Abstract
Improving the immunological potency, particularly the Ab response, is a serious hurdle for the protective efficacy and hence broad application of DNA vaccines. We examined the immunogenicity and protective efficacy of a hemagglutinin-based influenza DNA vaccine that was targeted to antigen-presenting cells (APCs) by fusion to CTLA4. The targeted vaccine was shown to induce an accelerated and increased Ab response (as compared with those receiving the nontargeted control) that was predominated by IgG1 and recognized conformationally dependent viral epitopes. Moreover, mice receiving the APC-targeted DNA vaccine had significantly reduced viral titers (100-fold) after a nonlethal virus challenge. The increased protective efficacy was most likely because of increased Ab responses, as cytotoxic T lymphocyte responses were not enhanced. Targeting was demonstrated by direct binding studies of CTLA4 fusion proteins to the cognate ligand (B7; expressed on APCs in vivo). In addition, a targeted protein was detected at 4-fold higher levels in draining lymph nodes within 2–24 h of administration. Therefore, this study demonstrates that targeting DNA-encoded antigen to APCs results in enhanced immunity and strongly suggests that this approach may be useful in improving the protective efficacy of DNA vaccines.
Immunization with DNA vaccines encoding antigen (Ag) has been used to induce both cellular and humoral immune responses and holds enormous potential for developing vaccines to a variety of pathogens. Recent reports of the first human clinical trials have shown that DNA vaccines were well tolerated, and the responses [especially cytotoxic T lymphocytes (CTLs)] were generally encouraging (1–3). However, there are still some questions concerning the potency of DNA vaccines, particularly with regard to Ab responses.
There is a low level of protein expression after DNA immunization (e.g., nanograms produced). We sought to enhance the effectiveness of this dose by delivering what small amount of Ag is made to the cells relevant to immune induction [i.e., Ag-presenting cells (APCs)]. We showed that increased Ab and T cell responses to a model Ag [human IgG (hIgG)] could be achieved by fusion of the Ag to CTLA4, which directs it to APCs through binding to the surface receptors B7-1 and B7-2 (4). Targeting APCs through the CD11c receptor has also been shown recently to enhance Ab responses (5). In this study, we sought to assess the efficacy of our targeting strategy in a viral challenge system. A plasmid expressing the influenza hemagglutinin (HA) molecule fused with CTLA4 was constructed and examined for its ability to enhance the immune response after DNA immunization and to reduce the amount of virus present in the lungs of mice after a nonlethal influenza challenge.
Materials and Methods
Plasmids.
The backbone of all the plasmids used for immunization was the same, namely pCI (Promega), which contains a human cytomegalovirus promoter plus intron. Influenza A/Puerto Rico/8/34 H1 (PR8) HA was fused to hIg or CTLA4-hIg [the nontargeted and targeted molecules, respectively, described by Boyle et al. (4)], whereby hIg has the leader sequence of CD5 to afford secretion and the hinge, CH2 and CH3 regions of hIgG1 and CTLA4 is the ectodomain of CTLA4. This fusion was done by creating a gly-gly-gly-gly-thr spacer at the 3′ NsiI site of hIg and adding the PvuII-BstYI fragment of HA (83–1609; GenBank accession no. J02143) 3′ of that spacer. This HA fragment represents the mature HA without the signal peptide and without the transmembrane domain. CTLA4-hIg-S was the control plasmid whereby HA was replaced by an irrelevant peptide (SIINFEKL) derived from chicken ovalbumin. HA plasmid encodes signal peptide and ectodomains of HA up to the same BstYI site above. The plasmids used for immunization are represented in Fig. 1d. DNA was prepared by the polyethylene glycol/Triton X-114 method (6), so that there was <0.05 ng of endotoxin per mg of DNA.
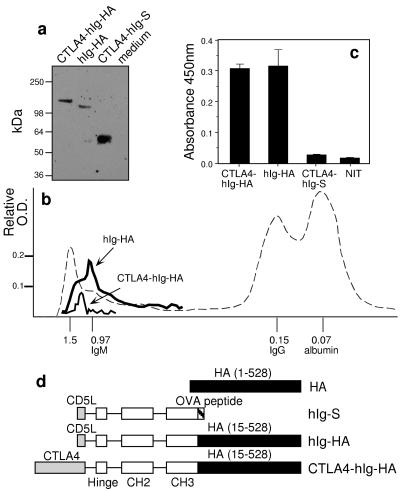
Figure 1.
Fusion molecules containing Ig-HA were secreted and dimerized. (a) Western blotting, after SDS/PAGE of proteins of transfected cells run under reducing conditions, by using peroxidase-conjugated goat anti-hIg and chemiluminescence. (b) Size chromatography of CTLA4-hIg-HA (thin line) and hIg-HA (thick line) from transfectant supernatants indicates that their Mr is 1.2 million and 1.0 million, respectively. Supernatants were concentrated and run at 24 ml/h on a Sephacryl S300 column (2.4 × 100 cm). The fractions were analyzed by capture ELISA for the presence of the fusion proteins. Only the positive fractions are shown. Blue dextran and sheep serum were run as size markers (shown as dashed line); numbers indicate the size in millions. (c) Capture ELISA of culture supernatants by using mAb E2.6 (which recognizes a conformationally dependent epitope) as capture Ab and peroxidase-conjugated goat anti-hIg crossadsorbed against mouse Ig as detecting Ab. (d) DNA constructs used for immunizations. All constructs were made on a pCI (Promega) backbone. Introns are indicated as lines connecting the exons of hIgG1 (hinge, CH2, and CH3) used. The amino acids of HA included are indicated in parentheses.
Virus.
Influenza virus A/PR8 (H1N1) was grown in the allantoic cavity of 10-day embryonated hens' eggs for 2 days at 35°C. The allantoic fluid was collected, clarified by centrifugation (2,000 × g, 15 min, 4°C), stored at −70°C, and used as a source of infectious virus for challenge of mice. Purified virus used in ELISAs and β-propiolactone-inactivated and sodium taurodeoxycholate-disrupted purified virus (split virus) for immunization of mice were obtained from CSL (Parkville, Victoria, Australia).
mAbs.
mAbs E2.6 and C4.2 are directed against the HA of A/PR8 virus and recognize conformation-dependent and conformation-independent epitopes on the viral HA, respectively. mAb 165 binds to carbohydrate on glycosylated egg-grown influenza viruses and therefore also recognizes a conformation-independent epitope.
ELISA.
The ELISA was performed as described (7) by using 96-well polyvinyl microtiter trays; each well was coated with 50 μl of a solution containing either purified intact virus (1,000 hemagglutinating units/ml) or split virus (5 μg/ml) per well. In ELISAs determining the levels of Abs recognizing denatured viral HA, the split PR8 virus was reduced and alkylated before coating. Anti-HA IgG subclass titers were determined by the method described (8). Briefly, bound Ab was detected after incubation with peroxidase-conjugated anti-mouse IgG1, IgG2a, or IgG2b Abs (Southern Biotechnology Associates) diluted in blocking buffer (5% skim milk powder in PBS) overnight at 4°C. Ab titers are expressed as the reciprocal of the Ab dilution giving an absorbance of 0.2 units.
Cytotoxic T-Cell Assay.
CTL assays were performed as described (9) by using P815 mastocytoma cells as targets.
Transfection, Western Blotting, Capture ELISA, and Molecular Sieving.
NIT cells (an insulinoma cell line) were electroporated with 20 μg of plasmid and cultured for 3 days; the protein products were analyzed by capture ELISA or by Western blotting. Capture ELISA used the PR8 HA-specific mAb E2.6 as the capture Ab on the microtiter tray. Culture supernatants were added to the tray wells, and bound Ag was detected with a peroxidase-conjugated goat anti-hIgG crossadsorbed against mouse Ig (Southern Biotechnology Associates). For Western blotting of proteins after SDS/PAGE with or without 2-mercaptoethanol, bands were detected with the peroxidase-conjugated anti-hIg followed by chemiluminescence. The relative size of the proteins was estimated by running concentrated supernatants on a Sephacryl S300 column (2.4 × 100 cm) at 24 ml/h. Fractions were collected and analyzed by capture ELISA for the presence of fusion proteins.
B7-Binding Assay.
Supernatants from Chinese hamster ovary cells transfected with the DNA expressing the CTLA4-hIg-HA, hIg-HA, or CTLA4-hIg-S plasmids were added to NIT cells expressing membrane-bound B7-1 or untransfected controls and bound protein detected with FITC-conjugated anti-hIg secondary Ab.
Immunization.
Groups of 10 female 6- to 8-wk-old BALB/c mice, bred in the animal facility of the Department of Microbiology and Immunology (University of Melbourne), were immunized i.m. in both quadriceps under ketamine/xylazine anesthesia with 100 μg of plasmid DNA in saline. As a positive control, mice were immunized s.c. in the scruff of the neck with split virus in PBS.
Intranasal Challenge of Mice, Preparation of Lung Extracts, and Assay for Infectious Virus.
Penthrane-anesthetized mice were challenged with 50 plaque-forming units of infectious virus by the intranasal route; each mouse received 50 μl of virus in the form of allantoic fluid diluted in PBS. At 5 days after challenge, mice were killed by cervical dislocation; lungs were removed and homogenized, and the supernatants were stored at −70°C before assay for infectious virus. Virus titers were determined by plaque assay on monolayers of Madin–Darby canine kidney cells as described (10).
Statistical Analysis.
Data were analyzed by using the nonparametric Mann–Whitney U test, which compares two sets of unpaired samples. The null hypothesis is that the two population medians are equal, and the resultant P value for particular comparisons is given.
Results and Discussion
Expression of HA Constructs in Mammalian Cells.
Mammalian expression plasmids were constructed to express HA under the control of the cytomegalovirus promoter. A targeted secreted fusion vaccine (CTLA4-hIg-HA) was made to encode CTLA4 at the N terminus for targeting, followed by the Fc of hIgG1 to promote dimerization for enhanced binding to B7 (11) and influenza HA at the C terminus. As controls, plasmid encoding a fusion molecule lacking the targeting ligand (hIg-HA) and another in which the HA was replaced with an irrelevant Ag sequence (CTLA4-hIg-S) were likewise made. Previously, we had shown that increased responses were achieved only when the Ag was fused with CTLA4 (4) and when a dimerization moiety (namely the Fc of IgG) was also present. The plasmids were first used to transfect cells in vitro. Western blotting confirmed the expected sizes of the reduced protein products to be 125 kDa and 145 kDa for hIg-HA and CTLA4-hIg-HA, respectively (Fig. 1a). Sephacryl chromatography revealed that the native structures were 1.0 and 1.2 million in size (Fig. 1b). These sizes are consistent with higher ordered structures (e.g., tetramers of dimers: the hIg acting as a dimerization moiety) (4). HA normally forms trimers, but higher-order structures may be formed from nonnative disulfide bonding or may be predisposed by the juxtaposition of HA by the hIg dimerization. In the context of this study, what is important is that both targeted and nontargeted molecules oligomerize similarly (the size ratio between subunit and native structure was similar for both), and thus differential oligomerization is not the reason for the differential Ab responses shown below. Furthermore, the protein levels after transfection were comparable between hIg-HA and CTLA4-hIg-HA. Secreted proteins in culture supernatants were also examined by capture ELISA by using an HA-specific mAb, and bound Ag was detected with peroxidase-conjugated Ab to hIgG. As expected, Ag was detected only in cultures transfected with plasmids encoding both HA and hIg sequences (Fig. 1c). Although the antigenic integrity of HA relies on the proper folding of the globular head domain (12), the mAb E2.6 used in the capture ELISA was a neutralizing Ab that recognizes a conformation-dependent epitope. This suggests that some conformational integrity has been maintained within the HA expressed from the hIg-HA and CTLA4-hIg-HA fusion vaccines. This was subsequently confirmed by the finding that the Ab responses obtained were largely against native HA (see below).
Targeting by the CTLA4 Ligand to Draining Lymph Nodes.
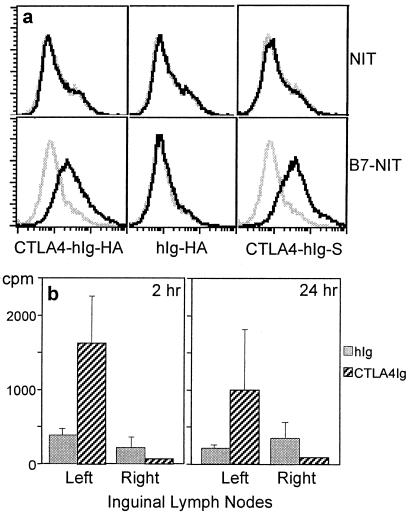
The binding of the fusion vaccine was confirmed by flow cytometric analysis, and significant binding was detected only with the CTLA4-Ig fusion proteins (Fig. 2a). Our hypothesis is that the effectiveness of this ligand targeting is mainly because of physical delivery to the anatomical sites of immune induction, either directly or through association with migrating dendritic cells that bear the B7 molecule. Direct injection of DNA vaccines into the lymph nodes (13) or spleen (14) elicits potent immune responses, and we have shown that spleen injections are, in fact, superior to i.m. and intradermal injections (14). The alternative possibility, that the targeting molecule is actually providing signals to the APC, is less likely considering l-selectin can also be used as a targeting moiety to enhance the immune response to certain Ags (4); it is difficult to envisage how any signaling to high endothelial venule cells would lead to enhancement of a specific Ab response. Coinjection of l-selectin-hIg or CTLA4-hIg with ovalbumin does not enhance any responses to ovalbumin (4); therefore, the effect cannot be a nonspecific effect on homing or migration. In Fig. 2b, we also confirm that the amount of protein fused to CTLA4 found in draining (ipsilateral) lymph nodes is 4-fold greater than that of nontargeted protein at 2 h or 24 h postinjection (P < 0.05; compare with the fact that no difference was found for contralateral nodes). This preferential accumulation of the targeted protein is consistent with the hypothesis that physical delivery would seem to be a major, if not the only, mechanism for immune enhancement.
Figure 2.
Fusion proteins bind B7 and accumulate in the draining lymph node. (a) Flow cytometric analysis shows that CTLA4-hIg-HA binds to B7. NIT cells expressing membrane-bound B7-1 or untransfected cells (B7-NIT and NIT, respectively) were incubated with supernatants from Chinese hamster ovary cells transfected with CTLA4-hIg-HA, hIg-HA, or CTLA4-hIg-S (black line). The gray line represents the NIT or B7-NIT cells incubated with untransfected Chinese hamster ovary cell supernatants. FITC-conjugated sheep anti-hIgG was used as the secondary Ab. (b) Enhanced localization of CTLA4-hIg in draining lymph nodes. BALB/c mice were injected in the left quadriceps with radioiodinated CTLA4-hIg or hIg (total cpm was 3 million; total protein was 5 μg). Lymph nodes were harvested at 2 h and 24 h after immunization. The mean and SEM of five mice per group are shown.
Enhancement of Ab Responses to DNA Immunization by Targeting.
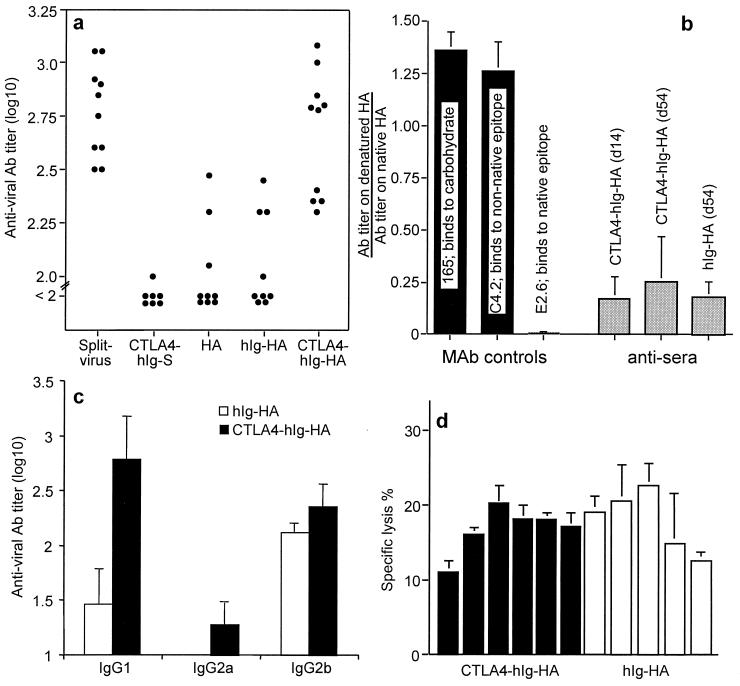
Groups of BALB/c mice (n = 9 or 10) were immunized on days 0 and 14 with 100 μg of the indicated DNA constructs in each quadriceps. As a positive vaccine control, an inactivated detergent-disrupted virus (split virus) that was produced in a manner similar to a current inactivated human influenza vaccine was injected. Sera were obtained at 2 and 4 weeks after initial immunization and assayed for antiviral Ab by ELISA. At 4 weeks, mice immunized with CTLA4-hIg-HA had produced the strongest Ab responses; these responses were significantly higher (P < 0.001) than those for the hIg-HA immunized controls (Fig. 3a). The Ab levels induced by CTLA4-hIg-HA immunization were comparable with those induced by the split virus control (P = 0.57; Fig. 3a). Mice receiving the DNA vaccine negative control (CTLA4-hIg-S) had no detectable antiviral Ab, whereas those that received the HA DNA mounted a low but detectable antiviral response (Fig. 3a). Interestingly, the 2- and 4-week responses to the DNA vaccines were similar, whereas the Ab levels in mice given split virus protein rose 4-fold after the second immunization (data not shown). This suggested that there was no requirement for, or benefit from, giving a second immunization of DNA (at 2 weeks) as was observed with split virus. Titers at 8 weeks were not significantly elevated from those at 2 or 4 weeks (data not shown); this is consistent with the observation that APC targeting speeds up the immune response for DNA vaccines (4, 5). To determine how much Ab elicited by the fusion DNA vaccines recognized the intact HA molecule, we compared the binding of Ab to native and denatured (reduced and alkylated) Ag (Fig. 3b). As controls, we used mAbs that recognized conformation-dependent and -independent epitopes. The relative binding for both hIg-HA and CTLA4-hIg-HA showed that most of the Ab elicited was against native epitopes, and that the relative binding was similar for both groups, suggesting that there was no difference in the way HA was folded between the two similarly fused proteins.
Figure 3.
Immune responses to the DNA vaccines. (a) Individual serum antiviral Ab titers after targeted DNA immunizations were greater than after nontargeted DNA immunization. Mice were immunized with 100 μg of plasmid DNA per leg in both quadriceps on days 0 and 14 and bled 2 weeks later or immunized with split virus as a control. (b) Antiviral Abs induced by CTLA4-hIg-HA and hIg-HA immunization recognize conformation-dependent epitopes on HA. Ab levels of mice at 14 and 54 days after priming were assayed by ELISA for binding to native split virus and denatured (reduced and alkylated) virus. For each mouse, the ratio of the titers of serum Ab recognizing denatured HA: native HA was calculated, and the mean and SD for each group were determined. As a control for relative coating levels and as an indicator of denaturation of PR8 HA, binding of mAbs 165 (recognizing a carbohydrate determinant), C4.2 (recognizing a conformation-independent determinant), and E2.6 (recognizing a conformation-dependent determinant) to native vs. denatured virus was also examined. It should be noted that the Ab levels of the untargeted DNA vaccine group on day 14 were too low for these relative binding data to be valid; therefore, these data are not shown. For the same reason, the data on day 54 for the untargeted group represent only three of 10 mice (the three highest responders). (c) IgG subclass responses. Groups of five female BALB/c mice were immunized with 100 μg of plasmid DNA; the IgG subclass titer was determined as the highest dilution to reach an OD of 0.2. The mean ± SEM for each group at 4 weeks is shown. (d) Targeted and nontargeted DNA vaccines elicit similar levels of virus-specific CTL activity. Mice were immunized i.m. with a single dose of 100 μg of plasmid DNA; after 4 weeks, individual spleens were taken, and single-cell suspensions were prepared. Cells were cultured for 5 days with virus-infected autologous spleen cells and subsequently tested in a 51Cr release assay for their ability to lyse virus-infected and uninfected P815 target cells. Each bar represents the cytolytic response of an individual mouse at an effector-to-target ratio of 100:1. The mean of triplicate cultures is shown, and background lysis measured on uninfected targets has been subtracted.
To examine further the form of the immune response, groups of five BALB/c mice were immunized with 100 μg of DNA; next, the relative HA-specific IgG subclass profile and, in a separate experiment, the CTL response were determined. All IgG subclasses tested (IgG1, IgG2a, and IgG2b) were elevated in the groups receiving the targeted vaccine, with the increase in IgG1 being the most substantial (Fig. 2c). There was no difference in the magnitude of the CTL response induced by the targeted and nontargeted vaccine (Fig. 2d). There have been several reports examining the mechanism of CTL induction after i.m. DNA immunization that indicate a role for crosspresentation of Ag through a bone marrow-derived APC (15, 16). As our targeting strategy may not affect crosspresentation, it is perhaps not surprising that CTLA4 targeting neither enhanced nor decreased CTL responses to influenza HA (Fig. 2d) and ovalbumin (data not shown).
Improved Protective Efficacy of the Targeted vs. Nontargeted DNA Vaccine.
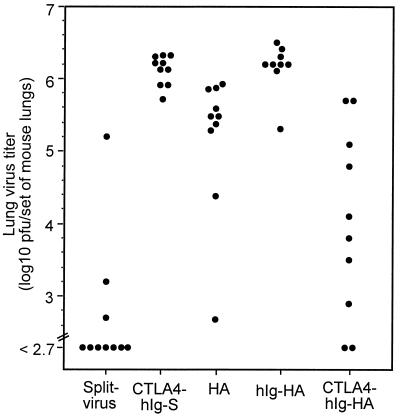
To determine the functional relevance of the immunity generated with the targeted vaccine, BALB/c mice were immunized with 100 μg of DNA and challenged 9 weeks later with infectious virus. The lung viral titers at 5 days after challenge were determined by plaque assay (Fig. 4). By comparing the geometric mean or the median titers in the control DNA vaccine-immunized group (hIg-HA), the viral load for the group that was immunized with CTLA4-hIg-HA was up to 100-fold lower (P = 0.0004) (Fig. 4). Given that the CTL responses were equivalent, we argue that the mechanism of enhanced viral protection is Ab based. Although significantly more IgG1 was induced with the targeted vaccine (Fig. 3c), it is likely that the overall titer of Ab, which correlated with viral neutralization (data not shown), is more relevant to protection than a particular subclass. In a repeat experiment, mice were immunized on days 0 and 14 with 200 μg of DNA and challenged with live virus 6 weeks after initial immunization. In this experiment, a similar enhancement of viral clearance with CTLA4-hIg-HA DNA to that shown in Fig. 4 was observed (data not shown). Incidentally, as with the Ab responses, the level of clearance observed with the targeted construct (CTLA4-hIg-HA) was not significantly different from that observed in mice given the split virus vaccine (P = 0.23), which was used as a positive vaccine control (Fig. 4). We feel, however, that this comparison is not germane, because the split virus contains HA and neuraminidase components; in addition, our strategy aims to improve on currently investigated DNA vaccine strategies through targeting so that they could be used, for example, where no current vaccine exists, rather than as a replacement for effective protein-based (or for that matter live virus) vaccines.
Figure 4.
Viral clearance responses induced by targeted DNA immunization were greater than those induced by nontargeted DNA immunization. Mice were immunized with 50 μg of plasmid DNA per leg in both quadriceps or with split virus as a control. After 65 days, the mice were challenged with a nonlethal dose of virus; 5 days later, the amount of infectious virus present in the lungs was determined by plaque assay. Titers for individual mice are shown.
DNA vaccines are considered to have potential advantages because of ease of construction, ability to induce long-lasting immune responses, high temperature stability of DNA, and low production cost. Ease and speed of production could be important, particularly for those vaccine Ags that differ from one epidemic to the next; as such, the vaccines need continual change. The potency of DNA vaccines has been improved in other systems with the inclusion of immunostimulatory sequences (17), DNA-encoding cytokines (18), or costimulator molecules (19) (reviewed in ref. 14). We have shown previously that the immune response to a model Ag fused to CTLA4 could be markedly enhanced (4). However, for the progression of DNA vaccines based on the targeting strategy described herein, it was important that the crucial issues of potency of immune responses and protective efficacy be addressed (20). Our results show that targeting influenza HA to APCs can be used to greatly increase the Ab response and, more importantly, the level of protection achieved. Thus, the targeting strategy described here may be useful as a generic strategy for improving the potency and efficacy of DNA vaccines.
Acknowledgments
We thank Prof. G. J. V. Nossal for reading the manuscript, Dr. Ian Barr for his adjutant role in this collaborative effort, and Joanne Pagnon for her technical assistance. This work was supported by the Cooperative Research Center for Vaccine Technology and by the National Health and Medical Research Council of Australia.
Abbreviations
- Ag
antigen
- CTL
cytotoxic T lymphocyte
- APC
antigen-presenting cell
- hIgG
human IgG
- PR8
influenza A/Puerto Rico/8/34 H1
- HA
hemagglutinin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120162497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120162497
References
- 1.Calarota S, Bratt G, Nordlund S, Hinkula J, Leandersson A C, Sandstrom E, Wahren B. Lancet. 1998;351:1320–1325. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- 2.MacGregor R R, Boyer J D, Ugen K E, Lacy K E, Gluckman S J, Bagarazzi M L, Chattergoon M A, Baine Y, Higgins T J, Ciccarelli R B, et al. J Infect Dis. 1998;178:92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 3.Wang R, Doolan D L, Le T P, Hedstrom R C, Coonan K M, Charoenvit Y, Jones T R, Hobart P, Margalith M, Ng J, et al. Science. 1998;282:476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 4.Boyle J S, Brady J L, Lew A M. Nature (London) 1998;392:408–411. doi: 10.1038/32932. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Griffiths M N, Burton D R, Ghazal P. Proc Natl Acad Sci USA. 2000;97:847–852. doi: 10.1073/pnas.97.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle J S, Brady J L, Koniaras C, Lew A M. DNA Cell Biol. 1998;17:343–348. doi: 10.1089/dna.1998.17.343. [DOI] [PubMed] [Google Scholar]
- 7.Jackson D C, Tang X L, Brown L E, Murray J M, White D O, Tregear G W. Virology. 1986;155:625–632. doi: 10.1016/0042-6822(86)90222-9. [DOI] [PubMed] [Google Scholar]
- 8.Boyle J S, Silva A, Brady J L, Lew A M. Proc Natl Acad Sci USA. 1997;94:14626–14631. doi: 10.1073/pnas.94.26.14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harling-McNabb L, Deliyannis G, Jackson D C, Gerondakis S, Grigoriadis G, Brown L E. Int Immunol. 1999;11:1431–1439. doi: 10.1093/intimm/11.9.1431. [DOI] [PubMed] [Google Scholar]
- 10.Tannock G A, Paul J A, Barry R D. Infect Immun. 1984;43:457–462. doi: 10.1128/iai.43.2.457-462.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene J L, Leytze G M, Emswiler J, Peach R, Bajorath J, Cosand W, Linsley P S. J Biol Chem. 1996;271:26762–26771. doi: 10.1074/jbc.271.43.26762. [DOI] [PubMed] [Google Scholar]
- 12.Wiley D C, Skehel J J. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 13.Lehner T, Wang Y, Cranage M, Bergmeier L A, Mitchell E, Tao L, Hall G, Dennis M, Cook N, Brookes R, et al. Nat Med. 1996;2:767–775. doi: 10.1038/nm0796-767. [DOI] [PubMed] [Google Scholar]
- 14.Boyle J S, Barr I G, Lew A M. Mol Med. 1999;5:1–8. doi: 10.1007/s008940050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doe B, Selby M, Barnett S, Baenziger J, Walker C M. Proc Natl Acad Sci USA. 1996;93:8578–8583. doi: 10.1073/pnas.93.16.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corr M, Lee D J, Carson D A, Tighe H. J Exp Med. 1996;184:1555–1560. doi: 10.1084/jem.184.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, Roman M, Tighe H, Lee D, Corr M, Nguyen M D, Silverman G J, Lotz M, Carson D A, Raz E. Science. 1996;273:352–354. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 18.Xiang Z, Ertl H C. Immunity. 1995;2:129–135. doi: 10.1016/s1074-7613(95)80001-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim J J, Bagarazzi M L, Trivedi N, Hu Y, Kazahaya K, Wilson D M, Ciccarelli R, Chattergoon M A, Challan A A, Agadjanyan M G, et al. Nat Biotechnol. 1997;15:641–646. doi: 10.1038/nbt0797-641. [DOI] [PubMed] [Google Scholar]
- 20.Langermann S. Nat Med. 1998;4:547–548. doi: 10.1038/nm0598-547. [DOI] [PubMed] [Google Scholar]