Abstract
We have developed a procedure for massively parallel resequencing of multiple human genes by combining a highly multiplexed and target-specific amplification process with a high-throughput parallel sequencing technology. The amplification process is based on oligonucleotide constructs, called selectors, that guide the circularization of specific DNA target regions. Subsequently, the circularized target sequences are amplified in multiplex and analyzed by using a highly parallel sequencing-by-synthesis technology. As a proof-of-concept study, we demonstrate parallel resequencing of 10 cancer genes covering 177 exons with average sequence coverage per sample of 93%. Seven cancer cell lines and one normal genomic DNA sample were studied with multiple mutations and polymorphisms identified among the 10 genes. Mutations and polymorphisms in the TP53 gene were confirmed by traditional sequencing.
Keywords: cancer analysis, high-throughput sequencing, multiplex amplification
Significant progress has been made in identifying the molecular genetic events underlying cancer. For nearly all malignancies, the cause of neoplastic development results from the accumulation of somatic mutations within specific genes, for example, the effect being inappropriate inactivation of tumor suppressors or constitutive activation of oncogenes (1). Not only is the accumulation of mutations causative for cancer in many cases, but it also contributes to cancer phenotype such as overall aggressiveness as seen in recurrence and resistance to molecular-targeted therapies. These cancer-related genes have a large number of functions, including growth regulation, adhesion, cell cycle control, DNA repair processes, and other cellular processes mediated by a variety of signal transduction pathways. More recently, mutations that lead to drug sensitivity or resistance have been discovered in specific kinases like the EGFR gene (2, 3). Undoubtedly, there are many other critical gene mutations to be discovered, and comprehensive mutation discovery from individual genomes will increase our understanding of the genetics underlying any individual tumor's phenotype. This mutation profile may translate into prognostic and predictive genetic biomarkers.
A recently published study examined the consensus coding sequences of a large number of human genes in colorectal and breast cancer (4). However, such large-scale surveys of candidate genes for mutations require preparation of thousands of individual PCRs followed by traditional Sanger sequencing using capillary-based automated instruments. The requirements for these projects include some degree of robotics to handle reagent processing of multiple samples, maintenance of capillary-based sequencers, and extensive bioinformatics infrastructure to handle the flow of data. As a result, high-throughput resequencing studies involving multiple genes are limited to relatively few genome centers and commercial companies that have the necessary extensive and expensive infrastructure. Even with such infrastructure in place, this sequencing approach incurs high cost for the analysis of multiple genes.
Significant efforts are being invested in developing a new class of massively parallel DNA sequencing technologies that have the potential to dramatically reduce cost and time required to carry out large-scale sequencing projects. Some of these technologies are available commercially, such as the sequencing-by-hybridization platform from Affymetrix Inc. (Sunnyvale, CA) (5, 6) and the sequencing-by-synthesis platform from 454 Life Sciences (7) (Bradford, CT). Other technologies and instruments are soon expected to become available, such as the sequencing-by-ligation platform from Applied Biosystems (8) (Foster City, CA) and the sequencing-by-synthesis platforms from Solexa (Hayward, CA) and Helicos Biosciences (9) (Cambridge, MA). These new technologies have proven to be useful in high-throughput de novo sequencing of microorganisms (10, 11) and sensitive mutation detection in single genes in heterogeneous cancer specimens (12). It also has been proposed that highly parallel resequencing can be used for large-scale mutation scans of a multitude of human genes simultaneously. However, efforts have been limited in resequencing candidate genes in cancer with these technologies. In part, this limitation is related to the need for traditional PCR amplification of sequences of interest, which require the same level of amplification reactions necessary for large-scale Sanger sequencing projects.
One approach to increase resequencing throughput and allow more efficient use of DNA samples is simultaneous amplification of many genomic DNA targets, which can be carried out by combining many specific PCR primer pairs in individual reactions (13, 14). However, one of the crucial problems with PCR is that when large numbers of specific primer pairs are added to the same reaction, undesired amplification products arise (15). Even with a careful primer design, PCR usually is limited to 10 simultaneous reactions before amplification yield is compromised by the accumulation of irrelevant products (16, 17).
As recently presented, the selector technology (18, 19) enables highly multiplexed amplification of specific DNA sequences while generating few amplification artifacts. The selector system requires one selector probe (≈80 nt in length) per amplification target and a general vector oligonucleotide (≈40 nt). Each selector probe has two single-stranded, target-complementary end sequences (≈20 nt each) that are linked by a general sequence motif, and the vector oligonucleotide is complementary to this motif. Combined with denatured restriction-digested DNA, each selector probe hybridizes to a specific target together with the vector oligonucleotide, resulting in a circular complex that can be covalently closed by DNA ligase. The general sequence that is introduced into the circularized fragments then allows PCR by using a single universal primer pair. Hundreds of individual selector constructs can readily be multiplexed in a single reaction volume.
By combining selector technology with high-throughput parallel sequencers, rapid resequencing can be accomplished from multiple genes with significantly less infrastructure needed compared with a traditional Sanger sequencing approach. In this proof-of-concept study, we have developed a selector assay that enables parallel sequencing of 10 genes involved in cancer development. We demonstrate that the integration of selector technology with massively parallel sequencing can be used to perform efficient resequencing analysis for discovery of somatic mutations and germ-line polymorphisms.
Results
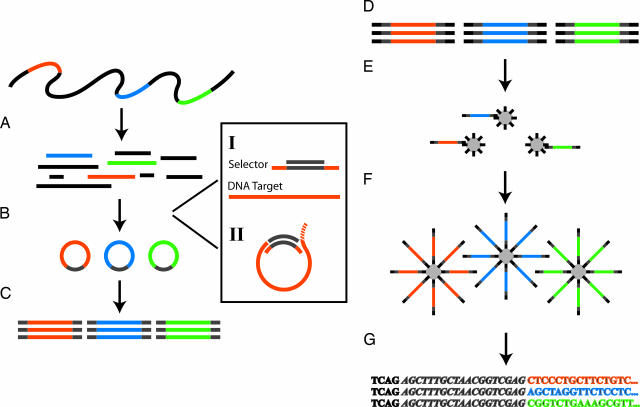
The general workflow for selector-based amplification and 454-sequencing procedure is illustrated in Fig. 1. The 454-system we used, GS-20, generates ≈20 million bases per run with average sequencing read lengths of ≈100 bases.
Fig. 1.
The selector and sequencing assay. (A) A DNA sample is digested to defined fragments by using restriction enzyme(s). The color bars represent the targets of interest. (B) Targeted circularization is performed by using selectors. (I) Selectors contain two oligonucleotides: a selector probe and a general vector oligonucleotide. The selector probe has two single-stranded target-complementary end sequences (orange) that are linked by a general sequence motif (gray) and the vector oligonucleotide that is complementary to the general sequence motif in the selector probes (gray). (II) The circularization reaction can be carried out by using two different approaches. Either both ends of the selected fragment connect to the vector oligonucleotide by hybridizing and ligation or the vector oligonucleotide forms a branched structure in an optional position at the 5′ end of the fragment. This latter structure is recognized and processed by the added endonucleolytic enzyme, forming ends suitable for ligation. (C) The circles are amplified in a multiplex PCR by using a primer pair complementary to the general vector sequence introduced in every circle. (D) The first step in the 454-sequencing procedure is to attach general, 454-optimized, adaptor sequences to each end of each PCR product. (E) The PCR products are separated into single strands and bound to beads in limiting dilutions, resulting in one unique fragment per bead. (F) The beads are clonally amplified in droplets of an oil-emulsion-based PCR, resulting in beads carrying millions of target sequences. (G) The beads are finally deposited into picoliter-sized wells, one bead per well, where solid-phase pyrosequencing is performed and monitored.
We initially designed a set of selectors targeting the coding exons and a portion of the adjacent introns of 10 genes (FRAP1, AKT1, AKT2, TGFBR2, TP53, KRAS, APC, SMAD4, EGFR, and MARK3). These genes where chosen based on their contribution to colorectal cancer development. In addition, we had a larger number of colorectal cancer cell lines that previously had been characterized for mutations in the TP53 gene (20). These genes comprised ≈49 kb of genomic DNA sequence that was targeted for amplification by a set of 425 selectors.
Multiplexed genomic circularization and amplification of the 10-gene set was first carried out on six different DNA samples (five colorectal cancer cell lines and one breast cancer cell line) and interrogated with 454-sequencing. Unlike Sanger sequencing, massively parallel sequencers such as the GS20 produce multiple sequence reads from the same individual amplicon. Therefore, to analyze any given region of interest for genetic variants, one needs to assemble a consensus sequence from these multiple reads. The consensus sequence quality depends on the depth of sequence reads from any given amplicon. We analyzed the sequence data by using software being developed specifically for this purpose (J.S., F.D., and H.J., unpublished data), as described in Materials and Methods. The average fraction of the region of interest for which there was at least one sequencing read was 74% for the six sequenced samples.
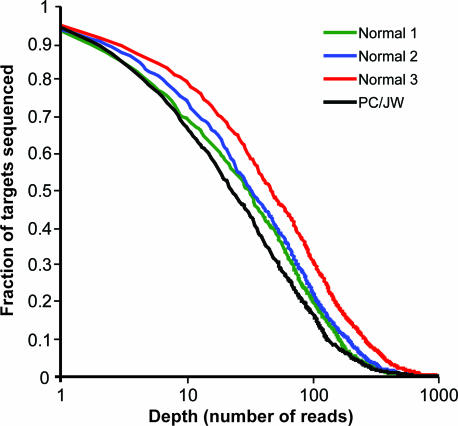
To increase the total sequence coverage, we designed another 83 selectors targeting genomic regions for which there were no sequencing reads in any of the samples analyzed in the first experiment. Using the combined set of 508 selectors, we performed the assay on a normal sample and on an additional colorectal cancer cell line sample. To determine the sequence quality, the normal sample was analyzed in triplicate reactions. For these four reactions, the average fraction of nucleotides in the target region covered by at least one sequencing read was 93%. The sequencing depth distributions for the four reactions are displayed in Fig. 2.
Fig. 2.
Sequencing depth. A normal sample, circularized and amplified in triplicate reactions, and a cancer cell line sample, PC/JW, were sequenced in one 454-experiment. The x axis shows number of reads (n), and the y axis shows the fraction of the target region with a sequencing depth of n or more.
The amount of sequence generated per sample varied significantly between the two experiments, depending on the use of different picotiter plate-loading gaskets. In the first sequencing experiment, the eight-lane loading gasket was used. In the second experiment, the four-lane gasket was used, resulting in more than twice the amount of sequence per sample. The total number of sequencing reads per sample, number of sequenced nucleotides per sample, and average sequence read lengths generated in the two experiments are presented in Table 1.
Table 1.
Sequence yield summary
| Samples | 454 Experiment 1 |
454 Experiment 2 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HTB-20D | SW1417 | VACO429 | COLO741 | C80 | RKO | Average | Normal replicate 1 | Normal replicate 2 | Normal replicate 3 | PC/JW | Average | |
| No. of reads | 33,922 | 33,068 | 18,940 | 17,080 | 9,543 | 19,322 | 21,979 | 48,512 | 54,933 | 78,700 | 55,102 | 59,312 |
| No. of nucleotides | 3,478,929 | 3,397,012 | 1,953,831 | 1,749,818 | 974,443 | 1,991,538 | 2,257,595 | 4,959,702 | 5,598,665 | 7,985,758 | 5,480,864 | 6,006,247 |
| Average read length | 103 | 103 | 103 | 102 | 102 | 103 | 103 | 102 | 102 | 101 | 99 | 101 |
| Coverage of ROI, % | 79 | 75 | 72 | 75 | 62 | 80 | 74 | 92 | 93 | 94 | 93 | 93 |
We investigated whether the increased coverage in the second experiment was generated by the additional selectors or by the increased sequencing output per sample. The data from the second experiment were analyzed excluding the reads generated by the additional 83 selectors. This analysis resulted in an average coverage of 88%, indicating that the additional selectors increased the covered region by ≈2,200 bases, whereas the increased number of sequenced reads per sample added ≈7,100 bases.
To determine the reproducibility of the sequence generated in our assay, we compared the consensus base calls of the three replicate reactions on a normal genomic DNA sample. Of the 43,730 nt that were sequenced with a depth of at least 5 reads in each of the three samples, we found that 99.72% yielded the same consensus base call in all replicate reactions. To investigate the accuracy of our assay, we compared the consensus base calls from all of the sequencing experiments with sequence generated from double-stranded Sanger sequencing of the TP53 gene exons amplified by simplex PCR. In the total of 7,805 nt of sequence covered by five reads or more in the 454 data, and for which there also was Sanger data, the sequence calls of the two methods were concordant to 99.94%.
When analyzing each of the 10 genes in all samples from the 454-experiments, with the same base-calling rules as above, we found a total of 437 positions where the consensus base call differed from the reference sequence. Among these, 158 indicated single-base substitutions, of which 104 were annotated in the dbSNP database (www.ncbi.nlm.nih.gov/projects/SNP). There were also 279 positions where insertions or deletions were indicated. On manual inspection of these variants, we found and discarded 237 that were located in homopolymer motif sequences (three or more consecutive nucleotides of the same type).
Liu and Bodmer (20) report six mutations in the colorectal cancer cell lines that we analyzed. One of these (in PC/JW) is located outside the region targeted in our assay. Three mutations (C80 codon 52, SW1417 codon 238, and VACO429 codon 58) were in locations not sequenced to a depth of 5 or more, which was our minimum requirement for assembling a consensus. In our data, the mutation in VACO429 codon 306 corresponded to the previously reported data, whereas the COLO741 codon 321 insertion was called heterozygous but was previously reported as homozygous by Liu and Bodmer (20).
In our analysis of the eight samples, we found nine additional variations in the TP53 gene. Four of these matched an entry in dbSNP (refSNP ID rs1042522), three were confirmed by Sanger sequencing, and two were contradicted by Sanger data. The number of sequence reads, location, nature, and effect of the TP53 variants are described in Table 2. The findings in the nine other genes remain to be confirmed.
Table 2.
TP53 Mutations and germ-line variants found in the eight samples analyzed
| Sample | Chromosome position | Ref. sequence | Observed genotype | Location | Codon | Effect | Sequence depth | 1st call | 2nd call | Note |
|---|---|---|---|---|---|---|---|---|---|---|
| N523 | 7520197 | C | C/G | Exon 4 | 72 | P→R | 95 | C (53) | G (41) | 1 |
| PC/JW | 7520197 | C | G/G | Exon 4 | 72 | P→R | 6 | G (6) | 1 | |
| RKO | 7520197 | C | C/G | Exon 4 | 72 | P→R | 7 | G (5) | C (2) | 1 |
| VACO429 | 7520197 | C | G/G | Exon 4 | 72 | P→R | 7 | G (7) | 1 | |
| SW1417 | 7519306 | A | A/— | Exons 5–27 | — | Intron | 8 | A (6) | — (2) | 4 |
| RKO | 7518341 | T | T/A | Exons 7–8 | — | Intron | 16 | T (10) | A (5) | 4 |
| HTB-20D | 7517810 | G | A/A | Exon 8 | 285 | E→K | 36 | A (33) | G (3) | 3 |
| VACO429 | 7517747 | C | C/T | Exon 8 | 306 | R→Stop | 154 | T (94) | C (59) | 2 |
| HTB-20D | 7517717 | G | C/C | Exons 8 + 29 | — | Intron | 394 | C (391) | G (2) | 3 |
| COLO741 | 7517610 | — | —/AA | Exon 9 | 321 | Frameshift | 14 | AA (10) | — (3) | 2* |
| RKO | 7517562 | A | A/G | Exons 9 + 18 | — | Intron | 29 | G (17) | A (12) | 3 |
Chromosome positions refer to sequence NC_000017.9. Sequences are presented in the TP53 coding strand polarity. Notes indicate the following: 1, Variation matches dbSNP entry rs1042522. 2, Mutation reported by Liu and Bodmer (20). 3, Confirmed by Sanger sequencing. 4, Contradicted by Sanger sequencing.
*This mutation is reported as homozygous by Liu and Bodmer (20).
Discussion
Given their importance for neoplastic development and phenotype, increasing effort is being placed on characterizing the mutations that are responsible for causing cancer and influencing its phenotype (21). There are several major efforts underway to create extensive catalogs of somatic cancer mutations from cancer cell lines and primary tumors (22). For example, Parsons et al. (23) selected 340 genes encoding tyrosine kinase from the human genome and resequenced them for mutations from primary colorectal carcinoma samples. They amplified individual exons by using PCR followed by Sanger sequencing. A total of 20 nonsynonymous point mutations, one insertion, and one splice-site alteration, were identified. A larger resequencing project involved the analysis of 13,023 genes in 11 breast and 11 colorectal cancers and identified 189 genes that were mutated at significant frequency (4). The majority of these genes were not previously known to be a frequent target of mutations. This project also relied on Sanger sequencing of simplex PCR products.
Herein, we present a strategy for large-scale resequencing of human genes by combining the recently developed selector technology with one of the currently available high-throughput sequencing technologies. This enables rapid resequencing from multiple genes with significantly less infrastructure required compared with a traditional resequencing procedure. We have applied this resequencing strategy for mutation identification from cancer cell lines.
To achieve cost-efficient high-throughput sequencing of multiplexed amplified sequences, it is essential that the target amplification step generates minimal artifacts and an even distribution of amplified target sequences. In our 10-gene experiments, an average of 90% of the generated sequence reads could be mapped to our reference sequence, illustrating the high specificity of the selector technology. The second 454-experiment generated ≈240,000 sequencing reads, and we were able to sequence four samples with average sequence coverage of 93%. However, because we required a sequencing depth of 5 or more to establish a consensus sequence, we only performed mutation analysis on an average of 81% of the total target sequence.
Improving sequencing coverage and depth is critical in the practical application of cancer genome resequencing and represents a limitation of the selector technology in its present form. Reasons for not obtaining full coverage may include poor digestion and/or denaturation of targets, inefficient circularization, and uneven amplification, which results in under- or overrepresentation of a given selector amplicon.
By performing a second iteration of selector design for target sequences that were not successfully sequenced in the first analysis, and adding the resulting selector probes to the existing set, we were able to increase the amount of sequence covered. This shows that the failure of one selector can be rescued by other selectors targeting the same region. This procedure could be repeated to further increase coverage. In addition, when developing new assays, a larger set of selectors could be designed initially, increasing the likelihood of success at any position. As more and larger sets of selectors are designed and used, we will learn to recognize sequence motifs that influence the probability of success. This knowledge then can be incorporated into the selector design procedure to increase the overall success rate, e.g., by designing a larger number of selectors for particularly difficult target regions.
In the present approach, some selectors generate more of their corresponding amplification product compared with others in the pool. This phenomenon decreases the overall sequence coverage by reducing the likelihood of sampling the underrepresented amplicons in the sequencing assay. A more even distribution of amplified targets will thus increase the overall sequence coverage. This could be achieved by increasing the concentration of individual selector probes that generate low amounts of amplification product and vice versa. Another potential approach to normalizing the distribution of amplified targets is to separate the pool of probes in one high-abundant and one low-abundant reaction, before PCR amplification. Furthermore, we recently developed an alternative sample preparation strategy, called “gene-collector,” which potentially generates a more uniformly distributed multiplexed amplification product compared with selector technology (24).
Our data show that the sequence coverage also can be increased by acquiring more sequence per sample. In the present study, each sample was sequenced by using approximately one-eighth and one-fourth of the GS20 instrument capacity in the first and second experiments, respectively. This increased sampling was the main contributor to the improved coverage in the second experiment. Improvements in parallel sequencing technologies have led to higher capacities, which can increase the sequence coverage, average sequence depth, and, ultimately, the number of genes targeted for analysis.
The selector design used in this study generates an amplification product with a size range of 138–238 bp, well suited for 454-analysis. If unspecific fragmentation of template DNA was performed before the sequencing reaction, the method would be less dependent on amplicon size. By selecting larger fragments with each selector, it would then be possible to decrease the number of selector probes required. We have shown previously that up to 1,000 bp fragments can be selected and amplified (19).
In the sequence data analysis, we identified a number of mutations and polymorphisms, including substitutions, deletions, and insertions, among the 10 genes. As a control, we used the Sanger method to sequence the TP53 gene in all of the samples. Double-stranded Sanger and 454-sequencing data were concordant to 99.94%, which agrees well with what has been reported for 454-seqeuncing (7). Where we had adequate sequence depth, we identified the previously characterized TP53 mutations, although the COLO741 AA insertion was called heterozygous instead of homozygous as described by Liu and Bodmer (20). In total, we confirmed 9 of the 11 variations we found. Furthermore, a number of genetic variants were found in the other nine genes and this represents an intriguing finding because these mutations may have functional effects on, e.g., kinase activity and sensitivity to inhibitors. We are pursuing additional studies to confirm these mutations and characterize their functional effects.
With the parameters used in our mutation screen, a large number of insertion/deletion variations were indicated. The vast majority of these represent the addition or removal of a single nucleotide at a position in or adjacent to a stretch of homopolymeric sequence containing that nucleotide. The 454-sequencing technology relies on a sequencing-by-synthesis process, pyrosequencing, well known to be susceptible to sequencing errors in homopolymer regions (25). The majority of the indicated insertion/deletions are thus likely to be artifacts from the pyrosequencing process. This type of error could be avoided by combining the selector technology with another sequencing platform. Also, as we refine our analysis algorithms and parameters, it will likely be possible to increase the fidelity of the consensus base-calling in these regions by using different base-calling criteria in different sequence contexts and analyzing the frequency of errors in a larger set of data. We currently are improving software to this end.
Massively parallel sequencing technologies have been proposed as means to carry out fast and cost-efficient mutation scans of complete human genomes. We propose to combine such technologies with methods for sequence-specific multiplex amplification to resequence genomic regions of particular interest, such as the coding sequences of cancer-related genes. For many applications, we believe this concept to have a number of advantages, including lower cost and greater sequencing depth per target than whole-genome sequencing.
Materials and Methods
Selector Design and Synthesis.
For each of the 10 target genes (FRAP1, AKT1, AKT2, TGFBR2, TP53, KRAS, APC, SMAD4, EGFR, and MARK3), all coding sequences including 50 adjacent nucleotides on either side were targeted for amplification. For each such target, the sequence and an additional 1,000 nt of sequence to either side was downloaded from the National Center for Biotechnology Information RefSeq database (26). Furthermore, dbSNP (27) was queried for known single-nucleotide polymorphisms in these regions, and the downloaded sequences were adjusted to reflect these polymorphisms by using the appropriate nucleotide degeneracy symbol.
The PieceMaker program (19) was used to select suitable restriction reactions and restriction fragments that fully covered the targeted regions, using a minimum fragment length of 100, a maximum fragment length of 200, and a maximum flap length of 500. The ProbeMaker software (28) was then used to design selector probe sequences for each of the selected restriction fragments.
All oligonucleotides were synthesized at the Stanford University Genome Technology Center. Selector probe sequences and their corresponding restriction enzymes, the vector sequence, and the PCR primer pair are described in supporting information (SI) Table 3.
Genomic DNA Samples.
Genomic DNA was extracted from six colorectal cancer cell lines (SW1417, VACO429, COLO741, C80, RKO1, and PC/JW) (20), one breast cancer cell line (HTB-20D), and one normal peripheral blood sample. Colorectal cancer cell lines were grown with 10% FBS (Autogen Bioclear, Wiltshire, U.K.) and 6 mM l-glutamine (CRUK). All cultures were mycoplasma-free and maintained in a humidified atmosphere with controlled CO2 content as indicated.
Genomic DNA was extracted from the colorectal cancer cell lines by using the DNeasy Tissue Kit (Qiagen, Crawley, U.K.) following the manufacturer's protocols. Genomic DNA was isolated from peripheral leukocytes by using the Gentra genomic DNA preparation kit (Minneapolis, MN). Genomic DNA from HTB-20D was obtained from the Coriell Institute for Medical Research (Camden, NJ).
Multiplex Amplification.
Five restriction digestion reactions were required to obtain full target sequence coverage. The enzymes used in the five reactions were FspBI/AluI, HpyCH4V, CviAII/BccI, DdeI/Bsp1286I, and MlyI/Hpy188I (New England Biolabs, Ipswich, MA). For each reaction, 10 units of each enzyme was used to digest the genomic DNA in recommended buffer and temperature for 1 h to a final concentration of 100 ng/μl. To ensure efficient denaturation of the digested DNA before the circularization reaction, the samples were heated to 105°C for 15 min by using a thermal cycler with heated lid (MJ Research, Waltham, MA). From each reaction, 250 ng of DNA was added to separate circularization reactions containing pooled selector probes in a total concentration of 10 nM, 100 nM of vector oligonucleotide, 1× Ampligase buffer (Epicentre, Madison, WI), 1 mM NAD, 5 units of TaqDNA polymerase (Invitrogen, Carlsbad, CA), 2 mM MgCl2, and 5 units of Ampligase (Epicentre) to a final volume of 20 μl. The circularization reaction was incubated at 95°C for 5 min, followed by 5 cycles of 95°C for 5 min, 75°C for 15 min, 65°C for 15 min, 55°C for 15 min, and 45°C for 15 min. Selector probes and vector oligonucleotides interfere with the PCR by generating a probe-dependent amplification artifact. To avoid this artifact, the uracil-containing probes were degraded by adding 10 μl of each circularization mix to individual 10-μl mixtures of 1× Uracil-Excision Buffer (Epicentre), 5 mM MgCl2, 0.01 μg/μl BSA, and 1 μl Uracil-Excision Mix (Epicentre) and incubated for 1 h at 37°C followed by 80°C for 20 min. Amplification was performed by adding 4 μl of each of the five uracil degraded circularization mixes to individual 21-μl mixes of 1× PCR buffer (Invitrogen), 0.25 mM dNTP, 3 mM MgCl2, 400 nM forward and reverse primers, respectively, and 0.02 units/μl Platinum Taq Polymerase (Invitrogen). Temperature cycling was performed as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The five PCR products finally were pooled and purified in a PCR purification column (Qiagen).
454-Sequencing.
The purified PCR products were analyzed according to the protocols described by Rothberg and coworkers (7), by using the GS 20 sequencing system (Roche, Indianapolis, IN). The large sequencing plate with either the four- or eight-lane gasket was used.
Sanger Sequencing.
Double-stranded Sanger sequencing on amplified exons was carried out on the TP53 gene for all samples described previously. Standard PCR and Sanger sequencing was performed similarly as presented in Liu and Bodmer (20). PCR primers are described in SI Table 4.
Sequence Data Analysis.
Sequence read data sets generated by 454-sequencing were reduced by grouping reads with identical sequence. Reference sequences for all regions targeted for amplification were downloaded from the National Center for Biotechnology Information RefSeq database (26). All unique reads were aligned to this set of reference sequences by using blastn (29) by executing the blastall program (version 2.2.15) with the default parameters except for gap open penalty 2, gap extend penalty 1, word size 16, and no filtering. If a sequence read generated multiple hits within the reference sequence set, the hit generating the highest blast score was used. For each position within the target regions, a sequencing depth was calculated as the sum of the sizes of all read groups with a hit covering that nucleotide position.
Consensus base-calling was performed for all positions with a sequence depth of 5 or more, by comparing the calls from all aligned hits at each position of the reference sequence. For each such position, the call of an individual aligned read could be either a single base, a gap (indicating loss of that base), or two or more bases if the alignment indicated an insertion of one or more bases between this position and the next. For positions where all reads yielded the same call, that base was immediately called. For positions with different calls from individual reads, the following rules were applied. If the second most common call was indicated in two or more reads, and in >20% of the total number of reads for that position, a heterozygote was called. In all other cases, the call most commonly made for the individual reads was used as the consensus call.
Supplementary Material
Acknowledgments
We thank Keith Anderson and Mike Jensen at the Stanford Genome Technology Center for oligonucleotide synthesis and Baback Gharizadeh, Roxana Jalili, and Shadi Shokralla for sequencing services. We thank Johan Banér for insightful comments on the manuscript. This work was supported by National Institutes of Health Grants 2P01HG000205 (to R.W.D., S.F., and K.W.) and 5K08CA96879–6 (to H.J.). Other support includes a Reddere Foundation grant (to H.J.), Liu Bie Ju Cha and Family Fellowship in Cancer (to H.J.), the CRUK program grant (to W.B. and D.B.), and the Wenner-Gren Foundation (F.D.).
Footnotes
Conflict of interest statement: M.N. and S.F. are cofounders of Olink AB and hold commercial rights to selector technology, and F.D and J.S. have business agreements with Olink AB.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702165104/DC1.
References
- 1.Bodmer WF. J Hum Genet. 2006;51:391–396. doi: 10.1007/s10038-006-0373-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, et al. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 6.Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 7.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 9.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, et al. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg SM, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, Halpern A, Khouri H, Kravitz SA, Lauro FM, et al. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, Laframboise T, Lee JC, Shah K, et al. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shigemori Y, Mikawa T, Shibata T, Oishi M. Nucleic Acids Res. 2005;33:e126. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan JB, Chee MS, Gunderson KL. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 16.Syvanen AC. Nat Genet. 2005;37(Suppl):S5–S10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 17.Broude NE, Zhang L, Woodward K, Englert D, Cantor CR. Proc Natl Acad Sci USA. 2001;98:206–211. doi: 10.1073/pnas.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenberg J, Dahl F, Landegren U, Nilsson M. Nucleic Acids Res. 2005;33:e72. doi: 10.1093/nar/gni071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Bodmer WF. Proc Natl Acad Sci USA. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varmus H, Stillman B. Science. 2005;310:1615. doi: 10.1126/science.310.5754.1615b. [DOI] [PubMed] [Google Scholar]
- 22.Vastag B. J Natl Cancer Inst. 2006;98:162. doi: 10.1093/jnci/djj062. [DOI] [PubMed] [Google Scholar]
- 23.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, et al. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 24.Fredriksson S, Baner J, Dahl F, Chu A, Ji H, Welch K, Davis RW. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronaghi M, Uhlen M, Nyren P. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenberg J, Nilsson M, Landegren U. BMC Bioinformatics. 2005;6:229. doi: 10.1186/1471-2105-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.