Abstract
Fluorescent protein (FP) variants that can be reversibly converted between fluorescent and nonfluorescent states have proven to be a catalyst for innovation in the field of fluorescence microscopy. However, the structural basis of the process remains poorly understood. High-resolution structures of a FP derived from Clavularia in both the fluorescent and the light-induced nonfluorescent states reveal that the rapid and complete loss of fluorescence observed upon illumination with 450-nm light results from cis–trans isomerization of the chromophore. The photoinduced change in configuration from the well ordered cis isomer to the highly nonplanar and disordered trans isomer is accompanied by a dramatic rearrangement of internal side chains. Taken together, the structures provide an explanation for the loss of fluorescence upon illumination, the slow light-independent recovery, and the rapid light-induced recovery of fluorescence. The fundamental mechanism appears to be common to all of the photoactivatable and reversibly photoswitchable FPs reported to date.
Keywords: crystallography, fluorescence, photoswitching, protein structure
Green fluorescent protein (GFP) from the jellyfish Aequorea victoria (1) and more recently, cyan, yellow, and red fluorescent proteins (FPs), isolated from coral reef organisms (2), have become standard research tools in cellular biology. Structural studies have revealed a universally conserved fold for FPs: an 11-stranded β-barrel with a central and axial α-helix (3, 4). A sequence of three amino acid residues on this central α-helix (Ser-65–Tyr-66–Gly-67 in Aequorea GFP) undergoes a series of autocatalytic posttranslational modifications to form the chromophore. Despite overall structural similarity across FPs from a variety of species and a variety of hues, members of the GFP family are quite diverse with respect to their photophysical properties. For example, properties such as emission color, extinction coefficient, quantum yield, and fluorescence lifetime can vary dramatically among FP variants. These properties are determined by both the influence of the protein matrix on the chromophore environment and the particular covalent chemical structure of the chromophore (5).
Another photophysical property common to all FPs is wavelength-dependent change in their fluorescence emission properties upon illumination. In most cases, these changes appear to result from irreversible covalent modification of the chromophore structure or its local protein environment. Examples of such permanent changes include light-induced transition from a fluorescent to a nonfluorescent state (photobleaching), transition from a nonfluorescent to a fluorescent state (photoactivation), or a change in excitation or emission wavelength maxima (photoconversion). Photobleaching, photoactivation, and photoconversion are well documented and can be advantageous for fluorescence imaging applications such as in vivo tracking of fusion protein movements (6–9). A less well documented process, occasionally described as “reversible photobleaching” (10) or photoswitching, is characterized by light-induced gain or loss of fluorescence followed by thermally driven recovery to the resting state. FPs capable of photoswitching are imaging tools that complement or exceed the performance of their irreversible brethren in many applications. For example, photoswitchable FPs such as asFP595/A143G (11), also known as the kindling FP (KFP), and Dronpa (12) have been used for imaging fast nucleocytoplasmic shuttling (12) and breaking the diffraction limit in fluorescence microscopy (13, 14).
Using a combination of rational mutagenesis and directed evolution, we recently developed monomeric teal FP (mTFP1), a bright and photostable version of the Clavularia cyan FP cFP484 (15). One of the intermediates obtained during the directed evolution process, a variant known as mTFP0.7, is brightly fluorescent (λemmax = 488 nm) but undergoes rapid photobleaching when illuminated with 458-nm light. Intriguingly, the fluorescence completely recovers at room temperature (RT) in a time span of minutes, indicating that this is a reversible photobleaching process. Furthermore, illumination of the bleached state at ≈360–400 nm accelerates the recovery of the fluorescent state. To elucidate the molecular basis for reversible photobleaching, we have determined high-resolution crystal structures of mTFP0.7 flash-frozen in the fluorescent and photobleached states. Structural differences between the fluorescent and photobleached states reveal the basis for reversible photobleaching and explain the slow thermal recovery of fluorescence from the bleached state and the rapid light-induced recovery.
Results
mTFP0.7 Spectral Properties and Photobleaching.
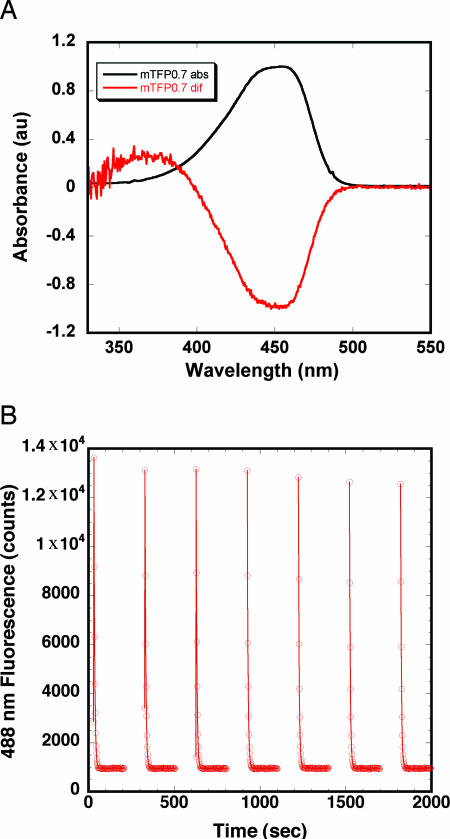
mTFP0.7 has a rather broad absorbance peak in the visible region centered at 453 nm (Fig. 1A). Excitation of this peak gives rise to cyan emission (λemmax = 488 nm). Under dilute conditions, such as those used in confocal fluorescence microscopy, intense illumination at the absorbance peak completely photobleaches mTFP0.7 (15). Fluorescence recovery is well fit by a single exponential but is strongly pH-dependent (Table 1). The recovery rate is 5-fold faster at pH 7.5 (τ ≈ 8 min) than at pH 4.3 (τ ≈ 46 min). Concomitant with the loss of fluorescence are spectral changes indicating a change in the protonation state of the chromophore (Fig. 1A). Absorbance at ≈450 nm decreases, whereas absorbance at ≈370 nm increases, indicating a shift in population from an anionic (fluorescent state) to a neutral, protonated (dark state) chromophore. Illumination within the absorbance band of the protonated chromophore (360–400 nm) dramatically accelerates the recovery of fluorescence in a pH-independent manner. Cycles of photobleaching and light-promoted recovery can be repeated with minimal loss in maximum fluorescence signal (Fig. 1B).
Fig. 1.
Spectroscopic response of mTFP0.7 to photobleaching. (A) mTFP0.7 ground-state absorbance spectrum (black) and postphotobleaching difference absorbance spectrum (red). Spectra are normalized at 450 nm to an absorbance of 1.0 and −1.0. (B) mTFP0.7 undergoing repeated cycles of photobleaching using unfiltered 458-nm LEDs (3 min, continuous portions of the plot) followed by photo-induced recovery using unfiltered 395-nm LEDs (2 min, discontinuous portions of the plot).
Table 1.
Fluorescence recovery time constants at 22°C
| Protein | pH | τ |
|---|---|---|
| mTFP0.7 | 4.3 | 46 ± 2 min |
| mTFP0.7 | 4.9 | 26 ± 2 min |
| mTFP0.7 | 5.2 | 8.7 ± 0.1 min |
| mTFP0.7 | 6.0 | 8.6 ± 0.2 min |
| mTFP0.7 | 6.5 | 7.8 ± 0.9 min |
| mTFP0.7 | 7.5 | 7.7 ± 0.2 min |
| mTFP0.7 | 10.7 | 4.1 ± 0.1 min |
| mTFP0.7/I161V | 7.5 | 68 ± 5 min |
| mTFP0.7/I161T (τ1)* | 7.5 | 2.5 ± 0.4 h |
| mTFP0.7/I161T (τ2)* | 7.5 | 70 ± 20 h |
*Fluorescence recovery in the I161T variant is biphasic.
Overall mTFP0.7 Fluorescent and Photobleached Structure.
Crystals of mTFP0.7 grew in space group P212121 with two molecules per asymmetric unit and a packing coefficient Vm = 2.0 Å3/Da (16). Statistics for data collection and refinement are given in supporting information (SI) Table 2. The final 1.79-Å resolution mTFP0.7 fluorescent state model (Rfactor = 0.186, Rfree = 0.252) contains all residues from Gly-6 through Tyr-221 for both protein chains, including the cyclized chromophore triads and 410 solvent molecules. Likewise, both chains in the 1.47-Å resolution photobleached state model (Rfactor = 0.192, Rfree = 0.261) contain residues Gly-6 through Tyr-221, including the chromophores, along with 450 solvent molecules and 1 acetate ion. PROCHECK analysis of both models found no residues in disallowed regions of the Ramachandran plot. Four residues in the fluorescent state model, Lys-102 and Glu-115 in both chains, and two residues in the photobleached state model, Glu-115 in both chains, were found in “generously allowed” regions. Inspection of the electron density maps for these residues indicates a good fit between the model and the maps (data not shown).
The overall fold of mTFP0.7 is the characteristic “β-can” that typifies the GFP family (4). Two independent monomers in the asymmetric unit of the crystal provide independent views of the fluorescent and dark states. The α-carbons of the noncrystallographic symmetry mates of the fluorescent and photobleached models superimpose with rmsd values of 0.13 and 0.16 Å, respec-tively. Overall, the structures of fluorescent and photobleached states are highly similar with α-carbon superposition of both chains giving a rmsd of 0.38 Å. Whereas mTFP0.7 was derived from the Clavularia sp. cFP484, sequence comparisons indicate that its closest relative with a known 3D structure is the green-to-red photoconvertable FP EosFP from Lobophyllia hemprichii (17). Despite differences in the oligomeric states of mTFP0.7 (monomeric) and EosFP (tetrameric), the α-carbons of the mTFP0.7 A and B chains in the fluorescent state superimpose very well on those of the EosFP A chain with rmsds of 0.32 and 0.38 Å, respectively.
The Chromophore and Environment.
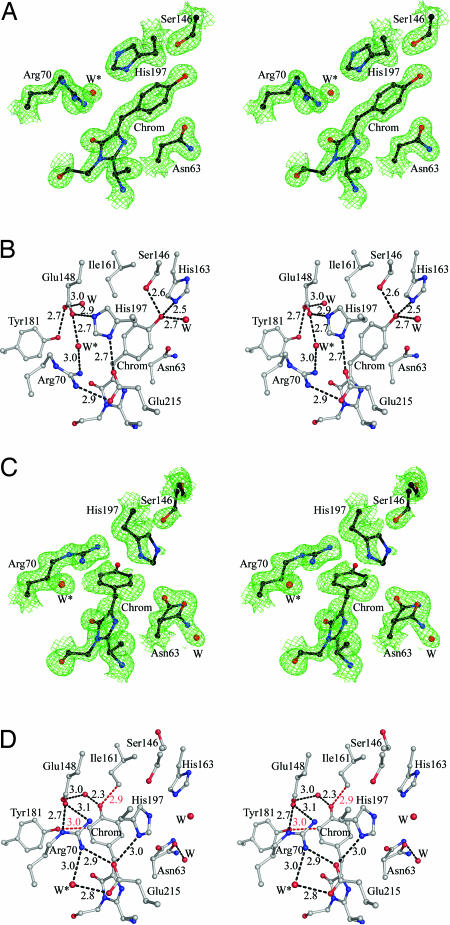
The mTFP0.7 chromophore forms from the tripeptide sequence -Ala-66–Tyr-67–Gly-68-. In the fluorescent state, the mTFP0.7 chromophore adopts a well ordered cis conformation with the aromatic rings slightly tilted from coplanarity (Fig. 2A and SI Fig. 3). The phenolate moiety is stabilized by hydrogen bonds to the side chains of Ser-146, His-163, and an internal water molecule (Fig. 2B). The side chain of His-197 stacks parallel to the phenolate ring and together with the side chains of Arg-70, Glu-148, Glu-215, and a second internal water molecule (W∗ in Fig. 2B) it participates in a quadrupole salt-bridge network that is commonly observed in the Anthozoa family of FPs (18, 19).
Fig. 2.
Stereoviews showing electron density and superimposed atomic models in the A monomers of mTFP0.7. Water molecules are represented as red spheres labeled W or W∗. The chromophores are labeled Chrom. (A) 2Fo − Fc maps (green) for the fluorescent-state model contoured at 1.8 σ. (B) The mTFP0.7 fluorescent-state model chromophore environment emphasizing hydrogen-bond partners and the quadrupole salt-bridge network adjacent to the chromophore. Hydrogen bonds are represented with black dashed lines (distances are given in angstroms). (C) 2Fo − Fc maps (green) for the photobleached dark state model, contour level 1.0 σ. (D) The chromophore environment of the photobleached state model (apparent close contacts are shown in red).
The structure of mTFP0.7 in the photobleached dark state reveals the chromophore to be in the trans isomeric state (Fig. 2C and SI Fig. 3). The electron density map indicates that the trans chromophore in both monomers is partially disordered and, on average, significantly nonplanar (angle ≈ 56°). Given the limitations of the diffraction resolution, we chose to represent the average configuration of the trans chromophore by a single atomic model. Refinement of the chromophore at full occupancy resulted in significant negative peaks in the Fo − Fc difference map around the phenol ring (data not shown). Refining the phenol rings of the A and B monomers with occupancies of 80% and 70%, respectively, eliminated these difference peaks. Residual positive features in an Fo − Fc difference electron density map suggest the presence of the cis isomer at low fractional occupancy and imply that photobleaching of the optically dense crystal was incomplete. In the dark state, the phenol moiety of the chromophore is positioned in a pocket bordered by the side chains of Glu-148, Ile-161, Tyr-181, and a water molecule (Fig. 2D), which in contrast to the chromophore, are well ordered.
The side-chain conformations of four residues adjacent to the chromophore (Asn-63, Arg-70, Ser-146, and His-197) and an internal water molecule undergo significant rearrangement between the fluorescent and dark states (Fig. 2). Upon photobleaching, the side chain of Asn-63 rotates to partially fill the void left by the isomerized chromophore. Ser-146 can no longer form a hydrogen bond to the phenolic oxygen of the chromophore and its major conformation has the hydroxyl group pointing away from the protein interior. Asn-63 and Ser-146 have alternative minor conformations consistent with those obtained in the fluorescent state. The side chain of His-197 is partially ordered in the dark state and was modeled with respective occupancies of 60% and 50% in the A and B monomers. Photobleaching destroys the quadrupole salt-bridge network, as the His-197 side chain rotates to partially fill the void created upon chromophore isomerization (Fig. 2) and thereby disrupts both the His-197 to Glu-148 ionic bond and the parallel stacking interaction of the imidazole ring with the chromophore. In turn, Arg-70 moves to a more extended conformation (modeled at 80% and 70% occupancy in the A and B monomers, respectively) to fill the void left by the His-197 side chain. Finally, a water molecule (W∗ in Fig. 2) fills the void left by the movement of Arg-70 and forms hydrogen bonds with Arg-70 and Glu-215. The combined effect of these dramatic rearrangements is to significantly alter the electrostatic environment and hydrogen-bonding pattern in the immediate vicinity of the chromophore (Fig. 2 B and D). To illustrate the conformational change, a video dramatization (SI Movie 1) has been produced by linear interpolation of the two end states.
Discussion
Structural Basis for Reversible Photobleaching.
Comparison of the mTFP0.7 structures led us to conclude that photoinduced chromophore isomerization has three major consequences, each of which is expected to diminish the fluorescent brightness. First, the trans isomer orients the phenolic ring such that there are few hydrogen-bond partners to stabilize the hydroxyl group in the anionic form (Fig. 2D). This, coupled with the close proximity of the phenolic oxygen to Cδ1 of Ile-161, should significantly shift the equilibrium toward the protonated phenol state, consistent with the observed increase in absorbance at 370 nm (Fig. 1A). Second, in contrast to FPs with high fluorescence quantum yields, which invariably display well ordered, relatively planar chromophore conformations (20), in the dark state the chromophore in mTFP0.7 is on average highly twisted and the two rings are noncoplanar. Third, the chromophore is partially disordered, presumably because the cavity shape does not preferentially stabilize any single configuration.
Experimental evidence suggests that any one of these three factors is sufficient to cause a complete loss of fluorescence. At neutral pH, the absorbance spectra of the overwhelming majority of FPs indicate the presence of a deprotonated, anionic chromophore. For most FPs, the chromophore becomes protonated and fluorescence is lost at low pH (21, 22). Thus, in mTFP0.7, isomerization to a more hydrophobic environment promotes chromophore protonation and loss of fluorescence efficiency. This conclusion is further supported by the greatly enhanced lifetime of the dark state at pH < 5.0 (τ ≈ 45 min at RT, pH 4.3; Table 1). An important exception to this general rule is wild-type GFP (and several other variants) that undergo excited-state proton transfer from the protonated chromophore to a conserved glutamic acid, leading to emission from the deprotonated form (23).
Large deviations in chromophore planarity dramatically reduce fluorescence efficiency, as first described for the blue chromoprotein Rtms5 (24) and later for the purple pigment KFP/A143G (25, 26). In both Rtms5 and KFP, the extremely low fluorescence quantum yield was attributed to a well ordered nonplanar chromophore present in the trans isomer. However, the trans isomer alone does not imply low fluorescence efficiency, as structural studies of the red FP eqFP611 revealed a well ordered trans planar chromophore (27). Presumably, the nonplanar chromophore configuration provides efficient pathways for nonradiative dissipation of excitation energy.
Finally, it is well established that the intact, rigid shell of GFP is essential for efficient fluorescence (28), presumably by suppressing vibrational or torsional modes of energy dissipation. Thus, the disordered state of the chromophore observed within photobleached mTFP0.7 is consistent with low fluorescence efficiency.
Structural Basis for Slow Thermal Recovery and Light-Induced Recovery.
The crystal structures of mTFP0.7 explain the slow, thermally driven dark recovery of fluorescence and rapid light-induced fluorescence recovery. Because of their tight confinement within the rigid β-barrel, side-chain rearrangements upon photobleaching are necessarily correlated and form “lock-and-key” interactions blocking recovery of the fluorescent-state conformation. Fluorescence recovery evidently requires concerted motions of the chromophore, as well as side chains Asn-63, Arg-70, Ser-146, and His-197 and at least one internal water molecule. Inspection of the best-fit model suggested that the side chain of Ile-161 could sterically crowd the chromophore hydroxyl group, destabilizing the trans isomer (Fig. 2D). To investigate this possibility, the I161V and I161T variants were constructed and purified. Although the spectral properties of I161V and I161T are essentially identical to those of mTFP0.7, indicating that the resting-state equilibrium still strongly favors the cis isomer, the lifetime of the photobleached state is dramatically lengthened (Table 1). Like its precursor, recovery of fluorescence in the I161V variant is monophasic (τ ≈ 1 h at RT, pH 7.5), whereas in the I161T variant recovery is biphasic (τ1 ≈ 2.5 h and τ2 ≈ 70 h at RT, pH 7.5). Thus, the protonated trans isomeric state is stabilized by both reduction of steric crowding and the addition of a hydrogen bond partner. Because the pKa of phenolic groups is known to decrease dramatically upon excitation (29), leading to excited-state deprotonation, it is not surprising that illumination at the absorbance maximum of the protonated chromophore should reverse the photoinduced dark state in all of the variants. Accordingly, illumination at intermediate wavelengths can be expected to have either effect (10).
Reversible Photobleaching vs. Photoswitching.
Reversible photobleaching very similar to that described here has been described for several GFP variants (10). In addition, the illumination-dependent spectroscopic changes observed for mTFP0.7 are remarkably similar to the behavior described as photoactivation of KFP (11) and as photoswitching of the FP Dronpa (12). The equilibrium state of KFP is nonfluorescent under physiological conditions with green light inducing photoactivation to a short-lived fluorescent state. By contrast, Dronpa is fluorescent at equilibrium and upon 488-nm illumination is converted to a long-lived nonfluorescent state. Transient-state lifetimes vary widely depending on the variant [t1/2 ≈ 50 s for KFP/A143G (11) and t1/2 ≈ 14 h for Dronpa (30)]. In both cases, absorbance spectra indicate that the photoswitched states of KFP and Dronpa are characterized by a shift in the protonation state of the chromophore, favoring the neutral protonated form. Illumination of the absorbance band of the protonated chromophore accelerates recovery to the resting state.
This similar spectroscopic behavior is echoed by striking structural similarities among these three proteins. Structural studies of KFP in the dark state have been conducted by three groups (25, 26, 31), revealing a significantly distorted trans chromophore stabilized by a hydrogen bond to the side chain of Ser-158 (corresponding to Ile-161 in mTFP0.7) and an internal water molecule not found in mTFP0.7. Andresen et al. (31) also described the structure of the putative activated state of KFP/A143S, which displays a cis chromophore isomer that is also significantly distorted. Although this observation supports the importance of trans to cis isomerization as a necessary step in the activation process, activated KFP/A143S has low quantum yield of fluorescence [≈0.12 (31)], thus the atomic model may reflect a mixture of fluorescent and nonfluorescent states (20).
More recently, crystal structures have been reported for Dronpa in the fluorescent state (30, 32). In each case, a slightly noncoplanar cis chromophore was observed, with conserved side chains forming an environment that was virtually superimposable upon that of the mTFP0.7 fluorescent state. Wilmann et al. (32) proposed a model for photoswitching of Dronpa that does not involve isomerization, but rather hinges on light-induced protonation of the cis isomer, which seems unlikely. On the other hand, Stiel et al. (30) proposed that photoswitching in Dronpa is caused by cis to trans isomerization, accompanied by conformational changes in the side chain of Arg-66 and possibly other residues. This proposal is consistent with the side-chain motion we observe for the analogous Arg-70, Asn-63, Ser-146, and His-197 residues of mTFP0.7. In Dronpa, mutation of Val-157 (corresponding to Ile-161 in mTFP0.7) to glycine increases the rate of photoswitching to the dark state ≈50-fold (30), purportedly by decreasing steric hindrance to chromophore isomerization. In KFP, the corresponding the S158V variant is permanently fluorescent with a relatively planar cis chromophore (31).
Although KFP, Dronpa, and mTFP0.7 are distantly related FP homologues, the similarities in their spectroscopic behavior and the chemical environment of their chromophore cavities suggest that the phenomena described as photoactivation, photoswitching, and reversible photobleaching share essentially the same structural basis. It also seems likely that similar processes underlie the previously described single-molecule on-off “blinking” and reversible photobleaching of GFP variants (10, 21, 33–36). Furthermore, these proteins have chemically different chromophores, as underscored by the distinctly different red- and cyan-emitting chromophores of KFP and mTFP0.7, respectively. In all cases, the protein must be able to accommodate cis–trans isomerization of the chromophore; however, it appears that in the resting state, the protein matrix determines whether the chromophore is fluorescent or nonfluorescent. In the case of KFP, the resting state enforces the distorted trans (hence nonfluorescent) conformation, whereas in Dronpa and mTFP0.7, the resting state stabilizes the cis-coplanar conformation. Chromophore planarity seems to be a key feature of efficient fluorescence; however, the structural elements within the protein that enforce the planar conformation are not yet understood. This research establishes a paradigm for the structural basis of reversible photobleaching and provides insight that will contribute to the design of novel photoswitchable genetic markers and improved research tools for cell biology.
Materials and Methods
Protein Expression, Purification, Crystallization, and Mutagenesis.
A pBAD/His B (Invitrogen, Carlsbad, CA) expression vector containing mTFP0.7 was transformed into Escherichia coli Top10 cells. A single colony was picked and used to inoculate 10 ml of LB media containing 0.1 mg/ml ampicillin (LB + amp), and the culture was grown overnight at 37°C. After dilution into 1 liter of LB + amp, cells were grown at 37°C to an OD600 ≈ 0.6, and expression was induced with the addition of 2 g of l-(+)-arabinose (Sigma, St. Louis, MO) to the culture. Cells were harvested after 16 h of growth at 30°C, centrifuged, and lysed by sonication. Protein was purified over Ni-NTA agarose columns (Qiagen, Valencia, CA) and concentrated by using centrifugal filters with a molecular mass cutoff of 10 kDa (Millipore, Billerica, MA) to a volume of 2.5 ml, and the concentration was estimated from theoretical molar extinction coefficients (37) at 280 nm.
Before crystallization, purified mTFP0.7 was digested for 24 h at RT with 1 mg α-chymotrypsin, type I-S (Sigma) per 50 mg of mTFP0.7 to remove the His tag. The sample was then buffer-exchanged (PD-10 sephadex column, Amersham Biosciences, Piscataway, NJ) into 50 mM Hepes, pH 7.9 + 0.3 M NaCl, passed back over the Ni-NTA column (collecting the flowthrough), and dialyzed against 1.2 liters of 50 mM Hepes, pH 7.9 + 0.3 M NaCl. Primitive orthorhombic crystals of mTFP0.7 grew in 1 week by hanging drop vapor diffusion. Drops included 1.8 μl of purified mTFP0.7 at 77.5 mg/ml in 50 mM Hepes, pH 7.9 + 0.3 M NaCl mixed with 1.8 μl of well solution (31% polyethylene glycol 3400, 0.2 M Li2SO4 and 0.1 M sodium acetate, pH 4.8) and 0.4 μl of 29 mM 1-s-Nonyl-β-d-thioglucoside (Hampton Research, Riverside, CA).
Site-directed mutagenesis was carried out (QuikChange; Stratagene, La Jolla, CA) using the following IDT primers: 5′-/5Phos/GTGCTGGTGGGCGATGTCAAACACAAG-CTGCTGC-3′ (I161V) and 5′-/5Phos/GTGCTGGTGGGCGATACCAAACACAAGCTGCTGC-3′ (I161T).
Spectroscopy.
Absorption and fluorescence spectra of mTFP0.7 were collected at RT in 50 mM Hepes, pH 7.9 + 0.3 M NaCl with a Hewlett–Packard (Palo Alto, CA) UV-visible spectro-photometer and PerkinElmer (Wellesley, MA) LS-55 fluorometer. Typical protein concentrations for absorbance and fluorescence measurement were 0.1 mg/ml and 20 μg/ml, respectively. The rate of thermal recovery from photobleaching was determined with a microspectrophotometer (4DX Systems, Uppsala, Sweden). The probe light was provided by a 75-W xenon lamp fitted with a 1-OD neutral density filter. Solutions of mTFP0.7 (≈1.2 mm3, A450 = 0.2 for a 1-mm pathlength) were placed in a 1-mm-diameter capillary tube and sealed with paraffin oil (Hampton Research) for data collection at 22°C. Photobleaching was essentially complete after 5 min of illumination from 458-nm light-emitting diodes (LEDs) at a flux of 7.5 mW/cm2. Photoinduced recovery was effected by using 362- or 395-nm LEDs at a flux of 4.0 mW/cm2.
Crystal Photobleaching and Data Collection.
For the fluorescent-state structure, a single crystal with dimensions ≈ 0.6 × 0.5 × 0.08 mm was swept through paratone-N oil, then flash-frozen at 77 K. Diffraction data were collected at 100 K by using the ADSC-Q315 detector on beam line 5.0.2 at the Advanced Light Source (Berkeley, CA). The resolution of the diffraction data set was limited by a suboptimal choice of detector position and not by the crystal. The data set and model will be replaced when higher-resolution data become available. Crystal photobleaching was performed with a 150-W halogen lamp fitted with a 450-nm broad band pass filter and bifurcated fiber optic bundle. A crystal with dimensions ≈ 0.4 × 0.35 × 0.15 mm was swept through paratone-N oil, mounted, and illuminated for 10 min with continual rotation before flash-freezing in liquid nitrogen. Diffraction data from the photobleached crystal were collected at 100 K by using the Quantum315 detector on beam line 14-BMC at the Advanced Photon Source (Argonne, IL).
Structure Determination and Refinement.
Diffraction images were indexed and processed with HKL2000 (HKL Research, Charlottesville, VA). The structure of mTFP0.7 in the fluorescent state was solved by molecular replacement using EPMR (38) and the coordinates for the A chain of EosFP (ref. 17; Protein Data Bank ID code 1ZUX) as the search model. Rigid body refinement (50–4.0 Å) was carried out with TNT (39) using a polyalanine skeleton derived from the molecular replacement solution. The program ARP/wARP (40) was used to build the initial model. Positional and B-factor refinement from 50 to 1.79 Å for the fluorescent state model was carried out with TNT using all data with no sigma or F cutoffs. The photobleached structure of mTFP0.7 was solved with EPMR using the refined mTFP0.7 fluorescent-state coordinates with the waters and chromophores removed. TNT was used for positional and B-factor refinement of the photobleached structure from 50 to 1.5 Å. In the final rounds of refinement, SHELXL (41) was used to include refinement of anisotropic displacement parameters for all data from 10 to 1.47 Å. The program Coot (42) was used for model building in between cycles of refinement, and the stereochemical quality of the final models was assessed with PROCHECK (43). Figures and rmsd calculations were made with Pymol (44). The angle between the two rings of the chromophore was determined by using GFPLANE (26).
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB 0417290 (to S.J.R.), National Institutes of Health Training Grant GM-07759 (to the Institute of Molecular Biology), the Natural Sciences and Engineering Research Council (R.E.C.), Alberta Ingenuity (R.E.C.), and the Canada Research Chairs Program (R.E.C.). SI Movie 1 was produced by M. W. Davidson of the National High Magnetic Field Laboratory, Florida State University, Tallahassee, FL. The Advanced Light Source is supported by Department of Energy Contract DE-AC02-05CH11231. Use of the Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by National Institutes of Health, National Center for Research Resources Grant RR07707.
Abbreviations
- FP
fluorescent protein
- KFP
kindling fluorescent protein
- mTFP
monomeric teal FP
- RT
room temperature
- LED
light-emitting diode.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2OTB and 2OTE).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700059104/DC1.
References
- 1.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 4.Yang F, Moss LG, Phillips GN. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 5.Remington SJ. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.van Drogen F, Peter M. Methods Mol Biol. 2004;284:287–306. doi: 10.1385/1-59259-816-1:287. [DOI] [PubMed] [Google Scholar]
- 7.Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Nat Cell Biol Suppl. 2003;5:S7–S14. [PubMed] [Google Scholar]
- 8.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 9.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 10.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 11.Chudakov DM, Belousov VV, Zaraisky AG, Novoselov VV, Straoverov DB, Zorov DB, Luykanov S, Lukyanov KA. Nat Biotechnol. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 12.Ando R, Mizuno H, Miyawaki A. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann M, Eggeling C, Jakobs S, Hell SW. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 15.Ai H, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 17.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Proc Natl Acad Sci USA. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 19.Henderson JN, Remington SJ. Proc Natl Acad Sci USA. 2005;102:12712–12717. doi: 10.1073/pnas.0502250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson JN, Remington SJ. Physiology (Bethesda) 2006;21:162–170. doi: 10.1152/physiol.00056.2005. [DOI] [PubMed] [Google Scholar]
- 21.Haupts U, Maiti S, Schwille P, Webb WW. Proc Natl Acad Sci USA. 1998;95:13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsliger MA, Wachter RM, Hanson GT, Kallio K, Remington SJ. Biochemistry. 1999;38:5296–5301. doi: 10.1021/bi9902182. [DOI] [PubMed] [Google Scholar]
- 23.Chattoraj M, King BA, Bublitz GU, Boxer SG. Proc Natl Acad Sci USA. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prescott M, Ling M, Beddoe T, Oakley AJ, Dove S, Hoegh-Guldberg O, Devenish RJ, Rossjohn J. Structure (London) 2003;11:275–284. doi: 10.1016/s0969-2126(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 25.Wilmann PG, Petersen J, Devenish RJ, Prescott M, Rossjohn J. J Biol Chem. 2005;280:2401–2404. doi: 10.1074/jbc.C400484200. [DOI] [PubMed] [Google Scholar]
- 26.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 27.Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 28.Ward WW, Bokman SH. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 29.Avigal I, Feitelson J, Ottolenghi M. J Chem Phys. 1969;50:2614–2617. [Google Scholar]
- 30.Stiel AC, Trowitzsch S, Weber G, Andresen M, Eggeling C, Hell SW, Jakobs S, Wahl MC. Biochem J. 2007;401:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andresen M, Wahl MC, Stiel AC, Grater F, Schafer LV, Trowitzsch S, Weber G, Eggeling C, Grubmuller H, Hell SW, Jakobs S. Proc Natl Acad Sci USA. 2005;102:13070–13074. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmann PG, Turcic K, Battad JM, Wilce MC, Devenish RJ, Prescott M, Rossjohn J. J Mol Biol. 2006;364:213–224. doi: 10.1016/j.jmb.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 33.Pierce DW, Hom-Booher N, Vale RD. Nature. 1997;388:338. doi: 10.1038/41009. [DOI] [PubMed] [Google Scholar]
- 34.Dickson RM, Cubitt AB, Tsien RY. Nature. 1997;388:356–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 35.Schwille P, Kummer S, Heikal AA, Moerner WE, Webb WW. Proc Natl Acad Sci USA. 2000;97:151–156. doi: 10.1073/pnas.97.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lounis B, Deich J, Rosell FI, Boxer SG, Moerner WE. J Phys Chem B. 2001;105:5048–5054. [Google Scholar]
- 37.Gill SC, von Hippel PH. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 38.Kissinger CR, Gehlhaar DK, Fogel DB. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 39.Tronrud DE, Ten Eyck LF, Matthews BW. Acta Crystallogr A. 1987;43:489–503. [Google Scholar]
- 40.Morris RJ, Perrakis A, Lamzin VS. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM, Schneider TR. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 44.DeLano WL. Pymol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.