Abstract
The molecular chaperone Hsp (heat-shock protein) 90 is critical for the activity of diverse cellular client proteins. In a current model, client proteins are transferred from Hsp70 to Hsp90 in a process mediated by the co-chaperone Sti1/Hop, which may simultaneously interact with Hsp70 and Hsp90 via separate TPR (tetratricopeptide repeat) domains, but the mechanism and in vivo importance of this function is unclear. In the present study, we used truncated forms of Sti1 to determine the minimal regions required for the Hsp70 and Hsp90 interaction, as well as Sti1 dimerization. We found that both TPR1 and TPR2B contribute to the Hsp70 interaction in vivo and that mutations in both TPR1 and TPR2B were required to disrupt the in vitro interaction of Sti1 with the C-terminus of the Hsp70 Ssa1. The TPR2A domain was required for the Hsp90 interaction in vivo, but the isolated TPR2A domain was not sufficient for the Hsp90 interaction unless combined with the TPR2B domain. However, isolated TPR2A was both necessary and sufficient for purified Sti1 to migrate as a dimer in solution. The DP2 domain, which is essential for in vivo function, was dispensable for the Hsp70 and Hsp90 interaction, as well as Sti1 dimerization. As evidence for the role of Sti1 in mediating the interaction between Hsp70 and Hsp90 in vivo, we identified Sti1 mutants that result in reduced recovery of Hsp70 in Hsp90 complexes. We also identified two Hsp90 mutants that exhibit a reduced Hsp70 interaction, which may help clarify the mechanism of client transfer between the two molecular chaperones.
Keywords: heat-shock protein, Hop, molecular chaperone, tetratricopeptide repeat domain
Abbreviations: DP, aspartic acid–proline repeat; 5-FOA, 5-fluororotic acid; GR, glucocorticoid receptor; GST, glutathione S-transferase; Hsc, heat-shock cognate protein; Hsp, heat-shock protein; TPR, tetratricopeptide repeat; WT, wild-type
INTRODUCTION
The abundant, essential molecular chaperone Hsp (heat-shock protein) 90 is required for the maturation and activation of diverse client proteins, including steroid hormone receptors and some oncogenic kinases. Since Hsp90 interacts with a number of oncogenic signalling proteins, including Akt, Raf-1, Bcr-Abl, mutant p53 and HER-2/Erb2, Hsp90 is a promising anti-cancer target and the Hsp90 inhibitor 17-AAG [17-(allylamino)-17-demethoxygeldanamycin] is currently in Phase II clinical trials [1]. In Saccharomyces cerevisiae the function of Hsp90 appears to be restricted to a subset of 200–400 cellular client proteins that have genetic or physical interactions with Hsp90 [2,3]. Little is known about what dictates the selectivity of these interactions, and the mechanism by which Hsp90 binds client proteins and mediates their folding and activation is also largely unknown [4–6].
Hsp90 function is dependent on multiple partner co-chaperone proteins that interact in an ordered, ATP-dependent pathway [4–6]. According to current models, Hsp90 client proteins first interact with the molecular chaperones Hsp70 and Hsp40 [7]. Subsequent interaction of Sti1/Hop allows formation of a complex between the client protein, Hsp70, Sti1 and Hsp90. In a poorly understood process, the client protein is transferred from Hsp70 to Hsp90, resulting in Hsp70/Hsp40 release [6,8,9]. Subsequent ATP binding by Hsp90 induces significant conformational changes, which result in a weakened Sti1 interaction along with interaction of Sba1/p23 and one of a class of large prolylisomerases, such as FKBP52/54 or Cyp40 (Cpr6 in yeast) [10–12]. ATP hydrolysis causes additional conformational changes in Hsp90 that result in Sba1/p23 and prolylisomerase release, client protein folding and activation [5,13,14].
The co-chaperone protein Sti1 of S. cerevisiae (Hop in mammalian cells) is an abundant, highly conserved protein that does not have chaperone activity on its own [15,16]. Sti1/Hop interacts with Hsp70 and Hsp90 through separate TPR (tetratricopeptide repeat) domains. TPR1 interacts with the C-terminal EEVD residues of Hsp70, and TPR2A interacts with the C-terminal EEVD residues of Hsp90 [17–19]. Both mammalian Hop and yeast Sti1 behave as a dimer in solution [20,21]. Sti1 is able to stimulate the ATPase activity of the Hsp70 Ssa1 [22] and inhibit the ATPase activity of Hsp90 [11,20], but the regions of Sti1 required for these functions are unknown. A recent study [9] demonstrated that Sti1 was required to mediate transfer of denatured luciferase from Hsp70 to Hsp90 in vitro, providing critical evidence for a role of Sti1 in the transfer of client proteins [23]. However, the mechanism of this process, and the in vivo importance of the interaction of Sti1 with Hsp70 and Hsp90 during this process is unclear since the amount of Hsp70 recovered in Hsp90 complexes was not reduced in yeast cells lacking Sti1 [24].
In a previous study, we used site-directed mutagenesis of the TPR domains combined with a genetic screen to isolate mutations that disrupt in vivo functions of Sti1 [25]. The most dramatic effects on Sti1 function were obtained upon deletion or mutation of the DP2 region (so named because of the presence of dipeptide repeats of aspartic acid and proline residues) [21,23]. Surprisingly point mutations in TPR1 or TPR2A designed to disrupt the Hsp70 or Hsp90 interaction had little effect on in vivo functions of Sti1. Our data, combined with data from other laboratories [21,23,26,27], suggested that TPR1 and TPR2B have overlapping or redundant functions in Hsp70 interactions, and that both the TPR2A and TPR2B domains contribute to the Hsp90 interaction.
The goal of the present study was to further elucidate the function of individual domains of Sti1. We assayed the effects of Sti1 truncation on the physical interaction with Hsp70 and Hsp90 in vivo and the ability to dimerize in vitro. We found that TPR1 is required for wild-type levels of Hsp70 interaction in vivo, but mutations in both TPR1 and TPR2B were required to disrupt the interaction of Sti1 with Hsp70 in vitro. The isolated TPR2A domain contained the minimal fragment able to dimerize in vitro, but was unable to interact with Hsp90 in vivo unless combined with the TPR2B domain. These studies provide evidence that TPR2B physically interacts with both Hsp70 and Hsp90. Our results also raise questions about the function of the critical DP2 domain, since loss of the DP2 domain did not affect the Hsp70 or Hsp90 interaction or dimerization. We also provide novel information regarding the role of Sti1 in promoting the in vivo interaction between Hsp70 and Hsp90.
MATERIALS AND METHODS
Strains and growth conditions
Standard yeast genetic methods were employed [28,29]. Yeast strains used for growth assays are isogenic to W303 and have been described previously [25]. These include strains JJ623 (sti1::MET2), JJ609 (sti1::MET2 ydj1::HIS3/pRS316-YDJ1), JJ816 (hsc82::LEU2 hsp82::LEU2/YeP24-HSP82) and JJ832 (hsc82::LEU2 hsp82::LEU2 sti1::MET2/YeP24-HSP82). An alternate yeast strain JJ882 (MATα pep4::HIS3 sti1::HIS3 leu2-3,112 ura3-52 trp1-Δ1 his3-11,15) was used for protein purification. This strain was constructed by mating strain CN11 (MATa sti1::HIS3) [30] to strain WY11 [MATα pep4::HIS3, a gift from Elizabeth Craig (Department of Biochemistry, University of Wisconsin, Madison, WI, U.S.A.)], followed by diploid sporulation and tetrad dissection.
Strains were grown at 30 °C in YPD [1% (w/v) yeast extract/2% (w/v) peptone/2% (w/v) glucose] or minimal medium lacking specific amino acids (0.67% yeast nitrogen base without amino acids, 2% dextrose, supplemented with all required amino acids). To analyse cell growth, cells were streaked onto selective medium plates or plates containing 5-FOA (5-fluororotic acid; Toronto Research Chemicals) and incubated at the indicated temperature for 2–4 days.
To test the ability of Sti1 truncation mutants to support viability of a sti1ydj1 strain, His-tagged versions of truncation mutants expressed in 415GPDHis–Sti1 were transformed into strain JJ609. Colonies were then streaked onto plates containing 5-FOA to counterselect for pRS316-YDJ1. To test the ability of Sti1 truncation mutants to support viability of an hsc82hsp82sti1 (where Hsc is heat-shock cognate protein) strain expressing various hsc82 alleles, the indicated constitutively expressed His-tagged versions of Hsc82 expressed from a HIS+ vector [12] was transformed into strain JJ832. Untagged WT (wild-type) or mutant Sti1 expressed from a LYS+ vector was then transformed into those strains and plated on selective medium. Resultant colonies were streaked on plates containing 5-FOA to counterselect for the plasmid expressing WT HSP82.
Plasmids and antibodies
WT or mutant STI1 was expressed under its own promoter from the low-copy LEU2 plasmid pRS315 [31]. To generate His–Sti1, an Nco1 restriction site was introduced at the start codon of Sti1 and cloned into pRSETHisB (Invitrogen) as an Nco1-HindIII fragment. His–Sti1 was subsequently cloned into plasmid 415GPD [32] for constitutive expression in yeast. Sti1 truncation mutants were generated using PCR-based methods and cloned into 415GPDHis-Sti1 (LEU+). Non-His-tagged versions of Sti1 truncation mutants were also cloned into the LYS2 plasmid pRS317. Plasmids expressing GST (glutathione S-transferase) fused to the 10 kDa fragment of Ssa1 or the 10 kDa fragment of Ssa1 lacking the terminal EEVD residues have been described previously [33]. Polyclonal antibodies were raised against peptides representing amino acids 91–108 of Sti1 and the C-terminus of Hsc82/Hsp82 [34]. Polyclonal antisera specific for Ssa1/2 was a gift from Elizabeth Craig. The anti-Xpress antibody (Invitrogen) was used to recognize His–Sti1.
Protein purification and size-exclusion gel chromatography
Plasmids expressing WT and mutant His–Sti1 proteins with an N-terminal histidine-tag were transformed into strain JJ882 (pep4sti1). Sti1 was purified using metal chelate affinity chromatography. Purified proteins were dialysed against a standard Tris buffer [20 mM Tris/HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2 and 1 mM DL-dithiothreitol]. The apparent molecular masses of His–Sti1 truncation and point mutants were determined using size-exclusion gel chromatography. Samples were loaded onto a Sephacryl 16/60 S-300 (Amersham Biosciences) column equilibrated with 20 mM Tris/HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2 and 1 mM DL-dithiothreitol. Blue dextran 2000 (void volume), thyrogloblin (669 kDa), apoferritin (443 kDa), B-amylase (200 kDa), ADH (alcohol dehydrogenase; 150 kDa), BSA (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), cytochrome c (23 kDa) and ATP (550 Da) were used as molecular mass references.
10 kDa-Ssa1 binding assays
The 10 kDa C-terminal fragment of Ssa1 (or Ssa1ΔEEVD) was expressed as a GST-fusion protein in Escherichia coli, purified on glutathione–agarose beads (Sigma) and released from GST by thrombin cleavage as described previously [35]. The 10 kDa fragment (or BSA as a control) was diluted into 0.1 M NaHCO3 and bound to 96-well EIA (enzyme-linked immunoassay) plates at a concentration of 0.1 μg/well. Wells were washed with PBS, blocked with PBS containing 0.5% fatty acid-free BSA (Sigma) and washed with PBST (PBS containing 0.05% Tween 20). WT or mutant Sti1 was serially diluted in PBST containing 0.2% BSA and added to wells for 1 h at room temperature (25 °C). After extensive washing with PBST, a 1:20000 dilution of polyclonal antiserum against Sti1 was added and incubated for 2 h. After additional washes, a 1:4000 dilution of goat-anti-rabbit HRP (horseradish peroxidase)-conjugated antiserum (Amersham) was added and incubated for 45 min. After washing, the reaction was developed using the TMB Peroxidase EIA Substrate Kit (Bio-Rad) according to the manufacturer's specifications. Alternatively, 5 μg of GST–10 kDa-Ssa1 was prebound to 20 μl of glutathione resin and then incubated for 30 min with 10 μg of purified WT or mutant His–Sti1. Glutathione resin was washed with 20 mM Tris/HCl (pH 8.0) containing 0.01% Nonidet P40, then boiled in SDS sample buffer and analysed by SDS/PAGE and Coomassie Blue staining.
Isolation of His–Hsc82 and His–Sti1 complexes
Plasmids expressing WT or mutant His–Sti1 were transformed into strain JJ609 (sti1::MET ydj1::HIS3/pRS316-YDJ1). Resultant colonies were grown overnight at 30 °C in selective medium to a D600 of 1.2–2.0. Cells were harvested, washed with water and resuspended in lysis buffer [20 mM Tris/HCl (pH 7.5), 100 mM KCl and 5 mM MgCl2 containing a dissolved protease inhibitor mixture tablet (Roche Applied Science)]. Cells were disrupted in the presence of glass beads with 8×30 s pulses. His–Sti1 complexes were isolated by incubation with nickel resin (1 h at 4 °C with rocking) followed by washes with lysis buffer plus 0.1% Tween 20 and 35 mM imidazole. Nickel resin was boiled in SDS/PAGE sample buffer and protein complexes were separated by gel electrophoresis followed by Coomassie Blue staining and immunoblot analysis using antibodies against Hsc82/Hsp82, Ssa1/2 or Sti1 as indicated. His–Hsc82 complexes were isolated in a similar manner from strain JJ832 (hsc82hsp82sti1/His-Hsc82) expressing WT or mutant Sti1 or from strain JJ816 (hsc82hsp82) expressing WT His–Hsc82, His–Hsc82-G309S or His–Hsc82-ΔMEEVD as the only Hsp90 in the cell [12].
RESULTS
Sti1/Hop independently interacts with Hsp70 and Hsp90 via separate TPR domains and is able to mediate formation of a ternary Hsp70–Sti1/Hop–Hsp90 complex in vitro [19,36,37]. For these and additional reasons, Sti1/Hop was proposed to play a critical role in the transfer of client proteins from Hsp70 to Hsp90 [23]. However, the importance of Sti1 in mediating the interaction between Hsp70 and Hsp90 in vivo is unclear since the absence of STI1 did not reduce the ability of Hsp70 to co-purify with Hsp90 [24]. The goal of the present study was to determine the minimal region of Sti1 required for in vivo functions, interaction with Hsp70 and Hsp90, and dimerization, and to clarify the function of Sti1 in mediating the in vivo interaction between Hsp70 and Hsp90.
Sti1(222–589), which mimics the form of Sti1 present in Caenorhabditis elegans, is able to support some, but not all in vivo functions of Sti1
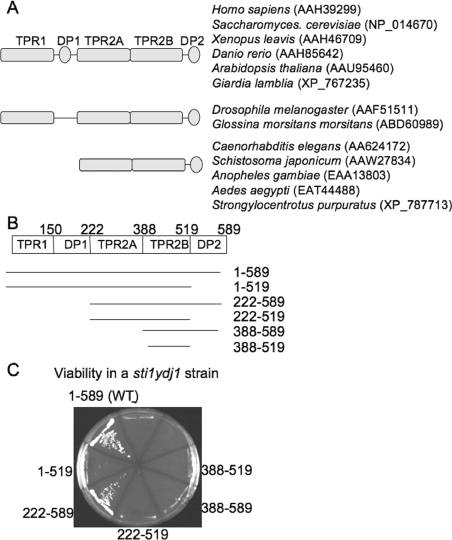
The domain structure of Sti1 is shown in Figure 1(A). Sti1 contains three separate TPR domains, TPR1, TPR2A and TPR2B, and two regions that contain aspartic acid–proline (DP) residue repeats: DP1 after TPR1 and DP2 after TPR2B [21,23]. The TPR1 domain of Hop (mammalian Sti1) co-crystallized with an octapeptide containing terminal EEVD residues of Hsp70, and the TPR2A domain of Hop co-crystallized with a peptapeptide containing the terminal MEEVD residues of Hsp90 [19].
Figure 1. Sti1 lacking the TPR1 and DP1 domains is able to support some functions of Sti1.
(A) Comparison of domain structure of various Sti1 orthologues. Some orthologues, such as those present in Drosophila or C. elegans, do not contain the DP1 or TPR1+DP1 domains respectively. Accession numbers of sequences used for comparison are indicated. (B) Schematic representation of Sti1 truncation mutants used in the present study. The amino acids present in each construct are indicated. (C) Ability of truncation mutants to rescue growth of a sti1ydj1/pRS316-YDJ1 strain. Plasmids expressing indicated His-tagged Sti1 truncation mutants were transformed into strain JJ609. Resultant colonies were grown in the presence of 5-FOA for 3 days at 25 °C to counterselect for the pRS316-YDJ1 plasmid.
A previous study compared the sequences of Sti1/Hop orthologues from different species and noted that the Drosophila homologue of Hop lacks the DP1 region [38]. We extended this analysis to include Sti1 orthologues from other species. We found that a number of Sti1orthologues, including those from C. elegans, Schistosoma japonicum, Anopheles gambiae, Aedes aegypti and Stronglyocentrotus purpuratus do not contain sequences corresponding to either TPR1 or DP1 domains (Figure 1A). This result suggests that the TPR1 and DP1 sequences arose as a duplication of TPR2B and DP2 or were subsequently lost. Either way, it suggests they may share redundant functions. Since prior studies determined that a fragment of Sti1 lacking TPR1 and DP1 was able to dimerize and inhibit the ATPase activity of Hsp90 [20,39], we examined whether TPR1 and DP1 are required for the in vivo functions of Sti1.
Yeast containing a deletion of STI1 exhibited only minor growth defects [30]. However, an STI1 deletion strain is unable to grow in the presence of the Hsp90 inhibitor radicicol, and a combined deletion of genes encoding Sti1 and the Hsp40 Ydj1 results in a lethal phenotype [25]. WT Sti1 containing an N-terminal 6× histidine tag (His–Sti1) was able to rescue the lethality of an sti1ydj1 strain as well as untagged Sti1 (results not shown and Figure 1C). We constructed His–Sti1 truncation mutants as shown in Figure 1(B) and tested their ability to support viability of a sti1ydj1/pRS316-YDJ1 strain in the presence of 5-FOA, which counterselects for the plasmid expressing WT YDJ1 (Figure 1C). Using an antibody specific for sequences in the His-tag, we demonstrated that all His–Sti1 fragments were expressed at similar levels in vivo (results not shown and Figure 3). Sti1(222–589), which lacks both TPR1 and DP1, was able to support viability of the sti1ydj1 strain similar to WT Sti1, while further deletion of the TPR2A or DP2 region renders truncation mutants unable to support viability. This result was consistent with prior studies that demonstrated that an sti1 strain expressing Sti1 lacking TPR1 was able to support viability of an sti1 strain in the presence of radicicol. However, Sti1 lacking TPR1 [26] or TPR1+DP1 (results not shown) exhibited defects in the activation of the heterologous Hsp90-dependent glucocorticoid receptor. These results indicate that TPR1 and DP1 are dispensable for some, but not all in vivo functions of Sti1.
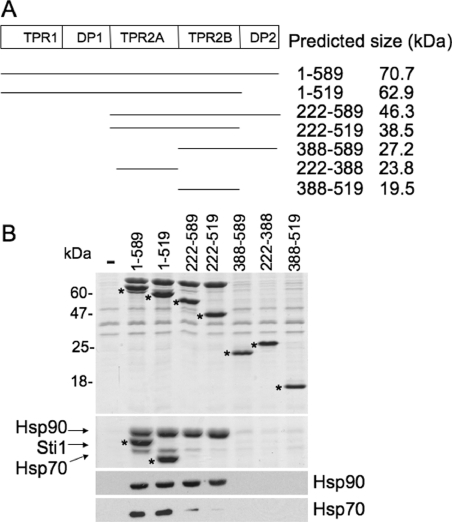
Figure 3. Effect of Sti1 truncation on in vivo interaction with Hsp70 and Hsp90.
(A) Schematic representation of Sti1 truncation mutants used in the present study. The predicted size of each construct is indicated. (B) WT or mutant His-tagged Sti1 was expressed in sti1 deletion strain JJ609. Cell extracts were prepared as described in the Materials and methods section and incubated with nickel resin for 1 h at 4 °C. Nickel resin-bound protein complexes were separated by SDS/PAGE followed by Coomassie Blue staining and immunoblot analysis. Upper panel, the Coomassie Blue stained 15% acrylamide gel showing migration of His–Sti1. Middle panel, the same samples run on 7.5% gel to maximize separation of Hsp90, Sti1 and Hsp70. The migration of His–Sti1 mutants is indicated by *. Lower panels, immunoblot analysis using antibodies against Hsc82/Hsp82 (Hsp90) or Ssa1/2 (Hsp70). −, No His-tagged protein was present in cell extracts.
TPR1 and TPR2B have similar functions in Hsp70 interaction
We previously found that single amino acid alterations in TPR1 and TPR2B residues predicted to contact the EEVD residues of Hsp70 [19] had only mild effects on Sti1 activity, but a combination of point mutations in TPR1 and TPR2B (R79A+R469A or K75E+R465E) completely disrupted the in vivo functions of Sti1 [25]. This result suggested that TPR1 and TPR2B may have overlapping or redundant functions in Hsp70 interaction. In order to assess directly whether these mutations in TPR1 and TPR2B disrupt the physical interaction of Sti1 with Hsp70, we examined the interaction of WT and mutant Sti1 with a C-terminal fragment of Hsp70 in vitro.
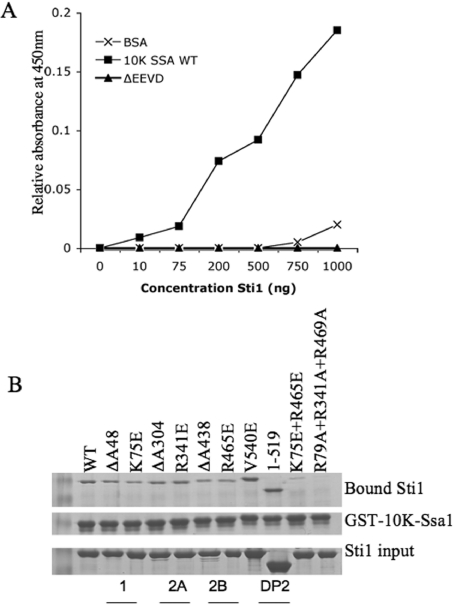
We purified WT and mutant His–Sti1 out of a protease-deficient sti1 strain (pep4sti1) using one-step standard metal-affinity chromatography. We monitored the interaction of purified Sti1 with the C-terminal 10 kDa fragment of the Hsp70 Ssa1 (10 kDa-Ssa1) using a modified and sensitive ELISA assay previously used to study interactions between 10 kDa-Ssa1 and the Hsp40 Sis1 [33]. 10 kDa-Ssa1 was expressed in E. coli as a fusion protein with GST (GST–10 kDa-Ssa1). The 10 kDa-Ssa1 fragment was released by thrombin cleavage [35]. Purified 10 kDa-Ssa1, 10 kDa-Ssa1 containing a deletion of the terminal EEVD residues (ΔEEVD) or BSA was bound to the wells of a microtitre plate. Increasing concentrations of purified WT His–Sti1 was added to the wells and the amount of Sti1 retained on the wells after multiple washes was determined using Sti1-specific antibodies. Full-length Sti1 exhibited significant binding to 10 kDa-Ssa1, while minimal interaction was observed for the control protein, BSA or 10 kDa-ΔEEVD (Figure 2A). This result indicated that the EEVD residues are required for stable Sti1 interaction.
Figure 2. Interaction of Sti1 with the C-terminal 10 kDa fragment of Hsp70.
(A) Binding of purified WT Sti1 to the 10 kDa fragment of Ssa1, 10 kDa-Ssa1ΔEEVD or BSA as a control. Serial dilutions of purified Sti1 (0–1000 ng) were incubated with 0.1 μg of 10 kDa-Ssa1, 10 kDa-Ssa1ΔEEVD or BSA immobilized in wells of a microtitre plate. Bound Sti1 was detected with an antibody specific for Sti1. (B) Purified WT or mutant His–Sti1 was incubated with GST–10 kDa-Ssa1 immobilized on glutathione resin. Proteins specifically bound to resin were analysed by SDS/PAGE and Coomassie Blue staining. The location of mutated residues (by domain name) is indicated.
Next we assayed the ability of Sti1 to be retained on glutathione resin in the presence of GST–10 kDa-Ssa1. Purified WT His–Sti1 was selectively retained by GST–10 kDa-Ssa1, but not GST–10 kDa-Ssa1ΔEEVD or GST alone (results not shown). WT or mutant His–Sti1 was incubated with GST–10 kDa-Ssa1 and the amount of bound His–Sti1 after washes was assessed by SDS/PAGE followed by Coomassie Blue staining. We analysed the effect of homologous mutations of basic residues in the EEVD-binding cleft of each TPR domain (K75E, R341E and R465E) and combinations thereof (K75E +R465E and R79A+R341A+R479A) or single amino acid deletions in each domain (ΔA48, ΔA304 and ΔA438) (Figure 2B). We also analysed Sti1ΔDP2 (1–519) and full-length Sti1 containing a mutation in a conserved residue within the DP2 domain (V540E) [25]. His–Sti1 binding to GST–10 kDa-Ssa1 was significantly disrupted only by combined mutations in TPR1 plus TPR2B (R79A+R469A) or all three TPR domains (R79A+R341A+R469A). This result indicates that the TPR1 and TPR2B domains of Sti1/Hop have redundant functions in the interaction with the C-terminus of Ssa1 in vitro and suggests that Hsp70 has similar contacts with the homologous EEVD-binding groove within TPR1 and TPR2B.
In vivo interaction of Sti1 truncation mutants with Hsp70 and Hsp90
In order to assess the contribution of individual Sti1 domains to the Hsp70 and Hsp90 interaction, we constitutively expressed His-tagged truncation mutants of Sti1 in an STI1 disruption strain. These constructs successively delete TPR1+DP1 [Sti1(222–589)], TPR1+DP1+TPR2A [Sti1(388–589)] or DP2(1–519). In addition, we tested constructs expressing only TPR2A(222–388), TPR2B(388–519) or TPR2A+TPR2B(222–519) (Figure 3A). Yeast expressing His–Sti1 constructs were lysed in a non-denaturing buffer. Lysates were bound to nickel resin, then washed with low levels of imidazole to reduce levels of non-specifically bound proteins. Proteins specifically bound to nickel resin were analysed by SDS/PAGE followed by Coomassie Blue staining and immunoblot analysis. We determined whether the truncated proteins were able to interact with Hsp90 (comprised of two isoforms, Hsc82 and Hsp82) and/or Hsp70 (comprised of two isoforms, Ssa1 and Ssa2). As shown in Figure 3(B), each of the truncation mutants was similarly retained by nickel resin. Sti1ΔDP2 domain(1–519) and Sti1ΔTPR1+DP1(222–589) each bound WT levels of Hsp90, indicating that TPR1, DP1 and DP2 are dispensable for Hsp90 interaction. Isolated TPR2A(222–388) or isolated TPR2B(388–519) failed to stably interact, but combined TPR2A+TPR2B(222–519) bound WT levels of Hsp90. This requirement of TPR2B for stable Hsp90 interaction was somewhat unexpected, since isolated TPR2A stably binds a peptide corresponding to the C-terminus of Hsp90 [19]. However, this result is consistent with our previous observation that deletion of an amino acid within TPR2B (ΔA438) disrupted the in vivo interaction between Sti1 and Hsp90, although a similar effect was not observed with the R465E point mutation in TPR2B (Figure 4 and [25]).
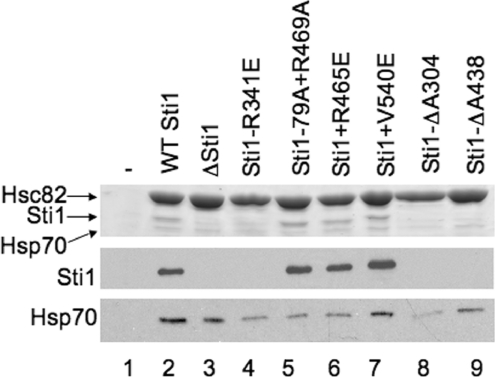
Figure 4. Effect of Sti1 mutation on ability of Hsp70 to co-purify with Hsp90.
WT or mutant (untagged) Sti1 was expressed in an hsc82hsp82sti1 strain expressing WT His-Hsc82 as the only Hsp90 in the cell. Cell extracts were prepared as described in the Materials and methods section and incubated with nickel resin for 1 h at 4 °C. Nickel resin-bound protein complexes were separated by SDS/PAGE followed by Coomassie Blue staining and immunoblot analysis. Upper panel, the Coomassie Blue stained 7.5% acrylamide gel showing migration of Hsp90, Sti1 and Hsp70. Lower panels, immunoblot analysis using antibodies specific for Sti1 or Ssa1/2 (Hsp70). Lane 1, control lane expressing untagged Hsc82; lane 2, WT Sti1; lane 3, strain was transformed with vector alone. For the remainder of the lanes the indicated mutant is listed.
The requirements for a stable Hsp70 interaction were distinct from the requirement for the Hsp90 interaction. Only Sti1(1–519) bound WT levels of Hsp70. Sti1(222–589) exhibited a reduced Hsp70 interaction, and the remaining truncation mutants failed to interact reproducibly with Hsp70. These results agree with our previous results that indicate that both TPR1 and TPR2B contribute to the Hsp70 interaction, but suggest that TPR1 is the primary site of interaction.
Identification of the minimal domain of Sti1 required for dimerization
Purified Sti1 or Hop behaves as a dimer in solution and previous studies have demonstrated that a fragment of Sti1(237–589) lacking TPR1 and DP1 was able to dimerize [20,21]. In order to determine which domain(s) of Sti1 are required for dimerization we monitored the migration of Sti1 truncation fragments using size-exclusion chromatography (Table 1). WT Sti1, Sti1 ΔDP2(1–519), ΔTPR1+DP1(222–589), or a fragment consisting only of TPR2A+TPR2B(222–519) migrated as a dimer. These results indicate that TPR1, DP1 and DP2 are dispensable for dimerization and suggest that TRP2A and/or TRP2B contains the regions required for this function of Sti1. We then monitored the migration of isolated TPR2A(222–388) or TPR2B(388–519). Isolated TPR2A migrated as a dimer, whereas isolated TPR2B migrated as a monomer. This result indicates that the TPR2A domain contains the sequences required for dimerization in addition to sequences required for Hsp90 interaction. Slight differences between predicted and apparent molecular masses may be due to differential post-translational modification of Sti1 fragments or conformational changes.
Table 1. TRP2A is necessary and sufficient for Sti1 dimerization.
WT and mutant His–Sti1 were purified out of yeast using one-step metal affinity chromatography, then analysed by gel filtration using a Sephacryl 16/60 S-300 column. A standard curve was generated for the column using molecular mass standards and the approximate retention volume and size for all Sti1 forms is indicated. Apparent stoichiometry was calculated by dividing the experimental mass by the deduced molecular mass.
| Domains present | |||||||
|---|---|---|---|---|---|---|---|
| TRP1+DP1 | TRP2A | TRP2B | DP2 | Experimental mass (kDa) (relative elution volume) | Deduced mass of monomer (kDa) | Apparent stoichiometry | |
| His–Sti1(1–589) | x | x | x | x | 160 | 70.7 | dimer (2.2) |
| His–Sti1(1–519) | x | x | x | 140 | 62.9 | dimer (2.2) | |
| His–Sti1(222–589) | x | x | x | 94 | 46.3 | dimer (2) | |
| His–Sti1(222–519) | x | x | 68 | 38.5 | dimer (1.8) | ||
| His–Sti1(222–388) | x | 46 | 23.8 | dimer (1.9) | |||
| His–Sti1(388–589) | x | x | 28 | 27.2 | monomer (1) | ||
| His–Sti1(388–519) | x | 15 | 19.5 | monomer (0.8) | |||
| His–Sti1(1–589)ΔA304 | x | x | x | x | 160 | 70.7 | dimer (2.2) |
| His–Sti1(1–589)R341E | x | x | x | x | 150 | 70.7 | dimer (2.1) |
| His–Sti1(1–589)ΔA438 | x | x | x | x | 170 | 70.7 | dimer (2.4) |
We next determined whether the three mutations that disrupt the Hsp90 interaction in vivo (R341E and ΔA304 in TPR2A and ΔA438 in TPR2B) [25] disrupted dimerization of full-length Sti1. His–Sti1 containing either of these mutations exhibited mobility similar to WT Sti1 (Table 1). This result indicates that although TPR2A contains the dimerization domain, mutations that target the EEVD binding cleft that are sufficient to disrupt Hsp90 interaction in vivo do not disrupt dimerization. This result is consistent with a previous study that demonstrated that mutations targeting the EEVD-binding cleft of TPR2A or TPR2B of Hop do not affect dimerization [21]. This result also suggests that the Sti1ΔA438 mutation is not grossly misfolded, even though it disrupts in vivo function [25].
Effect of Sti1 mutation on the interaction between Hsp90 and Hsp70
Purified mammalian Hsp70 and Hsp90 do not interact except in the presence of Hop, suggesting that Hop/Sti1 mediates the physical interaction between Hsp90 and Hsp70 [36,37]. However, a previous study indicated that deletion of STI1 did not dramatically reduce the ability of Hsp90 to interact with Hsp70 in vivo [24]. To determine the effect of STI1 deletion or mutation on the ability of Hsp90 to interact with Hsp70 in vivo, we examined His–Hsp90 complexes in cells expressing no Sti1, or untagged WT or mutant Sti1. We used an hsc82hsp82sti1 strain (JJ832) expressing His–Hsc82 as the only Hsp90 in the cell. Yeast cells expressing vector alone, WT Sti1, or the indicated mutant were disrupted in a non-denaturing buffer, incubated with nickel resin, and proteins bound to nickel resin after washes with lysis buffer containing low levels of imidazole were analysed by SDS/PAGE and immunoblot analysis (Figure 4).
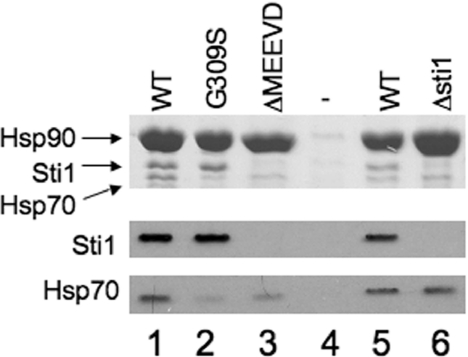
As shown in Figure 4, similar levels of Hsp90 were recovered from each strain. As expected, no Sti1 interaction was observed in the strain lacking STI1 (lane 3), or in yeast expressing the R341E and ΔA304 mutations within TPR2A or the ΔA438 mutation within TPR2B (lanes 4, 8 and 9) [25]. The remaining Sti1 mutants had wild-type levels of interaction with Hsp90. The main question was how loss or mutation of STI1 affected the in vivo interaction between Hsp90 and Hsp70. In the presence of WT Sti1 (lane 2), significant binding of Hsp70 was observed compared with cells that do not express His–Hsp90 (lane 1). In accordance with the previous report [24], deletion of STI1 does not significantly alter the level of Hsp70 observed in Hsp90 complexes (lane 3). Similarly, the level of Hsp70 recovered was not dramatically altered by the Sti1-V540E mutation, while the ΔA438 mutation caused an intermediate effect. Notably, the level of Hsp70 present in Hsp90 complexes was significantly reduced in the presence of Sti1-R341E, Sti1-R79A+R469A, R465E or Sti1-ΔA304 mutations.
The Hsc82-G309S and Hsc82-ΔMEEVD mutants exhibit a defect in Ssa1/2 interaction in vivo
The above results confirm that Sti1 is not essential for the in vivo interaction between Hsp70 and Hsp90, but indicate that Sti1 does contribute to formation of the ternary complex in vivo. In order to gain additional clues about the formation of Hsp90–Hsp70 complexes, we analysed mutations in Hsc82. The hsc82-G309S mutation caused a temperature-sensitive phenotype, but His–Hsc82-G309S did not exhibit defects in the interaction with Sti1, Sba1 or Cpr6 [12]. However, as demonstrated in Figure 5, Hsc82-G309S does exhibit a defect in the Hsp70 interaction. We isolated WT His-tagged Hsc82 and Hsc82 containing the G309S mutation from yeast expressing WT Sti1. We also isolated His–Hsc82-ΔMEEVD, which does not interact with Sti1 or Cpr6 due to deletion of the C-terminal MEEVD sequences [12,40]. Relative to WT Hsc82, G309S exhibits a marked reduction in Ssa1/2 interaction (Figure 5, compare lanes 1 and 2). The Hsc82-ΔMEEVD mutation (lane 3) also resulted in a similar reduction of the Hsp70 interaction compared with WT Hsc82, even though this mutation does not cause noticeable growth defects [41]. Since Hsc82-ΔMEEVD also exhibits a reduced Hsp70 interaction, this suggests that Sti1, or likely another protein that interacts with the C-terminus of Hsp90, mediates the in vivo interaction between Hsp90 and Hsp70. However, for unknown reasons, this effect is distinct from that observed in a STI1 disruption strain (lane 6).
Figure 5. The Hsc82-G309S mutant exhibits reduced Hsp70 interaction in vivo.
WT or mutant His–Hsc82 was expressed in an hsc82hsp82 strain as the Hsp90 in the cell. Cell extracts were incubated with nickel resin for 1 h at 4 °C. Nickel resin–bound protein complexes were separated by SDS/PAGE followed by Coomassie Blue staining and immunoblot analysis. Upper panel, the Coomassie Blue stained 7.5% acrylamide gel showing migration of Hsp90, Sti1 and Hsp70. Lower panels, immunoblot analysis using antibodies specific for Hsc82, Sti1 or Ssa1/2 (Hsp70). Lane 1, WT His–Hsc82; lane 2, His–Hsc82-G309S; lane 3, His–Hsc82ΔMEEVD; lane 4, control lane expressing untagged Hsc82. Lane 5 and lane 6, WT His–Hsc82 isolated out of hsc82hsp82sti1 strain expressing WT Sti1 (lane 5) or lacking Sti1 (lane 6).
The Hsc82-G309S mutation becomes inviable upon alteration of the TPR1 or TPR2B domains of Sti1
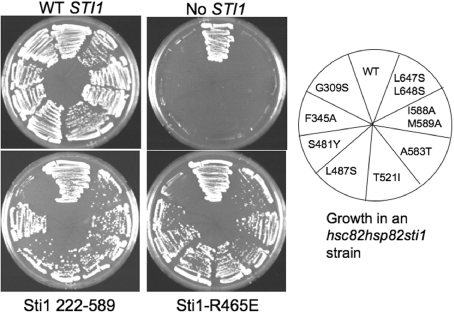
The above result suggests that the temperature-sensitive phenotype caused by the hsc82-G309S mutation is due to a specific defect in the Hsp70 interaction. To determine whether the function of Hsc82-G309S is further disrupted by mutations that disrupt the Sti1–Hsp70 interaction, we examined the genetic interaction between Hsp90 and Sti1. Deletion of both genes encoding Hsp90 (HSC82 and HSP82) is lethal [34]. We isolated a number of mutations in hsc82 that confer a temperature-sensitive phenotype when present as the only Hsp90 in the cell [12]. We examined the ability of these hsc82 mutant alleles to support viability in the absence of STI1. Plasmids expressing WT or mutant forms of HSC82 were transformed into strain JJ832 (hsc82hsp82sti1/YEp24-HSP82). Resultant colonies were streaked onto medium containing 5-FOA, which counterselects for the presence of the plasmid expressing WT HSP82 (YEp24-HSP82). Of 16 mutations tested, eight of the hsc82 mutants exhibited WT or near WT growth in the presence of STI1 (hsc82hsp82 strain) but were unviable in the absence of STI1 (Figure 6 and results not shown). The hsc82-ΔMEEVD mutation does not exhibit growth defects in the presence or absence of STI1 (results not shown). These Hsc82 mutants are located in the middle (G309S, F345A, S481Y, L487S and T521I) and C-terminal (A583T, I588A, M589A, L647S and L648S) domains of Hsp90 [10,42].
Figure 6. Genetic interaction between Sti1 and Hsc82-G309S.
Yeast strain JJ832 (hsc82hsp82sti1/YEp24-HSP82) expressing WT Hsc82, Hsc82-G309S, F345A, S481Y, L487S, T521I, A583T, I588AM589A or L647SL648S was transformed with plasmids expressing Sti1 WT, Sti1(222–589), Sti1-R465E or vector alone as indicated. Resultant colonies were grown for 3 days at 30 °C in the presence of 5-FOA, which counterselects for the presence of the YEp24-HSP82 plasmid.
Next we determined whether any of these hsc82 mutant alleles that are dependent on STI1 for viability are specifically affected by loss of the TPR1 and DP1 domains. We transformed a plasmid expressing Sti1(222–589) into the hsc82hsp82sti1/Yep24-HSP82 strain expressing the hsc82 mutant alleles shown in Figure 6. Transformants were then streaked onto medium containing 5-FOA to counterselect for the Yep24-HSP82 plasmid (Figure 6). In most cases, Sti1(222–589) was able to support growth. The exception was that the combination of hsc82-G309S with sti1(222–589) was lethal, suggesting that the Hsc82-G309S mutant is particularly dependent on the function of TPR1 and/or DP1. We then tested the combination of sti1-R465E with mutant hsc82 alleles as described above. Again, we observed that hsc82-G309S was the only mutant that was unviable in the presence of the sti1-R465E mutant. Since the Sti1-R465E mutation resulted in reduced interaction of Hsp70 with Hsp90 (Figure 4), these results suggest that further reduction in Hsp70 interaction disrupts the essential functions of Hsp90. Together, these results indicate that Hsc82-G309S either has a specific defect in Sti1-mediated Hsp70 interaction or has an enhanced requirement for Sti1-mediated Hsp70 interaction due to a defect in another mechanism that promotes in vivo interaction between the two molecular chaperones.
DISCUSSION
The present study further defines the domains of Sti1 required for in vivo interaction with Hsp70 and Hsp90 and identifies the region of Sti1 required for dimerization. We also provide novel evidence that mutation of Sti1 results in reduced interaction between Hsp90 and Hsp70, which is consistent with the proposed role for Sti1 in mediating the transfer of client proteins between Hsp70 and Hsp90 [9,23].
Interaction of Sti1 with Hsp70
TPR1 of Sti1/Hop has been recognized as the primary binding site for Hsp70 since an EEVD-containing peptide corresponding to the C-terminus of Hsp70 co-crystallized with isolated TPR1 [19]. However, other studies subsequently showed that mutations outside of TPR1 affected the Hsp70 interaction [21,23,27]. There is also evidence that suggests that Sti1/Hop has a contact site with Hsp70 in addition to the terminal EEVD residues, since a truncated form of Hsp70 lacking the C-terminal 38 amino acids maintained wild-type levels of Hop interaction [21], and Sti1 was able to stimulate the ATPase activity of the Hsp70 Ssa1 [22]. Our studies indicate that both TPR1 and TPR2B contribute to the Hsp70 interaction in vivo and in vitro, although loss of TPR1 has a much more dramatic effect on the interaction between Hsp70 and Sti1 in vivo. The C-terminus of Hsp70 likely has similar contacts with the EEVD-binding grooves of TPR1 and TPR2B, since homologous point mutations in both domains were able to completely disrupt the in vivo functions of Sti1 [25] and the in vitro interaction with 10 kDa-Ssa1. In contrast with previous studies with Hop [21,23], deletion of DP2 or a point mutation within DP2 did not inhibit the Hsp70 interaction. The reason for this difference is unknown and may reflect species-specific differences.
A previous study [38] and our results suggest that the TPR1 and/or DP1 sequences arose as a duplication event in species ranging from yeast to humans, or were lost in species such as C. elegans. What is the function of TPR1 and DP1? One possibility is that Sti1 has Hsp90-independent functions and that TPR1 and DP1 are critical for these functions. This possibility is supported by the observation that mutations in TPR1 specifically affect the regulation of Hsp70 function in yeast prion [PSI+] propagation [26]. We found that Sti1 lacking TPR1 and DP1 (222–589) was able to function in various growth assays, but was unable to function in the activation of the heterologous client protein, the GR (glucocorticoid receptor). Independent results suggest that both TPR1 and DP1 contribute to this function: Sti1 lacking TPR1, but not DP1, was also unable to function in GR activation [26], and when expressed in yeast, the Drosophila homologue of Hop (which lacks DP1) was able to bind both Hsp70 and Hsp90, but exhibited defects in GR activation [38]. Since GR activity is particularly sensitive to deletion of TPR1 and/or DP1, it is possible that activation of this heterologous client has enhanced requirements for Sti1 function. However, it is also possible that TPR1 and/or DP1 have a crucial role in co-ordinating the activity of Hsp70 and Hsp90 during transfer of client protein from one chaperone to the other. Additional evidence that TPR1 and TPR2B need to be within the same polypeptide to support GR function supports this hypothesis [43].
Interaction of Sti1 with Hsp90
TPR2A of Sti1/Hop has been recognized as the primary binding site for Hsp90 since an EEVD-containing peptide corresponding to the C-terminus of Hsp90 co-crystallized with isolated TPR2A [19]. Accordingly, we found that alterations in basic residues of TPR2A predicted to contact EEVD residues selectively disrupt Hsp90 interaction in vivo [25]. Although multiple lines of evidence suggest that there are additional sites of contact between Hsp90 and Sti1 [11,12,20,25,44], it was nonetheless surprising that the TPR2A domain did not stably interact with Hsp90 unless combined with TPR2B. TPR1 cannot replace this function of TPR2B since Sti1(1–387), which contains TPR1+DP1+TPR2A does not stably interact with Hsp90 (results not shown). This requirement for TPR2B is consistent with prior evidence from our laboratory and others that mutations in TPR2B disrupt Hsp90 binding [23,25,26]. Although the ΔA438 mutation in TPR2B disrupts the Hsp90 interaction, the R465E mutation targeting the EEVD-binding cleft did not affect the Hsp90 interaction. At this time we are unable to ascertain whether Hsp90 contacts a different site within TPR2B or whether Hsp90 does indeed contact the EEVD-binding cleft but the R465E mutation is not sufficient to disrupt the Hsp90 interaction due to altered specificity of the Hsp70 and Hsp90 interactions [27].
Dimerization of Sti1
Our results indicate that the isolated TPR2A domain contains the sequences required for dimerization. Of the multiple TPR-containing proteins that interact with Hsp90 [16], only Sti1 is known to function as a dimer and a dimer of Sti1 appears to interact with a dimer of Hsp90 [20]. The location of the dimerization domain within TPR2A is unknown, as neither the R341E nor the ΔA304 mutations disrupted the ability of full length Sti1 to migrate as a dimer in solution. At this time we do not know whether dimerization is required for Sti1 function in vivo since we are unable to distinguish between the functions of TPR2A in Hsp90 interaction and dimerization.
Role of Sti1 in mediating ternary complex formation
A ternary complex of purified Sti1/Hop, Hsp70 and Hsp90 occurs readily in an ATP-independent manner, but purified Hsp70 and Hsp90 interact only in the presence of Hop/Sti1 [6,36,37]. This finding led to the hypothesis that by promoting formation of the ternary complex, Sti1/Hop mediates the transfer of client proteins from Hsp70 to Hsp90 [23], and a recent report demonstrated that transfer of denatured luciferase from Hsp70 to Hsp90 in vitro was dependent on Sti1 [9]. Sti1 is likely to have a critical role in regulating the activity of both Hsp70 and Hsp90 during this process [11,20,22], but the regions of Sti1 required for this function remain unknown.
How does Sti1 mediate the interaction between Hsp70 and Hsp90? Our results combined with results from the Smith and Blatch laboratories [21,23,27] indicate that TPR1 and TPR2B both contact Hsp70, while both TPR2A and TPR2B contact Hsp90. Since Hsp70 binds Hop in a 2:1 molar ratio in the absence of Hsp90 but a 1:1 molar ratio in the presence of Hsp90 [8], it seems likely that Hsp90 binding displaces one monomer of Hsp70 from TPR2B, but additional studies are required to resolve this issue. Sti1/Hop is known to undergo conformational changes upon Hsp70 or Hsp90 interaction [20,45], and it is possible that the Hsp90–TPR2B interaction results in a conformational change that allows the Hsp90 interaction with TPR2A.
How important is Sti1 in mediating the in vivo interaction between Hsp90 and Hsp70? We identified Sti1 mutants that result in reduced recovery of Hsp70 in Hsp90 complexes (Figure 4). In two of these cases, R79A+R469A and R465E, the Sti1–Hsp90 interaction was unaffected but a reduced Hsp70 interaction was observed. Since these mutations alter the EEVD-binding cleft in TPR1 and TPR2B, the reduced Hsp70 interaction likely results from a defect in Hsp90-bound Sti1 to interact with Hsp70. The other mutants, R341E and ΔA304, result in loss of Sti1 interaction and reduced Hsp70 interaction. The reason that the R341E and ΔA304 mutations have an effect distinct from an STI1 null strain [24] is unknown. However, loss of STI1 resulted in enhanced transcription of heat-shock genes [30], and thus it is possible that a protein upregulated under these conditions functionally replaces Sti1 in mediating a stable Hsp70–Hsp90 interaction. In this scenario, the R341E and ΔA304 mutations may fail to upregulate the heat-shock response or may interfere with the other protein interacting with Hsp90. The nature of this other protein is unknown, although a number of other Hsp90 co-chaperones have been identified [2–4].
Our results raise additional questions about the transfer of client proteins from Hsp70 to Hsp90. The Hsc82-ΔMEEVD mutant exhibits reduced Hsp70 interaction, along with a reduced interaction of TPR-containing proteins, yet does not cause noticeable growth defects [12,41]. The most likely explanations are the presence of alternate pathways that promote Hsp70–Hsp90 interactions and/or that Hsc82-ΔMEEVD maintains in vivo interactions with both Hsp70 and/or co-chaperones but with reduced stability not detected in our assays. The Hsc82-G309S mutant should be very useful in understanding the mechanism and specificity of the client transfer between Hsp70 and Hsp90 since it appears to be specifically dependent on the ability of Sti1 to interact with Hsp70. In conclusion, our results answer some of the questions about the role of Sti1 in Hsp90 function, but questions of whether alternate pathways promote Hsp90–client interaction and whether all client proteins have similar interactions with Hsp90 and co-chaperone proteins remain unresolved.
Acknowledgments
We thank Elizabeth Craig (Department of Biochemistry, University of Wisconsin, Madison, WI, U.S.A.) for yeast strains and antibodies and Gary W. Daughdrill (Department of Microbiology, Molecular Biology and Biochemistry, University of Idaho, Moscow, ID, U.S.A.) for assistance in protein purification. This project was funded in part by the U.S. Department of Agriculture HATCH/CREES IDA01266 and additional funds from the University of Idaho Research Council. This publication was made possible by Grant Number P20 RR15587 from the NCRR (National Center for Research Resources), a component of the NIH (National Institutes of Health) and NIH Grant Number P20 RR016454 from the INBRE Program of the National Center for Research Resources. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
References
- 1.Whitesell L., Lindquist S. L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 2.Zhao R., Davey M., Hsu Y. C., Kaplanek P., Tong A., Parsons A. B., Krogan N., Cagney G., Mai D., Greenblatt J., et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Millson S. H., Truman A. W., King V., Prodromou C., Pearl L. H., Piper P. W. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryotic Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pratt W. B., Toft D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 5.Pearl L. H., Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 6.Wegele H., Muller L., Buchner J. Hsp70 and Hsp90–a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez M. P., Chadli A., Toft D. O. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez M. P., Sullivan W. P., Toft D. O. The assembly and intermolecular properties of the hsp70–Hop–hsp90 molecular chaperone complex. J. Biol. Chem. 2002;277:38294–38304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- 9.Wegele H., Wandinger S. K., Schmid A. B., Reinstein J., Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter K., Muschler P., Hainzl O., Reinstein J., Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the ATPase cycle. J. Biol. Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J. L., Halas A., Flom G. Nucleotide-dependent interaction of S. cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young J. C., Hartl F. U. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaughlin S. H., Sobott F., Yao Z. P., Zhang W., Nielsen P. R., Grossmann J. G., Laue E. D., Robinson C. V., Jackson S. E. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J. Mol. Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 15.Odunuga O. O., Longshaw V. M., Blatch G. L. Hop: more than an Hsp70/Hsp90 adaptor protein. BioEssays. 2004;26:1058–1068. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- 16.Smith D. F. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young J. C., Obermann W. M., Hartl F. U. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 18.Demand J., Luders J., Hohfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 1998;18:2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 20.Prodromou C., Siligardi G., O'Brien R., Woolfson D. N., Regan L., Panaretou B., Ladbury J. E., Piper P. W., Pearl L. H. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrigan P. E., Nelson G. M., Roberts P. J., Stoffer J., Riggs D. L., Smith D. F. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 2004;279:16185–16193. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- 22.Wegele H., Haslbeck M., Reinstein J., Buchner J. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- 23.Chen S., Smith D. F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 24.Chang H. C., Nathan D. F., Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol. Cell. Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flom G., Weekes J., Williams J. J., Johnson J. L. Effect of mutation of the tetratricopeptide repeat and aspartate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics. 2006;172:41–51. doi: 10.1534/genetics.105.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Masison D. C. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1) J. Biol. Chem. 2005;280:34178–34185. doi: 10.1074/jbc.M505420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odunuga O. O., Hornby J. A., Bies C., Zimmermann R., Pugh D. J., Blatch G. L. Tetratricopeptide repeat motif-mediated Hsc70–mSTI1 interaction. Molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 2003;278:6896–6904. doi: 10.1074/jbc.M206867200. [DOI] [PubMed] [Google Scholar]
- 28.Sherman F., Fink G. R., Hicks J. B. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1986. Laboratory course manual for methods in yeast genetics. [Google Scholar]
- 29.Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 30.Nicolet C. M., Craig E. A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 33.Aron R., Lopez N., Walter W., Craig E. A., Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 36.Johnson J., Corbisier R., Stensgard B., Toft D. The involvement of p23, hsp90, and immunophilins in the assembly of progesterone receptor complexes. J. Steroid Biochem. Mol. Biol. 1996;56:31–37. doi: 10.1016/0960-0760(95)00221-9. [DOI] [PubMed] [Google Scholar]
- 37.Chen S., Prapapanich V., Rimerman R. A., Honore B., Smith D. F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol. Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 38.Carrigan P. E., Riggs D. L., Chinkers M., Smith D. F. Functional comparison of human and Drosophila Hop reveals novel role in steroid receptor maturation. J. Biol. Chem. 2005;280:8906–8911. doi: 10.1074/jbc.M414245200. [DOI] [PubMed] [Google Scholar]
- 39.Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., Prodromou C. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J. Biol. Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 40.Abbas-Terki T., Donze O., Briand P. A., Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louvion J. F., Warth R., Picard D. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. Structural and functional analysis of the middle segment of hsp90. Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 43.Bredemeyer A. J., Carrigan P. E., Fehniger T. A., Smith D. F., Ley T. J. Hop cleavage and function in granzyme B-induced apoptosis. J. Biol. Chem. 2006;281:37130–37141. doi: 10.1074/jbc.M607969200. [DOI] [PubMed] [Google Scholar]
- 44.Chen S., Sullivan W. P., Toft D. O., Smith D. F. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrigan P. E., Sikkink L. A., Smith D. F., Ramirez-Alvarado M. Domain–domain interactions within Hop, the Hsp70/Hsp90 organizing protein, are required for protein stability and structure. Protein Sci. 2006;15:522–532. doi: 10.1110/ps.051810106. [DOI] [PMC free article] [PubMed] [Google Scholar]