Abstract
The haloarchaeal phototaxis receptor sensory rhodopsin I (SRI) in complex with its transducer HtrI delivers an attractant signal from excitation with an orange photon and a repellent signal from a second near-UV photon excitation. Using a proteoliposome system with purified SRI in complex with its transducer HtrI, we identified by site-directed fluorescence labeling a site (Ser155) on SRI that is conformationally active in signal relay to HtrI. Using site-directed spin labeling of Ser155Cys with a nitroxide side chain, we detected a change in conformation following one-photon excitation such that the spin probe exhibits a splitting of the outer hyperfine extrema (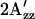 ) significantly smaller than that of the electron paramagnetic resonance spectrum in the dark state. The dark conformations of five mutant complexes that do not discriminate between orange and near-UV excitation show shifts to lower or higher
) significantly smaller than that of the electron paramagnetic resonance spectrum in the dark state. The dark conformations of five mutant complexes that do not discriminate between orange and near-UV excitation show shifts to lower or higher 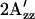 values correlated with the alterations in their motility behavior to one- and two-photon stimuli. These data are interpreted in terms of a model in which the dark complex is populated by two conformers in the wild type, one that inhibits the CheA kinase (A) and the other that activates it (R), shifted in the dark by mutations and shifted in the wild-type SRI-HtrI complex in opposite directions by one-photon and two-photon reactions.
values correlated with the alterations in their motility behavior to one- and two-photon stimuli. These data are interpreted in terms of a model in which the dark complex is populated by two conformers in the wild type, one that inhibits the CheA kinase (A) and the other that activates it (R), shifted in the dark by mutations and shifted in the wild-type SRI-HtrI complex in opposite directions by one-photon and two-photon reactions.
INTRODUCTION
The Halobacterium salinarum phototaxis receptor sensory rhodopsin I (SRI) is unusual among photoreceptors in that it produces opposite signals depending on the color of light stimuli (1). Absorption of orange light (λmax 587 nm) triggers an attractant response by the cell (inhibition of swimming reversals), whereas absorption of orange light followed by near-UV light (λmax 373 nm) triggers a repellent response (induction of swimming reversals). Both the one-photon attractant and two-photon repellent signals are transmitted through the membrane-embedded transducer protein HtrI, which exists in a complex with SRI (2).
One-photon and two-photon excitations of SRI produce cyclic photochemical reactions that have been characterized using kinetic laser flash spectroscopy (1,3,4) (earlier work reviewed by Hoff et al. (5)). Orange light converts the dark form of SRI into a series of photointermediate states, one of which is a long-lived (several seconds) near-UV light-absorbing species with maximal absorbance at 373 nm. This species, designated S373, is the attractant signaling state; i.e., its production suppresses swimming reversals. S373 is photoreactive, and its excitation by a near-UV photon returns it to the dark state SR587 through a reaction sequence that contains several species, predominantly 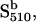 that send repellent signals. The net result is that in the wild-type SRI-HtrI complex, the one-photon reaction SR587 → S373 generates attractant signals, whereas the two-photon reaction path
that send repellent signals. The net result is that in the wild-type SRI-HtrI complex, the one-photon reaction SR587 → S373 generates attractant signals, whereas the two-photon reaction path 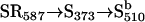 generates repellent signals. White light stimulation, approximating direct solar radiation, of the dark-adapted SRI state produces a mixture of one-photon and two-photon products. Because white light stimulation of the SRI-HtrI complex mediates a repellent response in H. salinarum cells, the latter evidently dominates the signaling.
generates repellent signals. White light stimulation, approximating direct solar radiation, of the dark-adapted SRI state produces a mixture of one-photon and two-photon products. Because white light stimulation of the SRI-HtrI complex mediates a repellent response in H. salinarum cells, the latter evidently dominates the signaling.
Behavioral mutants that have lost their color-sensing ability have been isolated by photobehavior selection schemes and their mutations mapped to SRI and HtrI residues in the complex, most on or near the predicted interface between the two proteins (6). Two different phenotypes are observed based on motility analysis of their phototaxis behavior: “Omniphobic” mutants of the SRI-HtrI complex deliver repellent signals on one-photon activation as well as to two-photon activation. Conversely, “omniphilic” mutants deliver attractant signals to both photostimuli. Combining omniphobic and omniphilic mutations restores a more wild-type motility behavior.
An equilibrium of two conformations has been invoked to explain the effects of protonation of the protonated Schiff base counterion Asp85 in BR and Asp73 in H. salinarum SRII. In the case of BR, substitution of Asp85 with Asn in bacteriorhodopsin (BR) produces a mixture of closed-channel and open-channel forms (7,8). In the case of SRII, the corresponding Asp73Asn mutant is partially constitutively active for phototaxis signaling, a property that was attributed to a mixture of the unactivated and activated conformations in the dark (9). The first suggestion of such an equilibrium mixture of two conformers of the dark state of the SRI-HtrI complex came from the observation that the corresponding residue Asp76 near the Schiff base is, unlike BR and SRII, naturally protonated in the dark complex (10,11). The suggestion is attractive because in a two-conformation equilibrium model, two receptor conformers, assumed to be similar to the well-established closed and open cytoplasmic channel conformers of BR, would be responsible for both attractant and repellent signals from SRI. Therefore, the model was proposed that SR587 exists in an equilibrium mixture of the two conformers, designated A (attractant) and R (repellent) (6). The equilibrium is assumed to be metastable in that different stimuli can induce a shift in either direction. The net effect of the one-photon cycle is to transiently shift the equilibrium to the A conformer, which suppresses reversals, whereas the two-photon cycle shifts the equilibrium to the R conformer, which induces reversals. The repellent effect of white light is explained as an overall shift toward the R conformer in the mixture of photoproducts.
The mutant phenotypes, showing typical two-state behavior, namely opposite effects that mutually cancel, support such a two-state equilibrium model. The omniphobic mutants D201N and H166S in SRI and E56Q in HtrI behave as if they are shifted extremely far into one of the conformers (the A conformer) in the dark, mediating repellent responses to both one-photon and two-photon activation (6). Conversely, the mutant R215W in SRI, isolated as a suppressor to the omniphobic mutant E56Q (6), is omniphilic; i.e., R215W mediates attractant responses to both one-photon and two-photon stimuli. Cells containing the R215W SRI-HtrI complex therefore behave as if the complex is shifted extremely far into the R conformer in the dark, exhibiting an attractant response to both one-photon and two-photon activation.
Our primary goals in this work were to assess conformations of the SRI-HtrI interface with an appropriately positioned spin-label probe and to use electron paramagnetic resonance (EPR) to test whether the wild type, omniphobic mutants, and omniphilic mutants exhibit the conformational differences predicted by a metastable conformational equilibrium model.
MATERIALS AND METHODS
Plasmid and strains
HtrI-free SRI, SRI-HtrI full-length fusion, and SRI-HtrI fusion truncated at position 147 in HtrI were expressed under the bop promoter in the plasmids pXP301, pXP6, and p147his, respectively (12). HtrI-free SRI and SRI-HtrI1-147 contain hexahistidine extensions at the C-termini. All the single-residue mutations were introduced by the two-step megaprimer PCR method with pfu turbo polymerase (13). H. salinarum strain Pho81Wr− (BR−HR−SRI−HtrI−SRII−HtrII− and restriction-deficient) (14) was used as the recipient in plasmid transformations.
Motility analysis
Motility responses to SRI photoactivation were assayed by computer-assisted cell tracking and motion analysis. Early stationary-phase cultures were diluted 1:13 in fresh medium for H. salinarum (CM) and incubated for 1 h at 37°C with agitation (2). Motility responses to orange, near-UV, and white light photostimuli were recorded with far red/infrared light (>700 nm). Swimming cells were subjected to four photostimuli: 2-s step up in 600 ± 20 nm (orange) light, 200-ms pulse of >350 nm (white) light, a 2-s step-down of orange light, and 2-s step-down of white light. The stimulus was delivered from a Nikon 100-W He/Xe short arc lamp. All stimuli were saturating for wild-type cells. Cell motility was captured in real-time AVI files using a Flashbus Spectrim Lite Video Capture PCI Card on a Dell Dimension 8500 that was running VirtualDub 1.6.14 (www.virtualdub.org) AVI encoder software for video capture. VirtualDub was set to record 10 frames per second during 5 s of cell swimming. After the first second, the stimulus was initiated using the appropriate filters. The cell-tracking and motion analysis software package Celltrak 1.2 Beta (Motion Analysis, Santa Rosa, CA) running on a Dell Dimension 9150 was then used to analyze cell swimming reversal frequency responses to photostimuli.
Membrane preparations
Membranes were prepared by sonication of H. salinarum cells grown to stationary phase in complex medium containing 1 μg/ml mevinolin as described (11). After low-speed centrifugation to remove unbroken cells and cell debris, the membranes were pelleted for 1 h at 147,000 × g in a Beckman OptimaTM L-100 XP ultracentrifuge and suspended in loading buffer (4 M NaCl, 25 mM Tris (pH 6.8)).
Purification and spin labeling of SRI and SRI-HtrI1-147 in detergent solution
The membranes were suspended with 1.2% dodecyl-β-maltopyranoside (DDM) in the binding buffer (4 M NaCl, 10 mM imidazole, 25 mM Tris (pH 6.8)) and allowed to solubilize overnight. The solubilized fraction was combined with QIAGEN Ni+-NTA agarose his-binding resin with an anticipated binding efficiency of ∼3.5 mg/ml (protein/resin) and gently shaken at 4°C for 2 h. The resin was washed with 10× bed-volume of 0.01% DDM in the binding buffer and subsequently suspended in the spin-labeling buffer containing 0.01% DDM, 200 μM (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (R1, Toronto Research Chemicals, North York, ON, Canada), 4 M NaCl, and 25 mM Tris (pH 6.8)) by gently shaking it at 4°C overnight. Excess labeling reagent was removed by washing with 10× bed-volume of the binding buffer containing 0.01% DDM. The purified and labeled hexahistidine-tagged proteins were eluted with the elution buffer containing 0.01% DDM (4 M NaCl, 250 mM imidazole, 25 mM Tris (pH 6.8)), and finally dialyzed to remove imidazole and change the buffer to pH 6.0 (4 M NaCl, 50 mM MES (pH 6.0)).
Labeling of monocysteine mutants with Lucifer yellow
Labeling with the thiol-reactive Lucifer yellow iodoacetamide (LY, Invitrogen) probe was done with purified SRI or SRI-HtrI1-147 (see above purification scheme), using excess LY (15× concentration) based on the SRI or SRI-HtrI1-147 concentration assessed by absorption spectroscopy using ɛ587 of 52,000 M−1cm−1. After overnight incubation of the LY labeling reaction at 4°C, excess LY was removed via overnight dialysis in a wash buffer (50 mM MES (pH 6.0), 4 M NaCl).
Spin labeling of monocysteine mutants in the native membrane
Before the spin-labeling of membranes, DTT was added to the samples (10 mM concentration) for 1 h. The samples were then washed via pelleting in a Beckman tabletop ultracentrifuge (147,000 × g for 15 min) four times with resuspension in 50 mM MES (pH 6.0), 4 M NaCl. Then excess (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (R1) (final concentration 200 μM) was added to the membrane suspension, and labeling was allowed to proceed overnight. The excess reagent was removed by pelleting and resuspending eight times in the same MES buffer using a Beckman tabletop ultracentrifuge.
Reconstitution of LY- and spin-labeled SRI and SRI-HtrI in proteoliposomes
LY- and R1-labeled SRI and SRI-HtrI1-147 were inserted into proteoliposomes by addition of purified detergent-solubilized (1.2% CHAPS) H. salinarum polar lipids (HPL) at a concentration of 5 mg HPL/1 A587 unit SRI in 1 ml buffer. Subsequent extensive dialysis removed the detergent and resulted in proteoliposomes, as assessed by turbidity of the sample. This dialysis process was accelerated by lowering the salt concentration of the wash buffer to 1 M NaCl, 50 mM MES (pH 6.0), for 3–5 days, after which turbidity developed. The samples were then dialyzed into a 4 M NaCl buffer (50 mM MES, pH 6.0) for 1 day.
Fluorescence spectroscopy
Fluorescence data were collected using a PTI (Photon Technologies International, Birmingham, NJ) spectrofluorometer. The software package used for data collection was PTI Felix32, with the acquisition set to collect time-based data with excitation and emission at 420 nm and 520 nm, respectively. To induce light-induced conformational changes of SRI, a Cole-Parmer 150-W tungsten-halogen illuminator was used with a 600 ± 20 nm interference filter.
EPR spectroscopy
EPR spectra of samples were measured in Los Angeles in a Bruker ELEXSYS 580 fitted with a high-sensitivity resonator in small volume aqueous flat cells (Wilmad Glass, Buena, NJ). Spectra were recorded at 100-G scan widths with an incident microwave power of 20 mW. Modulation amplitude (100 kHz) was optimized for maximum signal/noise ratio and was verified to produce minimal spectral distortions. Samples were equilibrated in the dark at room temperature for 30 min before the spectrum was collected. Typically, nine scans were signal-averaged per spectrum, and the total collection time did not exceed 5 min.
EPR spectra were recorded in Houston on a Bruker EMX spectrometer using a modulation amplitude of 2.0 G, a modulation frequency of 100 kHz, a microwave power of 1 mW, and a temperature between 270 and 318 K. The temperature was controlled with a heater and cold nitrogen gas provided through a silver-coated double-jacketed glass transfer line, and a BVT3000 temperature controller was used. Data acquisition was conducted using WinEPR.
RESULTS AND DISCUSSION
Motility behavior phenotypes
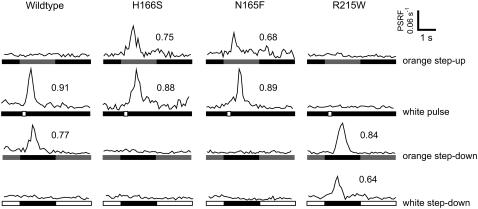
Phototaxis results from transient changes in the probability of cells reorienting their swimming direction as they encounter changes in light intensity (5). For example, if a cell swims down a spatial gradient of attractant orange light, the decrease in intensity induces it to reverse its swimming direction, thereby biasing its migration to higher intensities of orange light. Temporal changes in light intensity mimic spatial gradients and cause transient reversal frequency changes. For example, turning off an orange light induces swimming reversals for 2–3 s in a wild-type cell population. Such reversal frequency changes are transient because the cells adapt in several seconds to the new light condition. The transducer methylation/demethylation system rapidly modifies the Htr transducers to ensure that the cells exhibit reversal frequencies close to the value in the dark in constant light of any spectral composition, and the cells respond only to changes in light intensity. The SRI-HtrI complex produces attractant signals to one-photon activation by orange light. Therefore, when cells experience a temporal step-up in orange light intensity, their probability of reversing their swimming directions briefly decreases, and the cells adapt and return to near their dark swimming reversal probabilities (5). In their adapted state, a step down from orange light to darkness causes induction of reversals, followed by a return to the spontaneous reversal frequency in the dark. The wild-type strain used here exhibits a very low spontaneous reversal frequency (∼0.05/s), which is convenient because the step down reversal-induction response is a large and easily quantifiable measure of the attractant signal (i.e., its disappearance). The wild-type response to “orange step-down” results in the well-resolved peak of reversals in Fig. 1. The wild-type response to a brief pulse of white light is evident as a similar reversal-induction peak (Fig. 1) because white light causes two-photon activation of SRI and generates a repellent signal. The opposite stimuli, a step-up in orange light and a step-down in white light, have been shown to suppress reversal frequency in higher-reversal-frequency strains, but in the already low spontaneous reversal frequency of the strain used here, negligible suppression response is evident.
FIGURE 1.
Phototaxis responses of wild-type and behavioral mutants. Phototaxis responses of wild-type SRI-HtrI, the newly found omniphobic mutant N165F, and the previously described omniphobic H166S and omniphilic R215W are shown. One second after initiation of data collection, the cells were exposed from top to bottom to a 2-s stimulus of orange light (600 ± 20 nm), a 200-ms pulse of white light (>350 nm; heat-filtered only), a 2-s removal of continuous (30-s) orange light, or a 2-s removal of continuous (30-s) white light. The vertical axis is the population swimming reversal frequency (PSRF) calculated in the CellTrak 1.2 Beta software package as the Rate of Change of Direction (RCD)/Linear Speed (SPD), calibrated with values of the fraction of cells in the population that undergo swimming reversals assessed by individual cell tracking. For each reversal induction response, the area of the peak, which equals the fraction of the population reversing in the 2-s period following the stimulus, is shown.
The SRI mutants H166S and N165F exhibit the opposite response as wild-type cells to orange light: the step up in orange light results in reversal-induction peaks (Fig. 1). The mutants respond to a brief pulse of white light as a repellent stimulus similarly to wild-type. Therefore, both one-photon activation of SRI (by orange light) and two-photon activation (by white light) produce repellent signals in these mutants, which are therefore called “omniphobic” mutants. In contrast, the SRI R215W mutant, originally isolated as a suppressor of the omniphobic phenotype, exhibits similar responses as wild type to orange light step-up and step-down stimuli but shows inverted responses compared with wild type to white light stimuli. Both orange step-down and white step-down stimuli induced reversal peaks in the R215W mutant; i.e., both one-photon and two-photon stimuli produce attractant signals. Therefore, R215W is classified as “omniphilic.”
A possible explanation for the behavioral phenotypes is that the wild-type SRI-HtrI interface exists in an equilibrium mixture of two conformations, reversal-inducing (R) and reversal-suppressing (A), and omniphobic and omniphilic mutants have dark equilibria that are far biased toward one or the other of the conformations; in other words, they are constitutively activated for one-photon and two-photon signaling, respectively. If both conformations are present in photoproducts in the behavioral mutants, then an extreme bias in the dark toward A or R would imply that photoreactions would produce only repellent responses or attractant responses, respectively. A more mixed distribution of conformers in the photoproducts compared with the dark state would occur if the mutations exert a greater effect on the equilibrium in the less dynamic unphotolyzed state of the complex.
The experiments reported here test predictions from the two-conformation equilibrium model regarding the dark states of the omniphobic and omniphilic mutants. In order to probe the conformations of the mutant SRI-HtrI complexes, we sought to 1), develop a purified SRI-HtrI system that preserves the native structure of the complex, 2), identify a residue position in the wild-type complex sensitive to the conformation of the SRI-HtrI interface, and 3), attach a probe at this position to report on the interface conformation.
Development of a proteoliposome system for the SRI-HtrI complex
A criterion for native SRI interaction with HtrI is provided by the strong effect of HtrI on the pH dependence of the SRI photocycle (15). In this measurement and subsequent measurements using site-directed fluorescence labeling (SDFL) and site-directed spin labeling, a truncated version of the transducer (HtrI1-147) was used to reduce the protein content of the proteoliposome suspensions, unlike in the motility measurements, which required the full-length HtrI containing kinase-binding and methylation domains. HtrI1-147 contains the interaction sites with SRI that influence SRII absorption properties and photochemistry (16), and chimera analysis (17) shows that specific HtrI interaction sites with SRI are localized in the membrane and membrane-proximal domain, which are contained in HtrI1-147. A fusion complex in which the C-terminus of SRI is connected by a flexible linker to the N-terminus of HtrI1-147 was constructed and expressed in H. salinarum for purification. A similar fusion with full-length HtrI has been shown to mediate wild-type phototaxis responses and with full-length and truncated HtrI to exhibit known spectroscopically detectable effects of HtrI on SRI (12,18).
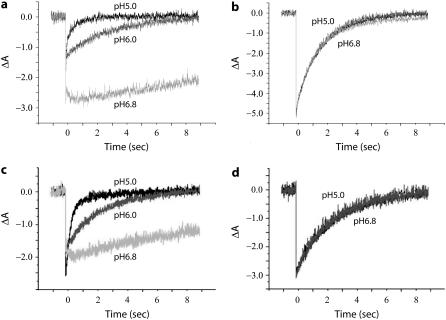
A proteoliposome system was designed to provide a lipid environment similar to the native H. salinarum lipid bilayer. The photocycle of free SRI in H. salinarum native membranes exhibits a ∼10-fold slower rate at pH 6.8 compared with pH 5.0 (Fig. 2 a), and when HtrI1-147 is bound to SRI in the cell membranes, the rate becomes nearly completely pH-independent (Fig. 2 b). In proteoliposomes, purified free SRI and SRI-HtrI1-147 complex mimic this behavior (Fig. 2, c and d), thereby meeting this criterion for fidelity of interaction of SRI and HtrI.
FIGURE 2.
Flash photolysis of HtrI-free SRI and SRI-HtrI1-147 fusion in membranes and proteoliposomes. Flash-induced absorption transients at 590 nm in (a) HtrI-free SRI in H. salinarum membrane vesicles, (b) SRI-HtrI1-147 fusion in H. salinarum membranes vesicles, (c) purified HtrI-free SRI in reconstituted proteoliposomes, and (d) purified SRI-HtrI1-147 fusion in reconstituted proteoliposomes. The flash was a 6-ns 532-nm laser pulse (40 mJ) triggered at time 0.
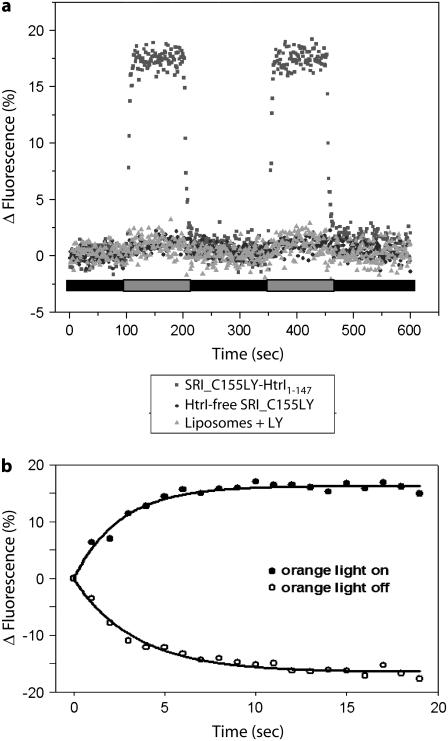
Fluorescent probes and the nitroxide side chain R1 show SRI position 155 in the SRI-HtrI complex to be a conformationally active site. In SRII, Ser154 on the cytoplasmic end of F was found to be conformationally active in the SRII-HtrII complex in detergent micelles by the light-dependent, HtrII-dependent effects on LY probe accessibility in the S154C SRII mutant (19). Ser154 was concluded to be part of the SRII/HtrII interface because S154C-Iaedans was found to be within Förster resonance energy-transfer distances of Trp residues introduced into HtrII in the membrane-proximal extension of the second transmembane helix (TM2) of HtrII (19). The corresponding residue Ser155 in SRI is similarly conformationally active as shown by the fluorescence changes of C155LY on photoactivation of SRI with >600-nm light only when HtrI1-147 is present (Fig. 3). The conformational change detected by LY at position 155 rises (t½ 2.6 s) and decays (t½ 2.4 s) with first-order rates matching the steady-state accumulation of S373 and return of SRI587, respectively, in the photocycle (Fig. 3 b), as expected from the prior identification of S373 as the one-photon cycle signaling state (14). Therefore, we proceeded to introduce the R1 nitroxide side chain (Fig. 4, inset) at position 155 to monitor the interface properties of the SRI-HtrI1-147 complex.
FIGURE 3.
HtrI-dependent light-induced fluorescence change in the SRI-HtrI complex. (a) Real-time course of >600-nm-light activated changes in fluorescence of LY-iodoacetamide labeled at residue S155C of SRI-HtrI1-147 and HtrI-free SRI, and liposomes with free LY probe. Transients were recorded from reconstituted HPL-liposomes at room temperature. (b) Kinetics of fluorescence changes of LY-labeled SRI C155-HtrI1-147 samples in proteoliposomes: open circles, orange light on; closed circles, orange light off.
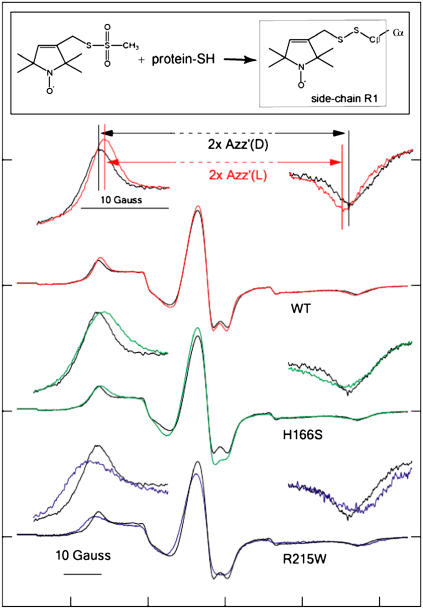
FIGURE 4.
EPR spectra of R1 nitroxide side chain labeled at residue S155C of SRI-HtrI1-147 wild type and mutants. The structure of the R1 probe (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate is shown in the inset. Spectra from wild-type complex in the dark state are shown in black for each pair of spectra. In the uppermost pair of full spectra, the spectrum from the light-activated state is shown in red. H166S (green) and R215W (blue) exhibit outermost peaks shifted in opposite directions relative to those of the dark wild-type complex. All spectra were recorded in reconstituted HPL-liposomes at ambient temperature. The outermost peaks are expanded above each spectrum. 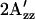 is defined as the distance between the outermost peaks.
is defined as the distance between the outermost peaks.
The EPR spectrum of C155R1-HtrI1-147 measured in the light has a splitting of the outer hyperfine extrema (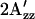 ; Fig. 4 a) significantly smaller than that of the EPR spectrum in the dark state. The decrease in
; Fig. 4 a) significantly smaller than that of the EPR spectrum in the dark state. The decrease in 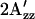 in the spectrum of the illuminated sample indicates mobility changes in R1, changes in local polarity, or both. In any case, a change in local structure near 155 is indicated. In terms of the model, the
in the spectrum of the illuminated sample indicates mobility changes in R1, changes in local polarity, or both. In any case, a change in local structure near 155 is indicated. In terms of the model, the 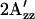 value provides us a measure of the relative amounts of A and R conformers, a smaller
value provides us a measure of the relative amounts of A and R conformers, a smaller 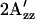 corresponding to shift of the conformer distribution toward A and a larger
corresponding to shift of the conformer distribution toward A and a larger 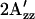 corresponding to a relative increase in R.
corresponding to a relative increase in R.
EPR spectra in omniphilic and omniphobic mutants of the SRI-HtrI complex
The key tenet of the model is that omniphobic and omniphilic mutants of the SRI-HtrI complex will be shifted in the dark into the A and R conformers, respectively. Therefore, the model predicts: 1), a smaller 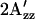 value in omniphobic mutants compared with wild-type in the dark; 2), a similar
value in omniphobic mutants compared with wild-type in the dark; 2), a similar 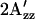 value in omniphobic mutants as in light-activated wild-type complex; 3), a larger
value in omniphobic mutants as in light-activated wild-type complex; 3), a larger 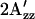 value in omniphilic mutants compared with wild-type in the dark; and 4), combining omniphilic and omniphobic mutations should return their shifted
value in omniphilic mutants compared with wild-type in the dark; and 4), combining omniphilic and omniphobic mutations should return their shifted 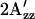 values to near that of wild type in the dark. Predictions 1–3 are confirmed in the EPR spectra of the omniphobic H166S and the omniphilic R215W (Fig. 4). The
values to near that of wild type in the dark. Predictions 1–3 are confirmed in the EPR spectra of the omniphobic H166S and the omniphilic R215W (Fig. 4). The 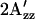 in H166S is reduced relative to WT similar to the change upon S373 formation in wild type, as expected if the conformation of H166S in the dark state is similar to that of the attractant signaling state in wild type. On the other hand,
in H166S is reduced relative to WT similar to the change upon S373 formation in wild type, as expected if the conformation of H166S in the dark state is similar to that of the attractant signaling state in wild type. On the other hand, 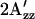 in R215W is increased relative to WT, suggesting that the conformational change caused by the R215W mutation is opposite of that occurring on the attractant signaling-state formation.
in R215W is increased relative to WT, suggesting that the conformational change caused by the R215W mutation is opposite of that occurring on the attractant signaling-state formation.
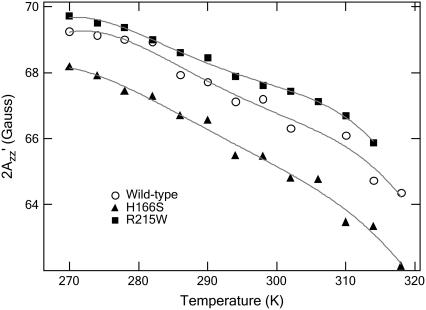
The relative differences of 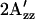 value among wild-type, H166S, and R215W are unchanged throughout the temperature range between 270 and 318 K (−3°C and 45°C) (Fig. 5), although the absolute values decrease for all of them at higher temperature because of the mobility increase with higher temperature expected from the increase in thermal fluctuations of the molecules. Our interpretation is that the differences among conformations of the three SRI-HtrI complexes are maintained throughout this temperature range. At the lowest temperature studied (270 K), where thermally activated motion is substantially reduced, differences in
value among wild-type, H166S, and R215W are unchanged throughout the temperature range between 270 and 318 K (−3°C and 45°C) (Fig. 5), although the absolute values decrease for all of them at higher temperature because of the mobility increase with higher temperature expected from the increase in thermal fluctuations of the molecules. Our interpretation is that the differences among conformations of the three SRI-HtrI complexes are maintained throughout this temperature range. At the lowest temperature studied (270 K), where thermally activated motion is substantially reduced, differences in 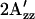 remain, suggesting that the origin of the effect has an important contribution from differences in polarity of the local environment.
remain, suggesting that the origin of the effect has an important contribution from differences in polarity of the local environment.
FIGURE 5.
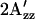 values measured at various temperatures, for the dark states of wild-type (○), H166S (▴), and R215W (▪). The temperature was controlled with a heater and a stream of cold N2 gas, the amount of which was regulated with a BVT 3000 temperature controller.
values measured at various temperatures, for the dark states of wild-type (○), H166S (▴), and R215W (▪). The temperature was controlled with a heater and a stream of cold N2 gas, the amount of which was regulated with a BVT 3000 temperature controller.
The EPR spectra of SRI C155R1 can be understood in terms of the location of that site at a contact region with the transducer, as indicated for the homologous Ser154 in the SRII-HtrII complex (19). Interaction with HtrI would explain the strongly immobilized component, whereas a more mobile component revealed by simulation (results not shown) can be accounted for by a second rotamer of the side chain. In the context of this model, the effect of the SRI mutations on the EPR spectra would be interpreted as resulting from a displacement of helix F in which Ser155 resides. This is plausible because, based on comparison of the SRI sequence to that of the SRII crystal structure (28), the mutations are in helix F or in contact with it. The EPR spectral changes apparently have a large component caused by polarity changes in the local environment, which also fits the model because a movement of the helix would give rise to a new local environment of Ser155 but not necessarily to changes in mobility because Ser155 would still be buried at an interface.
Some of the monocysteine SRI mutants (D201N, D201N/R215W, and H166S/R215W) prepared in this study are unstable in the detergent-solubilized conditions used in the process of proteoliposome reconstitution and, therefore, were labeled with R1 and examined in their native H. salinarum membranes; the results are summarized in Fig. 6. The EPR spectra of SRI C155R1 and the corresponding H166S and R215W SRI mutants in membrane preparations from H. salinarum exhibit the same opposite direction of changes of 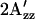 from wild-type as in proteoliposomes, although amplitudes of changes in the membrane samples are less reproducible in different preparations, possibly because of probes attached to other proteins.
from wild-type as in proteoliposomes, although amplitudes of changes in the membrane samples are less reproducible in different preparations, possibly because of probes attached to other proteins.
FIGURE 6.
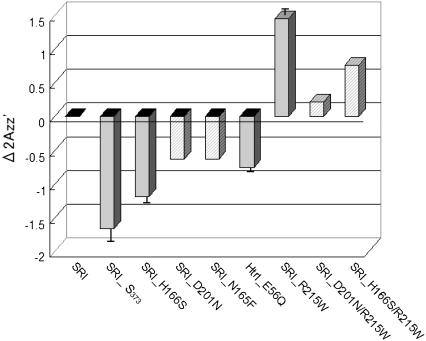
Shifts of 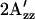 value relative to the SRI-HtrI1-147 wild-type in the dark. The light-activated wild-type SRI-HtrI1-147 complex, the omniphobic mutants H166S, D201N, and N165F of SRI, and E56Q of HtrI each show a decreased
value relative to the SRI-HtrI1-147 wild-type in the dark. The light-activated wild-type SRI-HtrI1-147 complex, the omniphobic mutants H166S, D201N, and N165F of SRI, and E56Q of HtrI each show a decreased 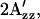 whereas the omniphilic mutant R215W and the double mutants R215W/H166S and R215/D201N of SRI, which exhibit the omniphilic phenotype, each show an increase in
whereas the omniphilic mutant R215W and the double mutants R215W/H166S and R215/D201N of SRI, which exhibit the omniphilic phenotype, each show an increase in 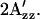 For wild-type, H166S, E56Q, and R215W (solid gray bars), proteoliposomes were used, whereas for D201N, N165F, R215W/H166S, and R215/D201N (hatch-marked bars) H. salinarum membrane vesicles were used (see text). The error bars indicate the standard deviation of two or three independent measurements.
For wild-type, H166S, E56Q, and R215W (solid gray bars), proteoliposomes were used, whereas for D201N, N165F, R215W/H166S, and R215/D201N (hatch-marked bars) H. salinarum membrane vesicles were used (see text). The error bars indicate the standard deviation of two or three independent measurements.
In Fig. 6, the relative 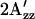 values at room temperature for other inverted mutants and double mutants of omniphobic and omniphilic mutants are compared with that of dark-state wild type in proteoliposomes. All of the omniphobic mutants investigated show decreases in
values at room temperature for other inverted mutants and double mutants of omniphobic and omniphilic mutants are compared with that of dark-state wild type in proteoliposomes. All of the omniphobic mutants investigated show decreases in 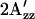 values (smaller hyperfine splitting) like the attractant signaling state of wild type, whereas addition of the mutation R215W, which suppresses the omniphobic behavioral phenotype in the omniphobic-mutant background, increases the
values (smaller hyperfine splitting) like the attractant signaling state of wild type, whereas addition of the mutation R215W, which suppresses the omniphobic behavioral phenotype in the omniphobic-mutant background, increases the 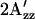 values relative to the dark-state wild type, confirming prediction 4.
values relative to the dark-state wild type, confirming prediction 4.
Light activation of the omniphilic and the omniphobic mutant receptors showed only very small changes in EPR spectra as compared with those of their dark states, and their 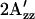 shifts were too small to be resolved (data not shown). However, the intensity of the central hyperfine lines, which increases and decreases when the probe becomes more mobile or immobile, respectively (see Fig. 4), exhibited changes in the opposite direction between R215W and H166S, indicating that the former becomes more mobile, whereas the latter becomes less mobile, as expected from the two-state model. The smaller change in the spectra during illumination of the mutant complexes as compared with that of wild-type complex may be a result of greater instability of the photoactivated conformations in the mutants, which would make them too short-lived to be accumulated under our illumination conditions.
shifts were too small to be resolved (data not shown). However, the intensity of the central hyperfine lines, which increases and decreases when the probe becomes more mobile or immobile, respectively (see Fig. 4), exhibited changes in the opposite direction between R215W and H166S, indicating that the former becomes more mobile, whereas the latter becomes less mobile, as expected from the two-state model. The smaller change in the spectra during illumination of the mutant complexes as compared with that of wild-type complex may be a result of greater instability of the photoactivated conformations in the mutants, which would make them too short-lived to be accumulated under our illumination conditions.
CONCLUDING REMARKS
The results reported above provide support for a two-state metastable equilibrium model in the dark state of the SRI-HtrI complex that can explain the mutation-induced altered behavioral responses of H. salinarum cells. The data obtained confirm specific predictions of structural differences between the wild-type complex and the mutants by monitoring the structure at a conformation-sensitive position in the complex. Because the data concern structure of the complex, the confirmation is completely independent of the cell behavior observations that the model was designed to explain and therefore provides a strong argument that the basic tenets of the model are correct.
Attractant responses and repellent responses in H. salinarum result from inhibition and activation, respectively, of the CheA histidine-kinase that binds to the cell's taxis transducers, HtrI in the case of SRI (20). Therefore, the two conformations of the model represent conformations of the SRI-HtrI interface corresponding to a CheA kinase-inhibiting structure (A) and a CheA kinase-activating structure (R). Photostimuli depending on their spectral composition as well as different mutations perturb the dark equilibrium so that the net change in kinase activity caused by one-photon or two-photon activation is positive, causing induction of swimming reversals, or negative, suppressing them.
In the proton pump BR from H. salinarum, light activation causes outward tilting of the cytoplasmic half of helix F as seen by cryo-electron-microscopy and EPR spectroscopy (21–25). Also in SRII from N. pharaonis, F-helix movement causes movement of helix F in the membrane boundary region, consistent with the conformational change in BR, according to spin-label studies (26,27). Therefore, different positions of the F helix cytoplasmic domain are likely to exist between the A and R conformers of SRI, and differing mobility of a probe at the 155 position at the cytoplasmic end of helix F, as we observe in the two conformers, is consistent with this expectation.
Based on comparison of SRI and HtrI with the crystal structures of the homologous sensory rhodopsin II (SRII) and the SRII-HtrII complex (28,29) and a model of the membrane proximal domain of the SRII-HtrII complex derived from site-directed spin labeling (30), the omniphobic and omniphilic mutation sites (except for D201N) are localized in the predicted SRI-HtrI binding interface, namely, on the cytoplasmic half of helices F (H166S, N165F) and G (R215W) of SRI and the HAMP domain of HtrI (E56Q), which may face SRI. The mutations may therefore alter the interface structure to produce the altered signaling phenotypes observed, in line with A and R conformers being characterized by two different conformations of the SRI-HtrI interface. Photoactivation of SRI is expected to perturb the interface structure in the process of signal relay from SRI to HtrI, which in terms of the model would destabilize the mutationally biased conformations of A and R conformers to induce transient shifts toward the other conformer, resulting in repellent and attractant phototaxis responses.
The two-state model proposed here is a simple model that fits both the behavioral phenotypes of the mutants and the shifts of the hyperfine lines in the EPR spectra of the mutants. An alternative explanation of the aberrant signaling by the mutant receptor-transducer complexes might be based on mutation-induced alterations in the photochemistry of SRI. In this alternative model, omniphilic and omniphobic mutants would accumulate different photointermediates, which deliver attractant and repellent signals, respectively, on photoexcitation. Two of the inverted mutants, H166S and D201N, exhibit greatly altered photocycles in which Schiff base deprotonation (i.e., M formation) is greatly reduced. However, the omniphilic R215W mutant and omniphobic SRI-N165F and HtrI-E56Q mutants all exhibit optically detected photocycle intermediates nearly identical to that of wild type, arguing against altered photochemistry causing the mutant signaling phenotypes. Because spectroscopic properties of the intermediate states are not correlated with the signaling phenotypes, explanations based on photointermediate states would need to further assume that only spectrally silent changes are responsible for the differences.
The EPR spectra demonstrate that the dark conformations of the mutants and wild-type receptor-transducer complexes are different, supporting the idea that the starting conformation in the dark determines the signal (attractant or repellent) generated by photoactivation. The model presented here assumes altered equilibria between only two conformers, A and R, in the dark state. However the data do not exclude the more complicated possibility of multiple conformations in the dark, with the wild-type complex having an average conformation intermediate between those of the attractant and repellent signaling forms, which may each contain multiple conformational states. Multiple conformations of the mutant complexes might explain the broader outermost bands of the EPR spectra for mutants compared with the wild type, which would be expected to be narrower if the wild-type dark state contains two conformers and the mutants contain primarily only one.
More comprehensive spin-labeling and other structural methods are needed to define the structural changes in the SRI-HtrI complex between the A and R conformations, and the omniphilic and omniphobic mutants, being biased toward one or the other conformer in the dark, provide valuable tools for such analysis.
Acknowledgments
This work was supported by National Institutes of Health Grants R37GM27750 (J.L.S.), R01EY05216 (W.L.H.), and R01GM56818 (A.L.T.), the Ford Bundy and Anne Smith Bundy Foundation (W.L.H.), a Jules Stein Professorship (W.L.H.), and a Robert A. Welch Distinguished Chair (J.L.S.).
Jun Sasaki and Brian J. Phillips contributed equally to this work.
References
- 1.Spudich, J. L., and R. A. Bogomolni. 1984. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 312:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao, V. J., and J. L. Spudich. 1992. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc. Natl. Acad. Sci. USA. 89:11915–11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz, T. E., I. Szundi, J. L. Spudich, and R. A. Bogomolni. 2000. New photointermediates in the two photon signaling pathway of sensory rhodopsin-I. Biochemistry. 39:15101–15109. [DOI] [PubMed] [Google Scholar]
- 4.Lutz, I., A. Sieg, A. A. Wegener, M. Engelhard, I. Boche, M. Otsuka, D. Oesterhelt, J. Wachtveitl, and W. Zinth. 2001. Primary reactions of sensory rhodopsins. Proc. Natl. Acad. Sci. USA. 98:962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoff, W. D., K.-H. Jung, and J. L. Spudich. 1997. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 26:223–258. [DOI] [PubMed] [Google Scholar]
- 6.Jung, K.-H., and J. L. Spudich. 1998. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J. Bacteriol. 180:2033–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kataoka, M., H. Kamikubo, F. Tokunaga, L. S. Brown, Y. Yamazaki, A. Maeda, M. Sheves, R. Needleman, and J. K. Lanyi. 1994. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J. Mol. Biol. 243:621–638. [DOI] [PubMed] [Google Scholar]
- 8.Spudich, J. L., and J. K. Lanyi. 1996. Shuttling between two protein conformations: the common mechanism for sensory transduction and ion transport. Curr. Opin. Cell Biol. 8:452–457. [DOI] [PubMed] [Google Scholar]
- 9.Spudich, E. N., W. Zhang, M. Alam, and J. L. Spudich. 1997. Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp73 intrahelical salt bridge. Proc. Natl. Acad. Sci. USA. 94:4960–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogomolni, R. A., W. Stoekenius, I. Szundi, E. Perozo, K. D. Olson, and J. L. Spudich. 1994. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc. Natl. Acad. Sci. USA. 91:10188–10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson, K. D., X.-N. Zhang, and J. L. Spudich. 1995. Residue replacements of buried aspartyl and related residues in sensory rhodopsin I: D201N produces inverted phototaxis signals. Proc. Natl. Acad. Sci. USA. 92:3185–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X., and J. L. Spudich. 2002. Demonstration of 2:2 stoichiometry in the functional SRI-HtrI signaling complex in Halobacterium membranes by gene fusion analysis. Biochemistry. 41:3891–3896. [DOI] [PubMed] [Google Scholar]
- 13.Chen, B., and A. E. Przybyla. 1994. An efficient site-directed mutagenesis method based on PCR. Biotechniques. 17:657–659. [PubMed] [Google Scholar]
- 14.Yan, B., and J. L. Spudich. 1991. Evidence that the repellent receptor form of sensory rhodopsin I is an attractant signaling state. Photochem. Photobiol. 54:1023–1026. [DOI] [PubMed] [Google Scholar]
- 15.Spudich, E. N., and J. L. Spudich. 1993. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J. Biol. Chem. 268:16095–16097. [PubMed] [Google Scholar]
- 16.Yao, V. J., E. N. Spudich, and J. L. Spudich. 1994. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J. Bacteriol. 176:6931–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, X.-N., J. Zhu, and J. L. Spudich. 1999. The specificity of interaction of archaeal transducers with their cognate sensory rhodopsins is determined by their transmembrane helices. Proc. Natl. Acad. Sci. USA. 96:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, X., and J. L. Spudich. 2004. Five residues in the HtrI transducer membrane-proximal domain close the cytoplasmic proton-conducting channel of sensory rhodopsin I. J. Biol. Chem. 279:42964–42969. [DOI] [PubMed] [Google Scholar]
- 19.Yang, C. S., O. Sineshchekov, E. N. Spudich, and J. L. Spudich. 2004. The cytoplasmic membrane-proximal domain of the HtrII transducer interacts with the E-F loop of photoactivated Natronomonas pharaonis sensory rhodopsin II. J. Biol. Chem. 279:42970–42976. [DOI] [PubMed] [Google Scholar]
- 20.Rudolph, J., and D. Oesterhelt. 1995. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 14:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam, S., M. Gerstein, D. Oesterhelt, and R. Henderson. 1993. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramaniam, S., and R. Henderson. 2000. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 406:653–657. [DOI] [PubMed] [Google Scholar]
- 23.Steinhoff, H. J., R. Mollaaghababa, C. Altenbach, K. Hideg, M. Krebs, H. G. Khorana, and W. L. Hubbell. 1994. Time-resolved detection of structural changes during the photocycle of spin-labeled bacteriorhodopsin. Science. 266:105–107. [DOI] [PubMed] [Google Scholar]
- 24.Vonck, J. 1996. A three-dimensional difference map of the N intermediate in the bacteriorhodopsin photocycle: part of the F helix tilts in the M to N transition. Biochemistry. 35:5870–5878. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, W., L. S. Brown, R. Needleman, J. K. Lanyi, and Y. K. Shin. 2000. Light-induced rotation of a transmembrane alpha-helix in bacteriorhodopsin. J. Mol. Biol. 304:715–721. [DOI] [PubMed] [Google Scholar]
- 26.Wegener, A. A., I. Chizhov, M. Engelhard, and H. J. Steinhoff. 2000. Time-resolved detection of transient movement of helix F in spin-labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 301:881–891. [DOI] [PubMed] [Google Scholar]
- 27.Wegener, A. A., J. P. Klare, M. Engelhard, and H. J. Steinhoff. 2001. Structural insights into the early steps of receptor-transducer signal transfer in archaeal phototaxis. EMBO J. 20:5312–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luecke, H., B. Schobert, J. K. Lanyi, E. N. Spudich, and J. L. Spudich. 2001. Crystal structure of sensory rhodopsin II at 2.4 A: insights into color tuning and transducer interaction. Science. 293:1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordeliy, V. I., J. Labahn, R. Moukhametzianov, R. Efremov, J. Granzin, R. Schlesinger, G. Buldt, T. Savopol, A. J. Scheidig, J. P. Klare, and M. Engelhard. 2002. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 419:484–487. [DOI] [PubMed] [Google Scholar]
- 30.Bordignon, E., J. P. Klare, M. Doebber, A. A. Wegener, S. Martell, M. Engelhard, and H. J. Steinhoff. 2005. Structural analysis of a HAMP domain: the linker region of the phototransducer in complex with sensory rhodopsin II. J. Biol. Chem. 280:38767–38775. [DOI] [PubMed] [Google Scholar]