Abstract
Elongation factor Tu (EF-Tu) exhibits significant specificity for the different elongator tRNA bodies in order to offset its variable affinity to the esterified amino acid. Three X-ray cocrystal structures reveal that while most of the contacts with the protein involve the phosphodiester backbone of tRNA, a single hydrogen bond is observed between the Glu390 and the amino group of a guanine in the 51–63 base pair in the T-stem of tRNA. Here we show that the Glu390Ala mutation of Thermus thermophilus EF-Tu selectively destabilizes binding of those tRNAs containing a guanine at either position 51 or 63 and that mutagenesis of the 51–63 base pair in several tRNAs modulates their binding affinities to EF-Tu. A comparison of Escherichia coli tRNA sequences suggests that this specificity mechanism is conserved across the bacterial domain. While this contact is an important specificity determinant, it is clear that others remain to be identified.
Keywords: minor groove interaction, RNA–protein interaction, base specific
INTRODUCTION
Elongation factor Tu (EF-Tu) is the bacterial G-protein responsible for catalyzing the efficient delivery of aminoacyl-tRNA (aa-tRNA) to the ribosome during translation. When complexed with GTP, EF-Tu binds elongator aa-tRNA with high affinity to form a ternary complex that subsequently binds the ribosomal entry site. EF-Tu binds all elongator aa-tRNAs with roughly equivalent affinities (Louie et al. 1984; Ott et al. 1990b). However, subsequent studies using misacylated tRNAs have shown that EF-Tu exhibits substantial specificity for both the tRNA body and the esterified amino acid (LaRiviere et al. 2001; Asahara and Uhlenbeck 2002; Dale et al. 2004). The affinities for the tRNA bodies and amino acids are arranged in such a way that correctly aminoacylated tRNA binds EF-Tu uniformly, presumably to compensate for the variable thermodynamic contribution of the esterified amino acids. Thus, the binding of EF-Tu to tRNA is sequence specific. At this point, the mechanism by which EF-Tu generates specificity for its wide range of elongator tRNA substrates is unknown.
The binding affinities of three Escherichia coli aa-tRNAs and one yeast aa-tRNA to 20 point mutations of the tRNA binding cleft of T. thermophilus EF-Tu revealed that the specificity of the protein for the different tRNA bodies was due to the varying thermodynamic contribution of only five amino acids (Sanderson and Uhlenbeck 2007b). A comparison of these data to the X-ray cocrystal structures of Thermus aquaticus EF-Tu bound to yeast Phe-tRNAPhe reveals that three of the five “specificity” amino acids contact sites on the tRNA that vary in sequence among the four test aa-tRNAs. This suggested that at least part of the mechanism of specificity involved a discrete set of amino acids that differentially bind certain tRNA sequence elements. The largest specificity effect is the Glu390Ala mutation, which caused a >2.8 kcal/mol range in binding affinity among the four aa-tRNAs tested. Interestingly, the cocrystal structures indicate that Glu390 can make a base-specific contact with a guanine in the minor groove of the 51–63 base pair (Nissen et al. 1995, 1999). The goal of this work is to establish the thermodynamic details of this contact to determine whether the 51–63 base pair contributes to the specificity of EF-Tu for all tRNA sequences.
RESULTS
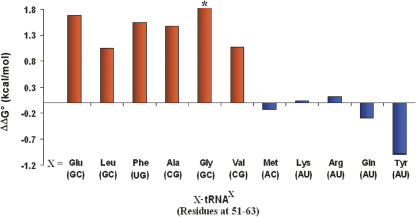
To extend the previous observations that Glu390 contributes to the specificity of the tRNA body, the binding affinities of 11 different E. coli aa-tRNAs were compared using both wild-type T. thermophilus EF-Tu and the corresponding Glu390Ala mutant. (Fig. 1; Table 1). For six of the 11 aa-tRNAs tested, the Glu390Ala mutation resulted in a 1.0 kcal/mol or greater decrease in the binding free energy. In contrast, four aa-tRNAs bound the Glu390Ala mutant with a free energy similar to native EF-Tu. Finally, in the case of Tyr-tRNATyr, the Glu390Ala mutation actually stabilized the binding by 1.0 kcal/mol compared to the wild-type protein. Among the 11 aa-tRNAs tested, mutation caused a total observed range of binding free energy of >2.8 kcal/mol. The magnitude of this effect is a substantial fraction of the 3.6 kcal/mol range of affinity observed among 19 different E. coli tRNAs acylated with valine (Asahara and Uhlenbeck 2002).
FIGURE 1.
The effect of the EF-Tu Glu390Ala mutation on the binding of cognate E. coli aa-tRNAs with (red) and without (blue) a guanosine in the 51–63 base pair. ΔΔG° is the ΔG° of wild-type EF-Tu minus the ΔG° for the Glu390Ala mutant protein measured in 0.5 M NH4Cl, 20 mM MgCl2, 50 mM HEPES (pH 7.0) at 4°C. Listed below the tRNA is the 51–63 base pair. The asterisk indicates where binding to the Glu390Ala protein was too weak to be accurately measured.
TABLE 1.
Binding of T. thermophilus EF-Tu to G51I E. coli Leu-tRNALeu
The sequences of the 11 E. coli aa-tRNAs that were tested reveal a strong correlation between the presence of a guanine in the 51–63 base pair and the effect of the Glu390Ala mutation on the binding affinity. The six tRNAs with either a G51–C63, C51–G63, or U51–G63 base pair all exhibited a decrease in binding affinity, while the five tRNAs with an A51–U63 or A51–C63 base pair showed either no effect or an improvement in binding affinity. Thus, Glu390 is capable of making a thermodynamically productive contact with a guanine on either side of the 51–63 base pair irrespective of whether it pairs with a cytosine or uracil. While this correlation is striking, it is clear that the results depend to some degree on elements outside the 51–63 base pair, since the magnitude of the effects are not uniform. For example, the ΔΔG° for tRNAs containing a G51–C63 pair range from 1.1 to >1.8 kcal/mol, and those containing an A51–U63 base pair ranged from 0 to −1.0 kcal/mol.
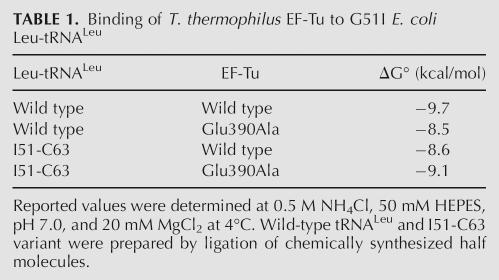
The available cocrystal structures of aa-tRNA with EF-Tu are consistent with the binding data. In the structure of T. aquaticus EF-Tu complexed with yeast Phe-tRNAPhe (Fig. 2A,B), the carboxylate of Glu390 extends into the minor groove of the T-stem to within 3.4 Å of the N2 amino group of G51, which could potentially form a hydrogen bond (Nissen et al. 1995). Indeed, in the unpublished structure of E. coli EF-Tu complexed with kirromycin and yeast Phe-tRNAPhe (PDB: 1OB2), Glu390 is within 2.7 Å of G51 (Fig. 2C). In addition, as shown in Figure 2D, the structure of T. aquaticus EF-Tu bound to E. coli Cys-tRNACys shows that Glu390 is within 2.9 Å of the N2 amino group of G63 (Nissen et al. 1999). Thus, this is a classic example of the sequence-specific recognition in the minor groove of a nucleic acid duplex by a protein by a mechanism that was initially predicted by Seeman et al. (1976). The amino group of guanine in a G–C base pair in a helix is nearly identical to its position in a C–G base pair, while the A–U and U–A pairs lack a functional group at this location.
FIGURE 2.
Crystallographic details of Glu390 contacting the 51–63 base pair. (A) General location and (B) detail of the contact of T. aquaticus EF-Tu•GDPPNP bound to yeast Phe-tRNAPhe (PDB:1TTT). (C) E. coli EF-Tu•GDPPNP complexed with kirromycin and yeast Phe-tRNAPhe (PDB: 1OB2). (D) T. aquaticus EF-Tu•GDPNP bound to E. coli Cys-tRNACys (PDB: 1B23). The distances are between the caboxylate oxygen of Glu390 nearest to the amino nitrogen.
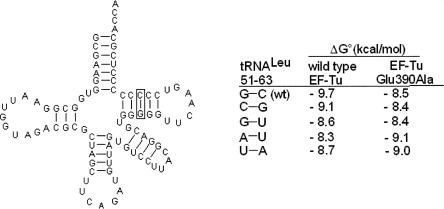
To further assess the contribution of the 51–63 base pair to the specificity of tRNA binding to EF-Tu, Leu-tRNALeu was chosen as an example of a tRNA that responds to the Glu390 mutation, but had not been cocrystallized with EF-Tu. The binding free energies of unmodified E. coli Leu-tRNALeu and four mutations of the 51–63 base pair were determined with native EF-Tu and with the Glu390Ala mutant (Fig. 3). The tRNA mutations correspond to the four other most common 51–63 pairs present in bacterial tRNAs. Compared to the G51–C63 base pair present in tRNALeu, all four mutant tRNAs showed weaker binding to EF-Tu. Interestingly, a simple reversal of the base pair to C51–G63 results in a ΔΔG° of 0.6 kcal/mol. Furthermore, as predicted by Nissen et al. (2003), the replacement of a wild-type pair with a G1–U63 wobble pair also reduces the free energy of binding (ΔΔG° = 1.1 kcal/mol). It appears that these differences are solely due to Glu390 since both base-pair mutations showed a similar ΔG° = −8.4 kcal/mol when bound to Glu390Ala protein. Presumably all three mutations retain the contact with Glu390 and the slightly weaker affinities result from subtle changes in the position of their amino groups. As would be expected, the tRNALeu mutants with an A51–U63 or U51–A63 base pair showed even weaker binding due to the absence of the amino group needed to interact with Glu390. Interestingly, the Glu390Ala mutation actually stabilized binding of the A51–U63 and U51–A63 by about 0.6 kcal/mol compared to the wild-type protein. This suggests that part of the weaker binding of the wild-type protein to the A51–U63 and U51–A63 tRNAs is due to a destabilizing contribution by Glu390. When the amino group is absent, the negatively charged glutamate may participate in electrostatic repulsion with the nearby phosphates and thereby act as a negative determinant.
FIGURE 3.
Cloverleaf representation of unmodified E. coli tRNALeu showing the effect of mutations of the 51–63 base pair on the binding to wild-type T. thermophilus EF-Tu and the Glu390Ala mutation. Binding experiments were performed in the same buffer as Figure 1.
As an alternative way to evaluate the contribution of the hydrogen bond on the stability of the complex, a variant of tRNALeu, where G51 was replaced by an inosine, was prepared by ligating two chemically synthesized tRNA half molecules (Sherlin et al. 2001). By replacing the amino group of G51 by a proton, the I51 derivative is predicted to bind less well. As shown in Figure 3, the affinity of the I51–C63 tRNALeu is 1.1 kcal/mol weaker than the wild-type tRNALeu, which is similar in magnitude to the U51–A63 and Glu390Ala mutations that also lack the hydrogen bond. As was observed for the A51–U63 and U51–A63 mutations, the I51–C63 tRNA binds slightly better to EF-Tu carrying the Glu390Ala mutation, again indicating the destabilizing effect of Glu390 when the amino group is absent.
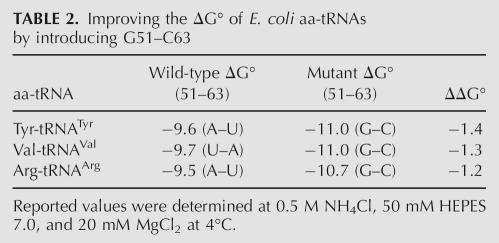
Based on the preceding experiments, it appears likely that the identity of the 51–63 base pair will affect the affinity of all E. coli tRNAs to EF-Tu. Thus, it should be possible to strengthen the binding of those tRNAs that have unfavorable (A–U or U–A) pairs by mutating them so that they contain the G51–C63 alternative. To test this, transcripts of both wild-type and G51–C63 versions of E. coli tRNAArg (UCU), tRNATyr (GUA), and tRNAVal (GAC) were prepared, and their binding affinities to EF-Tu were measured (Table 2). As expected, the replacement of the A51–U63 base pair in tRNATyr and tRNAArg and the U51–A63 base pair in tRNAVal by a G51–C63 base pair stabilized binding to EF-Tu. The ΔΔG° values were similar to those seen for the same base pair changes in tRNALeu (Fig. 3), supporting the idea that a G51–C63 base pair stabilizes the binding of EF-Tu to any tRNA by a similar amount.
TABLE 2.
Improving the ΔG° of E. coli aa-tRNAs by introducing G51–C63
DISCUSSION
Using a combination of protein and tRNA mutagenesis, we have established that the interaction between T. thermophilus EF-Tu and the tRNA body is significantly stabilized by a hydrogen bond observed in the X-ray structure between Glu390 and the amino group of guanine in the minor groove at the 51–63 base pair in the T-stem. Hydrogen bonds between the side chains of glutamate or aspartate with the amino group of guanine are found in numerous protein–nucleic complexes (Allers and Shamoo 2001; Jones et al. 2001; Treger and Westhof 2001; Cheng et al. 2003). For example, the carboxylate of Asp235 of E. coli glutamanyl tRNA synthetase contacts the amino group of the C2–G70 base pair of tRNAGlu (Sherlin et al. 2000) and Glu188 of yeast asparaginyl tRNA synthetase contacts the N2 amino group of the single-stranded G34 residue of E. coli tRNAAsp (Cavarelli et al. 1993). Considering that both glutamate and aspartate are both commonly observed interacting with the N2 amino group of guanine, it is interesting that while Glu390 in EF-Tu is universally conserved in bacteria, the corresponding amino acid in the orthologous proteins in archae and eukaryotes is always an aspartate. Thus, it appears likely that a similar sequence-specific contact occurs with tRNA in all three kingdoms.
The most critical feature of the Glu390 contact is that the amount it contributes to the overall free energy of binding depends upon the identity of the 51–63 base pair. A range of 1.4 kcal/mol was observed among the binding free energies of different 51–63 base pairs in Leu-tRNALeu. When the tightest G–C pair was introduced into three E. coli tRNAs containing weaker A–U or U–A pairs their binding free energy was strengthened by 1.2–1.4 kcal/mol. Such a variable thermodynamic response makes this contact an important specificity element for “tuning” the affinity of different tRNA bodies for the protein. Since the free energy of binding of 20 different valylated E. coli tRNAs ranged from −8.1 to −11.7 kcal/mol (Asahara and Uhlenbeck 2002), the Glu390 contact could potentially be responsible for nearly one half of this “tuning.”
It is instructive to compare the sequences of individual E. coli tRNAs at positions 51 and 63 with the measured affinities of their bodies. Of 46 E. coli tRNAs, 26 have a guanine at this base pair, which is expected to give tighter binding. For the most part, these are indeed the tRNA bodies that bind tightly. For example, the three tightest E. coli tRNA bodies; Glu, Asp, and Gly, all contain a G51–C63, the tightest binding base pair. Meanwhile, the two weakest binding E. coli elongator tRNAs, Tyr and Gln, both have the weakest binding A51–U63 base pair. Additionally, tRNASec contains an A51–U63 pair, which may help explain why it does not bind EF-Tu in vivo (Baron et al. 1990; Ott et al. 1990a). However, there are clear exceptions, where weak binding tRNAs such as tRNATrp and tRNAfMet have the very stable G51–C63 base pair. This suggests that these tRNAs must contain other sequence determinants that weaken EF-Tu binding affinity and thereby offset the stabilizing contribution of the 51–63 pair. One such determinant could be the 1–72 pair, which makes extensive contacts with EF-Tu. Indeed in the case of tRNAfMet, which does not bind EF-Tu in vivo, the unusual mismatched C1–A72 base pair has been shown to be an important “antideterminant” that weakens EF-Tu binding (Fischer et al. 1985; Seong and RajBhandary 1987). When this base pair is changed to C1–G72 or U1–A72, tRNAfMet binds EF-Tu much better, undoubtedly aided by the stabilizing 51–63 base pair. In the case of tRNATrp, which must bind EF-Tu only weakly in order to offset its tight binding amino acid, it may be its A1–U72 pair that serves as the weakening element, an hypothesis that can be tested by mutagenesis experiments. It is clear that the discovery of the first tRNA specificity element for EF-Tu presented here will greatly aid in the identification of additional tRNA specificity elements.
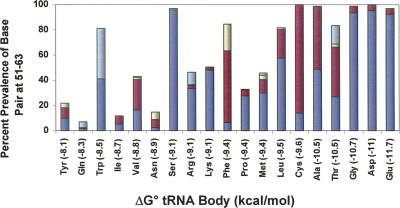
Since Glu390 in EF-Tu is universally conserved among bacteria, it is interesting to consider whether the contact is used by all bacterial EF-Tu to modulate their affinities to their corresponding tRNAs. One way to approach this is to examine the sequence of the 51–63 base pair among bacterial tRNAs specific for a given amino acid. If it is assumed that the hierarchy of tRNA binding affinities for all bacterial EF-Tu resembles that determined for T. thermophilus EF-Tu with E. coli tRNAs (Asahara and Uhlenbeck 2002), then the tighter binding tRNAs should contain a preponderance of G residues in the 51–63 base pair. As shown in Figure 4 for the tRNAs from 129 complete bacterial genomes, this trend is clearly observed. Of the seven tightest tRNA bodies (Glu, Asp, Gly, Thr, Ala, Cys, and Leu), >84% contain a G in the 51–63 pair while <18% of three of the four weakest tRNA bodies (Tyr, Gln, and Ile) have a G at this position. While this correlation supports the view that Glu390 “tunes” the affinity of EF-Tu for tRNA in all bacteria, the correlation is not perfect. For example, there are a limited number of bacteria where tRNAGlu does not contain a G at 51 or 63 or where tRNATyr does contain either a G51–C63 or C51–G63 pair. This could either mean that in these bacteria either Glu390 is not a specificity determinant, or the hierarchy of tRNA binding affinities is dramatically different. However, it seems more likely that the tRNAs in these organisms have evolved their sequences to use one or more of the other specificity determinants to set their appropriate binding affinities. This would partially explain why tRNA sequences are much more phylogenetically variable among bacteria than the EF-Tu sequences to which they bind.
FIGURE 4.
The correlation of the prevalence of guanosine in the 51–63 base pair in 129 bacterial tRNA isoacceptors with the position of the tRNA on the EF-Tu binding hierarchy. Listed in parentheses on the horizontal axis is the EF-Tu binding affinity of the tRNA body as determined for E. coli tRNAs (Asahara and Uhlenbeck 2002). Green (G51-U63), yellow (U51-G63), red (C51-G63), and blue (G51-C63).
MATERIALS AND METHODS
EF-Tu mutagenesis and purification
The initial plasmid containing the T. thermophilus tuf1 gene was provided by Anidya Banerjee and Marvin Mackinen (University of Chicago). A sequence coding for (His)6-TEV cleavable linker was appended to the 5′ end of the EF-Tu gene by PCR and cloned into a pET-3a vector between NdeI and BamHI sites. The N-terminal amino acid sequence of the resulting protein was MH6GNKYFQA so that cleavage by TEV after the glutamine resulted in the native N-terminal alanine residue. Mutagenesis and purification of EF-Tu was performed as described previously (Sanderson and Uhlenbeck 2007a).
tRNA preparation
DNA templates for wild-type and mutant variants of E. coli tRNALeu (CAG), tRNAVal, tRNATyr, and tRNAArg were prepared by primer extension using overlapping DNA oligonucleotides (IDT). In vitro transcription was performed in reactions containing 40 mM Tris-HCl (pH 8.0), 10 mM dithiothreitol, 20 mM Mg(OAc)2, 25 mM NaCl, 2 mM spermidine, 2.5 mM each NTP, 20 mM GMP, 50 μg/mL bovine serum albumin (BSA), and 30 μg/mL T7 RNA polymerase. Transcribed tRNAs were purified on a 10% denaturing polyacrylamide gel. Wild-type tRNALeu (CAG) and a variant containing the G51I mutation were prepared by joining tRNA half molecules using T4 tRNA ligase (Bruce and Uhlenbeck 1982; Sherlin et al. 2001). A 100 μL reaction containing 45 μM each of a 34 nucleotide (nt) 5′ half and a 53 nt 3′ half, were combined in 10 mM HEPES (pH 7.5) and 10 mM KCl, heated to 90°C, and allowed to cool to 40°C over a period of 15 min. Subsequently, the annealed half molecules were ligated at position 34 to form a full-length tRNA by the addition of 10 mM DTT, 20 μg/mL BSA, 10 mM ATP, and 0.02 μM of T4 RNA ligase to give a final volume of 112.5 μL. Ligation reactions were incubated at 37°C for 2 h. After ligation, the full-length tRNA product was purified in a 10% denaturing polyacrylamide gel. All tRNAs were aminoacylated as described by Dale et al. (2004).
EF-Tu assays
The dissocation rate constants of aa-tRNAs from the EF-Tu•GTP complexes were determined as described previously (Sanderson and Uhlenbeck 2007a).
Bioinformatic analysis
Aligned tRNA sequences from 129 bacterial genomes were downloaded from the publicly available Web site, tRNA DataMart (http://trnamart.uoregon.edu/). The downloaded tRNA sequences represented all of the major bacterial phyla that have been identified to date. Duplicate, truncated, and/or misaligned sequences were removed. The acquired tRNA sequences were then separated according to their amino acid acceptor identity. The composition of the 51–63 base pair was then analyzed using Microsoft Excel.
ACKNOWLEDGMENTS
This grant was supported by NIH grant GM37552 to O.C.U. and also by Grant Number 1C06RR018850-01 (National Center for Research Resources [NCCR], a component of the National Institutes of Health [NIH]). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCCR or NIH.
Footnotes
Abbreviations: EF-Tu, elongation factor Tu; aa-tRNA, aminoacyl-tRNA.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.485307.
REFERENCES
- Allers, J., Shamoo, Y. Structure-based analysis of protein–RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- Asahara, H., Uhlenbeck, O.C. The tRNA specificity of Thermus thermophilus EF-Tu. Proc. Natl. Acad. Sci. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, C., Heider, J., Bock, A. Mutagenesis of selC, the gene for the selenocysteine-inserting tRNA-species in E. coli: Effects on in vivo function. Nucleic Acids Res. 1990;18:6761–6766. doi: 10.1093/nar/18.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, A.G., Uhlenbeck, O.C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982;21:855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Cavarelli, J., Rees, B., Ruff, M., Thierry, J.C., Moras, D. Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993;362:181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Cheng, A.C., Chen, W.W., Fuhrmann, C.N., Frankel, A.D. Recognition of nucleic acid bases and base pairs by hydrogen bonding to amino acid side chains. J. Mol. Biol. 2003;327:781–796. doi: 10.1016/s0022-2836(03)00091-3. [DOI] [PubMed] [Google Scholar]
- Dale, T., Sanderson, L.E., Uhlenbeck, O.C. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004;43:6159–6166. doi: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- Fischer, W., Doi, T., Ikehara, M., Ohtsuka, E., Sprinzl, M. Interaction of methionine-specific tRNAs from Escherichia coli with immobilized elongation factor Tu. FEBS Lett. 1985;192:151–154. doi: 10.1016/0014-5793(85)80062-4. [DOI] [PubMed] [Google Scholar]
- Jones, S., Daley, D.T., Luscombe, N.M., Berman, H.M., Thornton, J.M. Protein–RNA interactions: A structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere, F.J., Wolfson, A.D., Uhlenbeck, O.C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Louie, A., Ribeiro, N.S., Reid, B.R., Jurnak, F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., Nyborg, J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Thirup, S., Kjeldgaard, M., Nyborg, J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Nyborg, J., Clark, B.F.C. Translational elongation. In: Lapointe J., Brakier-Gingras L., editors. Translation mechanisms. Kluwer Academic/Plenum Publishers; New York: 2003. pp. 322–339. [Google Scholar]
- Ott, G., Jonak, J., Abrahams, I.P., Sprinzl, M. The influence of different modifications of elongation factor Tu from Escherichia coli on ternary complex formation investigated by fluorescence spectroscopy. Nucleic Acids Res. 1990a;18:437–441. doi: 10.1093/nar/18.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, G., Schiesswohl, M., Kiesewetter, S., Forster, C., Arnold, L., Erdmann, V.A., Sprinzl, M. Ternary complexes of Escherichia coli aminoacyl-tRNAs with the elongation factor Tu and GTP: Thermodynamic and structural studies. Biochim. Biophys. Acta. 1990b;1050:222–225. doi: 10.1016/0167-4781(90)90170-7. [DOI] [PubMed] [Google Scholar]
- Sanderson, L.E., Uhlenbeck, O.C. Directed mutagenesis identifies amino acid residues involved in elongation factor Tu binding to yeast Phe-tRNAPhe . J. Mol. Biol. 2007a;386:119–130. doi: 10.1016/j.jmb.2007.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson, L.E., Uhlenbeck, O.C. Exploring the specificity of elongation factor Tu for different tRNA bodies. Biochemistry. 2007b doi: 10.1021/bi602548v. (in press). [DOI] [PubMed] [Google Scholar]
- Seeman, N.C., Rosenberg, J.M., Rich, A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, B.L., RajBhandary, U.L. Mutants of Escherichia coli formylmethionine tRNA: A single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. 1987;84:8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlin, L.D., Bullock, T.L., Newberry, K.J., Lipman, R.S., Hou, Y.M., Beijer, B., Sproat, B.S., Perona, J.J. Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases. J. Mol. Biol. 2000;299:431–446. doi: 10.1006/jmbi.2000.3749. [DOI] [PubMed] [Google Scholar]
- Sherlin, L.D., Bullock, T.L., Nissan, T.A., Perona, J.J., Lariviere, F.J., Uhlenbeck, O.C., Scaringe, S.A. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- Treger, M., Westhof, E. Statistical analysis of atomic contacts at RNA–protein interfaces. J. Mol. Recognit. 2001;14:199–214. doi: 10.1002/jmr.534. [DOI] [PubMed] [Google Scholar]