sedding D., Daniel J. M., Muhl L., Hersemeyer K., Brunsch H., Kemkes-Matthes B., Braun-Dullaeus R. C., Tillmanns H., Weimer T., Preissner K. T., Kanse S. M. The G534E polymorphism of the gene encoding the factor VII-activating protease is associated with cardiovascular risk due to increased neointima formation. J. Exp. Med. 2006;203:2801–2807. doi: 10.1084/jem.20052546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi-Miura N. H., Takahashi K., Yoda M., Saito K., Mazda T., Tomita M. Proteolytic activation and inactivation of the serine protease activity of plasma hyaluronan binding protein. Biol. Pharm. Bull. 2001;24:448–452. doi: 10.1248/bpb.24.448. [DOI] [PubMed] [Google Scholar]
- 5.Kannemeier C., Feussner A., Stohr H. A., Weisse J., Preissner K. T., Romisch J. Factor VII and single-chain plasminogen activator-activating protease: activation and autoactivation of the proenzyme. Eur. J. Biochem. 2001;268:3789–3796. doi: 10.1046/j.1432-1327.2001.02285.x. [DOI] [PubMed] [Google Scholar]
- 6.Nakazawa F., Kannemeier C., Shibamiya A., Song Y., Tzima E., Schubert U., Koyama T., Niepmann M., Trusheim H., Engelmann B., Preissner K. T. Extracellular RNA is a natural cofactor for the (auto-)activation of Factor VII-activating protease (FSAP) Biochem. J. 2005;385:831–838. doi: 10.1042/BJ20041021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altincicek B., Shibamiya A., Trusheim H., Tzima E., Niepmann M., Linder D., Preissner K. T., Kanse S. M. A positively charged cluster in the epidermal growth factor-like domain of Factor VII-activating protease (FSAP) is essential for polyanion binding. Biochem. J. 2006;394:687–692. doi: 10.1042/BJ20051563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roemisch J., Feussner A., Nerlich C., Stoehr H. A., Weimer T. The frequent Marburg I polymorphism impairs the pro-urokinase activating potency of the factor VII activating protease (FSAP) Blood Coagul. Fibrinolysis. 2002;13:433–441. doi: 10.1097/00001721-200207000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Romisch J. Factor VII activating protease (FSAP): a novel protease in hemostasis. Biol. Chem. 2002;383:1119–1124. doi: 10.1515/BC.2002.121. [DOI] [PubMed] [Google Scholar]
- 10.Bouton M. C., Richard B., Rossignol P., Philippe M., Guillin M. C., Michel J. B., Jandrot-Perrus M. The serpin protease-nexin 1 is present in rat aortic smooth muscle cells and is upregulated in L-NAME hypertensive rats. Arterioscler. Thromb. Vasc. Biol. 2003;23:142–147. doi: 10.1161/01.atv.0000047867.98019.2d. [DOI] [PubMed] [Google Scholar]
- 11.Kim J. A., Tran N. D., Li Z., Yang F., Zhou W., Fisher M. J. Brain endothelial hemostasis regulation by pericytes. J. Cereb. Blood Flow Metab. 2006;26:209–217. doi: 10.1038/sj.jcbfm.9600181. [DOI] [PubMed] [Google Scholar]
- 12.Baker J. B., Low D. A., Simmer R. L., Cunningham D. D. Protease-nexin: a cellular component that links thrombin and plasminogen activator and mediates their binding to cells. Cell. 1980;21:37–45. doi: 10.1016/0092-8674(80)90112-9. [DOI] [PubMed] [Google Scholar]
- 13.Willnow T. E., Nykjaer A., Herz J. Lipoprotein receptors: new roles for ancient proteins. Nat. Cell. Biol. 1999;1:E157–E162. doi: 10.1038/14109. [DOI] [PubMed] [Google Scholar]
- 14.Lillis A. P., Mikhailenko I., Strickland D. K. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. J. Thromb. Haemost. 2005;3:1884–1893. doi: 10.1111/j.1538-7836.2005.01371.x. [DOI] [PubMed] [Google Scholar]
- 15.Nykjaer A., Conese M., Christensen E. I., Olson D., Cremona O., Gliemann J., Blasi F. Recycling of the urokinase receptor upon internalization of the uPA–serpin complexes. EMBO J. 1997;16:2610–2620. doi: 10.1093/emboj/16.10.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone S. R., Brown-Luedi M. L., Rovelli G., Guidolin A., McGlynn E., Monard D. Localization of the heparin-binding site of glia-derived nexin/protease nexin-1 by site-directed mutagenesis. Biochemistry. 1994;33:7731–7735. doi: 10.1021/bi00190a028. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen M. S., Nykjaer A., Warshawsky I., Schwartz A. L., Gliemann J. Analysis of ligand binding to the α2-macroglobulin receptor/low density lipoprotein receptor-related protein. Evidence that lipoprotein lipase and the carboxyl-terminal domain of the receptor-associated protein bind to the same site. J. Biol. Chem. 1995;270:23713–23719. doi: 10.1074/jbc.270.40.23713. [DOI] [PubMed] [Google Scholar]
- 18.Willnow T. E., Rohlmann A., Horton J., Otani H., Braun J. R., Hammer R. E., Herz J. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. EMBO J. 1996;15:2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 19.Sorkin A., Westermark B., Heldin C. H., Claesson-Welsh L. Effect of receptor kinase inactivation on the rate of internalization and degradation of PDGF and the PDGFβ-receptor. J. Cell. Biol. 1991;112:469–478. doi: 10.1083/jcb.112.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tingstrom A., Reuterdahl C., Lindahl P., Heldin C. H., Rubin K. Expression of platelet-derived growth factor-β receptors on human fibroblasts: regulation by recombinant platelet-derived growth factor-BB, IL-1, and tumor necrosis factor-α. J. Immunol. 1992;148:546–554. [PubMed] [Google Scholar]
- 21.Willeit J., Kiechl S., Weimer T., Mair A., Santer P., Wiedermann C. J., Roemisch J. Marburg I polymorphism of Factor VII–activating protease: a prominent risk predictor of carotid stenosis. Circulation. 2003;107:667–670. doi: 10.1161/01.cir.0000055189.18831.b1. [DOI] [PubMed] [Google Scholar]
- 22.Ireland H., Miller G. J., Webb K. E., Cooper J. A., Humphries S. E. The Factor VII activating protease G511E (Marburg) variant and cardiovascular risk. Thromb. Haemost. 2004;92:986–992. doi: 10.1160/TH04-05-0275. [DOI] [PubMed] [Google Scholar]
- 23.Kanse S. M., Chavakis T., Al-Fakhri N., Hersemeyer K., Monard D., Preissner K. T. Reciprocal regulation of urokinase receptor (CD87)-mediated cell adhesion by plasminogen activator inhibitor-1 and protease nexin-1. J. Cell. Sci. 2004;117:477–485. doi: 10.1242/jcs.00861. [DOI] [PubMed] [Google Scholar]
- 24.Rohlena J., Kolkman J. A., Boertjes R. C., Mertens K., Lenting P. J. Residues Phe342-Asn346 of activated coagulation factor IX contribute to the interaction with low density lipoprotein receptor-related protein. J. Biol. Chem. 2003;278:9394–9401. doi: 10.1074/jbc.M209097200. [DOI] [PubMed] [Google Scholar]
- 25.Bovenschen N., van Stempvoort G., Voorberg J., Mertens K., Meijer A. B. Proteolytic cleavage of Factor VIII heavy chain is required to expose the binding-site for low-density lipoprotein receptor-related protein within the A2 domain. J. Thromb. Haemost. 2006;4:1487–1493. doi: 10.1111/j.1538-7836.2006.01965.x. [DOI] [PubMed] [Google Scholar]
- 26.Stefansson S., Muhammad S., Cheng X. F., Battey F. D., Strickland D. K., Lawrence D. A. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low-density lipoprotein receptor-related protein. J. Biol. Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- 27.Crisp R. J., Knauer D. J., Knauer M. F. Roles of the heparin and low-density lipid receptor-related protein-binding sites of protease nexin 1 (PN1) in urokinase-PN1 complex catabolism: the PN1 heparin-binding site mediates complex retention and degradation but not cell surface binding or internalization. J. Biol. Chem. 2000;275:19628–19637. doi: 10.1074/jbc.M909172199. [DOI] [PubMed] [Google Scholar]
- 28.Li X., Herz J., Monard D. Activation of ERK signaling upon alternative protease nexin-1 internalization mediated by syndecan-1. J. Cell. Biochem. 2006;99:936–951. doi: 10.1002/jcb.20881. [DOI] [PubMed] [Google Scholar]
- 29.Sarafanov A. G., Ananyeva N. M., Shima M., Saenko E. L. Cell surface heparan sulfate proteoglycans participate in Factor VIII catabolism mediated by low density lipoprotein receptor-related protein. J. Biol. Chem. 2001;276:11970–11979. doi: 10.1074/jbc.M008046200. [DOI] [PubMed] [Google Scholar]
- 30.Strickland D. K., Medved L. Low-density lipoprotein receptor-related protein (LRP)-mediated clearance of activated blood coagulation co-factors and proteases: clearance mechanism or regulation? J. Thromb. Haemost. 2006;4:1484–1486. doi: 10.1111/j.1538-7836.2006.01987.x. [DOI] [PubMed] [Google Scholar]
- 31.Rovelli G., Stone S. R., Preissner K. T., Monard D. Specific interaction of vitronectin with the cell-secreted protease inhibitor glia-derived nexin and its thrombin complex. Eur. J. Biochem. 1990;192:797–803. doi: 10.1111/j.1432-1033.1990.tb19293.x. [DOI] [PubMed] [Google Scholar]
- 32.Loukinova E., Ranganathan S., Kuznetsov S., Gorlatova N., Migliorini M. M., Loukinov D., Ulery P. G., Mikhailenko I., Lawrence D. A., Strickland D. K. Platelet-derived growth factor (PDGF)-induced tyrosine phosphorylation of the low density lipoprotein receptor-related protein (LRP): evidence for integrated co-receptor function between LRP and the PDGF. J. Biol. Chem. 2002;277:15499–15506. doi: 10.1074/jbc.M200427200. [DOI] [PubMed] [Google Scholar]
- 33.Boucher P., Liu P., Gotthardt M., Hiesberger T., Anderson R. G., Herz J. Platelet-derived growth factor mediates tyrosine phosphorylation of the cytoplasmic domain of the low-density lipoprotein receptor-related protein in caveolae. J. Biol. Chem. 2002;277:15507–15513. doi: 10.1074/jbc.M200428200. [DOI] [PubMed] [Google Scholar]
- 34.Newton C. S., Loukinova E., Mikhailenko I., Ranganathan S., Gao Y., Haudenschild C., Strickland D. K. Platelet-derived growth factor receptor-β (PDGFR-β) activation promotes its association with the low density lipoprotein receptor-related protein (LRP): evidence for co-receptor function. J. Biol. Chem. 2005;280:27872–27878. doi: 10.1074/jbc.M505410200. [DOI] [PubMed] [Google Scholar]
- 35.Boucher P., Gotthardt M., Li W. P., Anderson R. G., Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 36.Boucher P., Gotthardt M. LRP and PDGF signaling: a pathway to atherosclerosis. Trends Cardiovasc. Med. 2004;14:55–60. doi: 10.1016/j.tcm.2003.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.