Abstract
The association between large tumor size and metastatic risk in a majority of clinical cancers has led to questions as to whether these observations are causally related or whether one is simply a marker for the other. This is partly due to an uncertainty about how metastasis-promoting gene expression changes can arise in primary tumors. We investigated this question through the analysis of a previously defined “lung metastasis gene-expression signature” (LMS) that mediates experimental breast cancer metastasis selectively to the lung and is expressed by primary human breast cancer with a high risk for developing lung metastasis. Experimentally, we demonstrate that the LMS promotes primary tumor growth that enriches for LMS+ cells, and it allows for intravasation after reaching a critical tumor size. Clinically, this corresponds to LMS+ tumors being larger at diagnosis compared with LMS− tumors and to a marked rise in the incidence of metastasis after LMS+ tumors reach 2 cm. Patients with LMS-expressing primary tumors selectively fail in the lung compared with the bone or other visceral sites and have a worse overall survival. The mechanistic linkage between metastasis gene expression, accelerated tumor growth, and likelihood of metastatic recurrence provided by the LMS may help to explain observations of prognostic gene signatures in primary cancer and how tumor growth can both lead to metastasis and be a marker for cells destined to metastasize.
Keywords: cancer, genomics, oncogenesis
The consistent association of large tumor size, rapid growth rate, and metastatic risk in a majority of cases of clinical cancer suggests that the molecular bases of these phenomena may be linked (1–3). However, the nature of this link remains unresolved. Conventional models of metastasis envision rare metastatically competent variants emerging by chance as primary tumors grow, causally linking growth with likelihood of metastatic relapse (4, 5). In this view, genes that control primary tumor growth operate independently of stochastically acquired metastasis genes. Alternative models posit that prometastatic gene expression events are acquired early during tumorigenesis and may overlap with the genes that promote primary tumor growth, making tumor size a marker for metastatic risk (6). These alternative models form a teleological basis for using gene expression signatures from primary tumors to forecast whether patients are at high risk for micrometastatic disease. However, despite several reports on the success of gene signatures from primary tumors to predict development of distant spread (7–12), tumor size remains an independent prognostic factor on multivariate analysis (9). Thus, to what degree conventional versus alternative models can explain the acquisition of a metastatic phenotype remains unclear.
One of the difficulties in addressing the fundamental question on how metastasis gene expression events are acquired relates to the genetically complex nature of the phenotype itself. It has long been believed that there are numerous genes that control metastatic behavior due to the multiple steps required for distant growth and the observation that tumors demonstrate tissue tropism (4, 5). However, despite the description of multiple microarray gene signatures that are derived by statistical approaches, the statistical association of genes with the complex genetics behind a “poor prognosis” clinical phenotype result in genes that primarily track with metastatic propensity but are of unknown relevance as mediators of metastatic behavior (13, 14). The absence of experimental evidence that these gene signatures are actual mediators of metastasis makes it difficult to test predictions on the basis of the conventional versus alternative hypothesis of metastasis. In contrast, functional screens for genes that mediate metastasis can yield gene sets that are functionally relevant to the metastatic phenotype that they control (15, 16). Based on such gene sets, questions can be asked about if and when metastasis gene expression events are acquired by the primary tumor and the relationship of these genes to parameters such as tumor size.
We previously used the human breast cancer cell line MDA-MB-231 in a mouse xenograft model to select cell subpopulations that are highly metastatic to lung (15). These populations share a gene expression signature that is associated with and mediates lung metastasis in a mouse model. Furthermore, these genes promote tumor growth within the mouse mammary gland, providing evidence that metastagenicity can be linked to primary tumorigenicity. A subset of these genes, called the “lung metastasis gene-expression signature” (LMS), is expressed in a subgroup of human primary breast tumors with a pattern resembling the canonical expression profile of the lung metastatic cell lines. When tested on a cohort of 82 primary human breast cancers, a multigene classifier derived from the LMS predicted for patients at high risk for selective distant relapse to the lungs, but not to the bones. The expression of these genes in primary human tumors suggested a role both in primary tumor growth and as mediators of lung colonization from the circulation. By seeking further clinical validation that the LMS predicts for patients at high risk for lung metastasis, we have now uncovered experimental and clinical evidence that may explain how metastasis gene expression can be selected at the primary site. With this validation that the LMS can both mediate and predict lung metastasis, the influence of primary tumor growth in the context of conventional and alternative models of metastasis is addressed.
Results
Validation That the LMS Predicts for Development of Lung Metastasis.
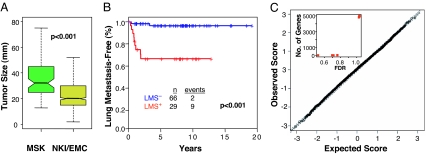
To validate that LMS expression by primary breast cancers can predict for risk of lung metastasis, two large cohorts of early stage breast cancer patients (NKI-295 and EMC-344) (9, 11) along with an expanded cohort (MSK-99) from an original series of tumors (15) were analyzed. The MSK cohort was more locally advanced compared with either the NKI-295 or the EMC-344 series (91% T2–T4 and 66% node positive compared with 47 and 49%, and 51 and 0%, respectively). This represents 738 primary tumors subjected to gene expression profiling across two microarray platforms. Hierarchical clustering with the 18 most univariately significant genes in the LMS classifier (15) shows their expression pattern among primary breast tumors [see supporting information (SI) Fig. 6]. Tumors classified as LMS+ show more uniform expression of these genes in a manner that resembles the cell line signature (weighted average Pearson correlation of LMS+ tumors to cell line signature is 0.27). These data demonstrate repeated observations of LMS expression by a subgroup of primary breast tumors.
Several statistical learning methods to classify tumors on the basis of LMS status performed similarly in predicting that LMS+ tumors are at high risk for developing lung metastasis (15). For simplicity and nonredundancy, here, we use only the nearest centroid classifier method. Results from this classifier demonstrate that LMS+ tumors from the MSK cohort have a significantly higher risk for lung metastasis compared with LMS− tumors (Fig. 1A).
Fig. 1.
Expression of the LMS predicts for increased risk of distant failure selectively in the lung and is associated with other markers of poor prognosis. (A) Kaplan–Meier curves representing the probability of cumulative lung metastasis-free survival for the MSK (Left) and combined NKI-295/EMC-344 (Right) cohorts. (B) Distribution of site(s) of first distant failure (simultaneous metastasis sites included) in the NKI-295/EMC-344 cohort according to LMS status. The P value for the difference in distribution for lung metastasis is shown and was calculated by a χ2 test. Patients with LMS+ primary tumors are shown in red, and LMS− tumors are in blue. (C) Hierarchical clustering was performed on the NKI-295 cohort with the indicated pathological and genomic markers consisting of the 70-gene prognosis signature (9, 10) (NKI 70), 16-gene recurrence score (18) (Rec score), molecular subtype (19) (Subtype), and wound response signature (17) (Wound). The legend for the color codes for each marker is shown. The luminal A, luminal B, and normal-like subtypes were grouped together as “Other” and consist of 5.8, 1.9, and 7.7% of LMS+ tumors, respectively.
Of a combined total of 639 patients from the NKI-295 and EMC-344 series, 231 patients developed distant metastases; of these, 70 patients developed lung metastases after a median follow-up of 8.7 years. Compared with the MSK cohort, the prevalence of lung metastasis in both the NKI-295 and EMC-344 groups is expected to be low because most tumors were removed at an early stage. Nonetheless, LMS expression is significantly associated with a worse lung metastasis-free survival in the individual (data not shown) and combined NKI-295/EMC-344 cohort (Fig. 1A), regardless of whether the analysis was done on cumulative lung metastasis events (Fig. 1A) or site of first distant failure (SI Fig. 7A). Multivariate analysis revealed the predictive ability of the LMS was independent of other standard prognostic markers (Table 1). Patients with LMS+ tumors selectively failed in the lung compared with bone and other visceral sites, including liver, and pleura (SI Fig. 7A). Although this was not associated with a higher rate of distant relapse in general or the likelihood that first distant failure involved multiple organs (data not shown), it was associated with a worse overall survival (SI Fig. 7B). An analysis of all sites of first distant failure revealed that compared with LMS− tumors, LMS+ tumors are more likely to relapse in lung than in bone, liver, or pleura (Fig. 1B). Analysis with the NKI-295 cohort showed that LMS+ tumors mostly were estrogen receptor-negative (73%), grade 3 (69%), and of poor prognosis (92%) on the basis of either a previously described 70-gene expression signature (9, 10), wound response signature (17), or 16-gene Recurrence Score (18) (Fig. 1C). The majority (65%) belonged to the basal-like molecular subtype with a smaller fraction (19%) classified as the ERBB2 subtype (19). Thus, consistent with our preliminary findings (15), LMS+ primary breast cancers form a subgroup of tumors with other poor prognosis molecular markers and have higher rates of distant metastasis to the lung compared with most other distant sites.
Table 1.
Multivariate Cox proportional hazard regression model for lung metastasis-free survival
| Variable | HR (95% C.I.) | P value |
|---|---|---|
| LMS (pos. vs. neg.) | 2.05 (1.1 to 3.83) | 0.024 |
| ER negative (yes vs. no) | 2.04 (0.91 to 4.58) | 0.082 |
| Tumor size (per mm) | 1.02 (1.00 to 1.04) | 0.037 |
| Lymph nodes (per node) | 1.07 (0.89 to 1.29) | 0.500 |
| Age (per year) | 0.97 (0.95 to 1.00) | 0.048 |
| Chemotherapy (yes vs. no) | 0.67 (0.23 to 1.93) | 0.450 |
| Endocrine therapy (yes vs. no) | 1.37 (0.40 to 4.75) | 0.620 |
The covariates shown in bold are significant, with a P value of <0.05. Grade was excluded in this analysis due to 108 missing values out of 639 patients in the NKI-295/EMC-344 cohort. The final model contains 632 patients (seven deleted due to missing values for other covariates).
The LMS Can Promote Primary Tumor Growth Resulting in Its Selection at the Primary Site.
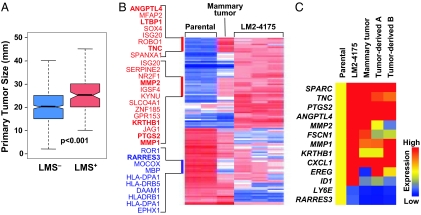
LMS genes can promote experimental tumor growth within the orthotopic mouse mammary gland (15). If clinically relevant, primary human breast cancers that are LMS+ will be larger at the time of diagnosis compared with LMS− counterparts. Analysis of the median primary tumor size demonstrated that LMS+ tumors are larger than LMS− tumors (Fig. 2A). In the absence of serial biopsies from the same patient, it is difficult to know whether LMS expression evolved as the tumor grew, resulting in LMS− tumors becoming LMS+. To experimentally determine whether LMS genes can confer a selective advantage that results in an LMS− tumor becoming LMS+ during tumor growth, the mouse model system was used. Mammary gland tumors were established from the parental MDA-MB-231 cell line, which harbors a small LMS+ subpopulation (< 1%, data not shown). Gene expression profiling (Fig. 2B) and quantitative RT-PCR analysis for LMS genes (Fig. 2C and SI Fig. 8) demonstrated an enrichment for LMS gene expression that includes genes that mediate lung metastasis, both in tumors generated from the parental population and in cell cultures derived from these tumors. These observations suggest that LMS+ cells within a breast cancer population have a growth advantage that can be efficiently selected during primary tumor progression.
Fig. 2.
LMS+ primary tumors are larger and can be experimentally selected for during primary tumor growth. (A) A box-and-whisker plot comparing the size distributions for LMS− and LMS+ primary tumors in the NKI-295/EMC-344 cohort. Shown are the medians, 25th and 75th percent quartiles, and 1.5 times the interquartile range. P values for the box plots were calculated by using a Wilcoxon rank sum test. (B) Transcriptomic microarray profiling was performed on parental MDA-MB-231 cells, the LMS+ in vivo selected lung metastatic LM2 subpopulation (LM2–4175) (15), and cells derived from xenografted parental MDA-MB-231 mouse mammary fat pad tumors. Mammary tumor denotes a sample from which in vivo mRNA expression was assessed directly from a fresh frozen mouse mammary tumor. The heatmap corresponds to relative gene expression levels for 95 previously identified lung metastasis genes (113 probe sets), with red being high and blue indicating low expression. Gene labels are provided for gene clusters of interest due to partial selection during outgrowth of a parental MDA-MB-231 mammary tumor. Genes highlighted in bold are functionally validated mediators of lung metastasis or are included in the 18-gene LMS. (C) Confirmation of microarray-based gene expression levels by using quantitative RT-PCR analysis. Expression values for representative lung metastasis genes were normalized to parental MDA-MB-231 expression levels and displayed graphically as a heatmap. Tumor-derived A and B represent in vitro analyses of independent isolates of cells purified from dissociated MDA-MB-231 mammary tumors.
Tumor Size Influences LMS-Related Metastasis.
The ability of LMS+ tumor cells and metastatic mediators of the LMS to both promote primary tumor growth and be selected within the primary site suggest that tumorigenesis and metastasis can be mechanistically linked and tumor size alone would not independently influence LMS-related metastasis. However, previous work has shown that when LMS+ cells are grown within the mouse mammary gland and then excised after reaching a size of 300 mm3, only about half of the mice develop lung metastasis, despite robust development of lung metastasis after direct inoculation of 2,000 cells into the mouse circulation (15). These results suggest that the primary site may be a significant barrier against distant dissemination even though the tumor is metastatically competent. To directly test this, LMS+ cells were grown as mammary tumors and circulating tumor cells were quantified by ex vivo colony forming assay. These results revealed that LM2 tumor cells do not initially intravasate into the circulation; however, progressive growth of the primary tumor leads to cells readily moving into the circulation (Fig. 3A). Once in the circulation, as few as 200 cells can form lung metastases (Fig. 3B). These results suggest that tumor growth may not necessarily contribute to metastasis solely by allowing for accumulation of metastasis genes, but rather may be required for intravasation of preexisting metastatically competent cells.
Fig. 3.
LMS+ primary tumors intravasate upon reaching larger tumor sizes. (A) Mice bearing parental or LM2 tumors grown in the mammary gland to various sizes were killed and bled via cardiac puncture. Mouse blood was lysed and plated in culture. Tumor cell colonies were scored and plotted as a function of tumor volume. (B) A suspension of 200 LM2 or parental MDA-MB-231 cells expressing luciferase were injected into the tail vein of mice. The development of lung metastasis was monitored over the indicated time course by using noninvasive luciferase bioluminescence imaging and quantitated by measuring photon flux. Values are the mean of four animals ± SEM. Representative mice from each group are shown.
To determine whether the experimental results are paralleled by clinical observation, the link between LMS expression, tumor size, and metastasis was investigated with the NKI-295/EMC-344 cohort. To avoid model assumptions such as linearity and to control for standard prognostic markers and potential interactions, a random survival forest analysis (20) was used to examine how tumor size is predicted to influence metastasis among LMS+ primary tumors (Fig. 4A and SI Fig. 9). In this analysis, the estimated risk for metastasis is determined after controlling for standard prognostic markers and interactions. The estimated risk for each patient with the indicated tumor size reveals that for the LMS+ patients there is a sharp rise in risk with tumor sizes >2 cm. In contrast, LMS− primary tumors showed no clear threshold for metastasis risk but rather display a steady rise beginning with tumors that are <1 cm. Similar results were observed when analysis was restricted to lung metastasis (data not shown). Stratification of LMS+ primary tumors into stage T1 (≤2 cm) versus T2 and larger (>2 cm) revealed that lung metastases are infrequent for the smaller LMS+ tumors (data not shown).
Fig. 4.
LMS+ primary tumors show a marked rise in metastatic risk after reaching ≥2 cm. (A) Factors that influence the risk of metastasis for patients from the NKI-295/EMC-344 cohort were determined by a random survival forest analysis. Clinical and pathological variables that include tumor size, patient age, histological grade, estrogen receptor status, and the number of positive lymph nodes were simultaneously entered into the model. This method is virtually free of model assumptions and involves constructing survival trees from bootstrap samples by using randomly selected covariates for tree splitting to deliver an ensemble cumulative hazard estimate for metastasis-free survival. The expected frequency of patients developing metastasis from the 128 patients with LMS+ tumors (Left) and the 511 patients with LMS− tumors (Right) is obtained from the ensemble estimate and plotted for each covariate. Shown are the results for tumor size. Results for other covariates are shown in SI Fig. 9. Patients that actually developed metastasis are indicated in red along with a lowess regression line through these points shown in magenta. (B) A concordance index from a random survival forest analysis modeling the influence of the LMS, tumor size, and other breast cancer prognostic gene expression signatures on the risk for lung metastasis was calculated (indicated by “All”) by using the NKI-295 cohort. This was then repeated with each of the indicated gene signatures or tumor size omitted from the full model (indicated above the blue bracket “Variable Removed”). The results from 50 runs are shown as a box-and-whisker plot. Nonoverlapping notches are considered significant. Both lung metastasis (Left) and overall survival (Right) were separately analyzed.
The association with larger tumor size and genes that promote primary tumor growth suggest that LMS+ tumors have high proliferative capacity. Proliferative capacity likely influences metastatic propensity. Indeed, the basal-like molecular subtype and the wound response signature are two gene profiles that are associated with proliferation-related gene expression events (21, 22) (SI Fig. 10). Because most LMS+ tumors are basal-like and/or express the wound response signature (refer to Fig. 1C), it was important to ensure that the increased risk of lung metastasis in LMS+ tumors is not merely due to the LMS being a marker for high proliferative capacity or for other metastasis gene expression signatures. To this end, the LMS and tumor size were combined with previously reported prognostic gene expression signatures in a multivariate model. Even after controlling for other gene signatures, tumor size, and potential interactions between them, LMS status remains strongly prognostic for lung metastasis (SI Fig. 11). Likewise, the ability to predict for lung metastasis is also significantly improved by the addition of the LMS to a model that already contains tumor size and other prognostic gene signatures that include the wound response signature and molecular subtypes. This is demonstrated by a decrease in the concordance index (the proportion of patients predicted to have a worse outcome that actually do have a worse outcome) with the omission of the LMS from the full model (Fig. 4B). For lung metastasis, the wound response and the NKI 70 gene signatures contribute less to prediction. In contrast, the LMS has the smallest influence on prediction for overall metastasis risk when compared with other signatures (Fig. 4B). In total, these results argue that the LMS and the interaction with tumor size do not merely overlap with markers for cellular proliferation or with other gene expression signatures. Both the LMS and tumor size add to the predictive accuracy for lung metastasis beyond what is achieved by other breast cancer prognostic signatures.
This influence of tumor size on LMS-related lung metastasis may partly explain the larger discriminatory effect of the LMS on lung metastasis in the MSK cohort. The mean primary tumor size in the MSK cohort was significantly larger compared with the combined NKI-295/EMC-344 series (Fig. 5A). When tumors smaller than the median MSK tumor size (≈3 cm) were excluded from the NKI-295/EMC-344 cohort, there was a larger separation in the lung metastasis survival curves between LMS+ and LMS− tumors (Fig. 5B), which approached the results from the MSK cohort (refer to Fig. 1A). The persistent separation of these lung metastasis survival curves even after >10 years of follow-up provides additional evidence that the LMS does not merely track with proliferation. In aggregate, these data suggest that although LMS+ primary tumors express genes that mediate lung metastasis, tumor size remains a strong influence on metastatic risk.
Fig. 5.
Tumor size influences the probability of lung metastasis for LMS+ tumors in a manner that is independent of discernable uniform changes in gene expression. (A) Box-and-whisker plot comparing tumor size distributions between the MSK and NKI-295/EMC-344 cohorts. The P value was calculated by using a Wilcoxon rank sum test. (B) Lung metastasis-free survival for patients with LMS+ tumors (red) compared with LMS− tumors (blue) in the NKI-295/EMC-344 cohort after excluding patients with a tumor size <3 cm (the approximate median size for the MSK cohort). P value was calculated by using the log-rank test. (C) Gene expression changes in tumors <2 cm in size and tumors ≥2 cm in size among LMS+ primaries from the NKI-295 cohort were compared by using SAM. Shown is a Q–Q plot with the Δ value corresponding to a median FDR of 10% marked by the dashed line. Genes that lie outside of this dashed line are deemed significant. In this case, no genes are determined to be significant at an FDR of 10%. The inset shows the number of significant genes as a function of increasing FDR.
One possible explanation for the prognostic influence of tumor size on metastasis among LMS+ tumors is that undiscovered genes that control metastasis may depend on tumor growth for expression. To investigate this, gene expression changes that are associated with LMS+ tumors >2 cm compared with ≤2 cm were analyzed with either the NKI-295 (Fig. 5C) or the EMC-344 (data not shown) cohorts. By significance analysis of microarrays (SAM) (23), even after allowing for a median false discovery rate (FDR) of 50%, no genes were found to be significantly associated with T1 versus T2 and larger tumors (Fig. 5C). Increasing the median FDR to >70% did not change the results (Fig. 5C Inset). An analysis using tumor size as a quantitative response variable, and an analysis for genes associated with lung metastasis-free survival among LMS+ tumors gave similar results (data not shown). Thus, among LMS+ primary breast cancers, tumor size contributes to metastatic potential in a manner that appears to be independent of appreciable uniform changes in the expression of other genes.
Discussion
Our data are consistent with the idea that the LMS confers a selective growth advantage for tumor cells at the primary site. Without the need for additional genetic alterations, the functions encoded by the LMS can drive the expansion of a pool of metastatically competent cells for continuous selection within the primary tumor. The consequent linkage between metastasis-promoting gene expression, accelerated tumor growth, and the likelihood of metastatic recurrence may help to explain the repeated observation of prognostic gene expression signatures in primary malignancies.
Tumor size is an important and often independent variable associated with metastasis in clinical studies and studies on poor-prognosis gene-expression signatures (9) (Fig. 4B). Because of clinical implications, it has long been debated whether tumor growth leads to metastasis or whether aggressive growth is a marker for cells destined to metastasize. Here, we show both can be true. Because LMS+ cells can be enriched from a predominantly LMS− population, unabated tumor growth can contribute to metastasis by selecting for metastatically competent cells. In this situation, early intervention may prevent expansion of LMS+ cells in the primary tumor. Interestingly, both our experimental and clinical analysis suggest that even after a primary tumor has reached full LMS+ status and is metastatically competent, progressive growth to attain larger tumor sizes is still needed to allow for intravasation. Local intervention during this window may still prevent distant spread. In contrast, once LMS+ tumors grow beyond this critical size, the ability of the LMS to promote accelerated growth and the resultant larger tumor sizes now become a marker for metastasis. In this situation, local modalities become insufficient for cure. Clinical analysis suggests that the critical size after which LMS+ tumors markedly increase the likelihood of intravasation and distant spread is ≈2 cm, which provides a biological basis for the observed importance of this size delineation in the breast cancer staging system.
Clinical validation that the LMS couples metastasis with primary tumor growth in a way that involves a biological interaction with tumor size might be confounded by the general influence that proliferation has on metastatic propensity. Indeed, the observation that poor prognosis gene expression signatures and molecular subtypes are associated with proliferation-related features, argues that general proliferative ability can contribute to metastasis. Because LMS+ tumors are both larger at the time of diagnosis compared with LMS− tumors and also express other poor prognosis gene expression signatures, it is important to ensure that the LMS is not simply a marker for proliferation. To this end, we demonstrate that molecular subtypes, other gene expression signatures, and proliferation-related transcriptomic changes are insufficient to fully account for the predictive utility of the LMS. Furthermore, the large predictive influence of tumor size and the lack of appreciable gene expression changes to explain the marked rise in metastasis of LMS+ tumors after 2 cm are also consistent with how tumor size plays a role independent of gene expression changes. Taken together, the LMS highlights the potential of combining clinical, pathological, and experimentally derived genomic markers in prognostic modeling. There are important covariates that cannot be captured by measuring gene expression changes alone, and gene expression signatures derived from functional screenings can provide complementary information (14, 24).
The mechanism by which LMS+ primary tumors acquire the ability to intravasate after reaching a certain size is currently under experimental investigation. Recent work was revealed that the LMS genes EREG, COX2, MMP1, and MMP2 constitute a vascular remodeling program that is coopted by tumor cells for the induction of tumor antiogenesis, intravasation, and lung extravasation (25). We note that despite the absence of a detectable change in gene expression as a function of either tumor size or lung metastatic risk in LMS+ tumors, we cannot exclude rare or diverse genetic or epigenetic changes that could contribute to metastatic behavior. In fact, experimental evidence has been provided for the existence of such gene expression events in regards to “metastasis virulence genes,” which principally control the rate rather than the likelihood of metastasis (15, 16).
The mode of progression to metastasis in LMS+ tumors may not apply to metastasis mediated by other genes or to other sites. Analysis of LMS− tumors demonstrates a steady rise in risk as a function of tumor size instead of a critical threshold size as seen in LMS+ tumors. Because LMS− tumors predominantly metastasize to bone, bone metastases gene-expression signatures may not correlate with tumor size despite correlating with clinical bone metastases and shorter survival. Thus, the present paradigm may only apply to fast-growing basal type tumors that metastasize to the lungs but not other more indolent but still lethal breast cancer syndromes. These issues highlight the complex genetic basis for metastasis and the historical difficulties in breaking ground on the credibility of different models. By providing confirmation of the biological and clinical relevancy of the LMS, this study leverages the known biology of a metastasis gene signature to address long-standing questions on how metastasis genes are engaged and to put forth new paradigms for future hypothesis testing.
Materials and Methods
Tumor Gene Expression Profiles.
Maintenance of cell lines and xenografting of immunocompromised mice has been described (15). RNA extraction, labeling, and hybridization for DNA microarray analysis of cell lines and primary breast tumors have been described for the MSK-99 and NKI-295 cohorts (9, 15). The EMC-344 cohort consists of 286 samples previously described (11) and an additional 58 estrogen receptor-negative samples and is available from the Gene Expression Omnibus (accession no. GSE5327). For quantitative RT-PCR studies, cDNA was synthesized from 1 μg of total RNA. Gene expression levels were determined by using Taqman and the ABI Prism 7900HT (Applied Biosystems). β2-Microglobulin was used as an endogenous control.
Data Analysis.
New tumors from each cohort (MSK, EMC-344, NKI-295) were classified as LMS+ or LMS− based on a nearest centroid classifier as previously described (15). There were 61 patients in the NKI-295 that overlapped with the 78 patients used to define LMS class membership. For these patients, their original class assignments were used. Hierarchical clustering of normalized data with median centering of gene expression values was done by using the 18 most univariately significant genes of the LMS using TIGR MultiExperiment Viewer 3.1 (26). Kaplan–Meier survival analysis and Cox proportional hazards regression modeling was performed with the “survival” package 2.26 in the R statistical package 2.3.1 (www.r-project.org). Random survival forest analysis was performed with the “RandomSurvivalForest” package 1.0.0 for R by H. Ishuaran and U. B. Kogalur. SAM was done with the “samr” 1.20 package. All relevant clinical information for patients included in the cohorts has previously been published (11, 13, 17), or is provided in SI Table 1.
Intravasation.
Parental or LM2 cells (15) were infected with pBabe retrovirus expressing a puromycin marker and injected into mammary glands of immunodeficient mice. Tumor volumes were measured as previously described (15). Animals bearing palpable tumors were killed, and 1–2 ml was bled through cardiac puncture. Red blood cells were removed by using ammonium chloride lysis buffer (Cambrex), and remaining cells were plated. After 48 h, adherent cells were grown in media with puromycin. After 10 days, colonies were stained with crystal violet blue and scored.
Supplementary Material
Acknowledgments
We thank A. Sieuwerts for RNA isolation and M. Meijer-van Gelder for data collection; L. Norton, A. Chiang, P. Bos, and P. Gupta for insightful discussions; and C. Arteaga, K. Polyak, and J. Nevins for critical reading of the manuscript. A.J.M. is supported by a grant from the American Society of Therapeutic Radiology and funds from The Ludwig Institute for Cancer Research. M.v.d.V., B.K., and D.N. are funded by Dutch Cancer Society Grant NKB2002-2575. J.M. is funded by National institutes of Health (NIH) Grant P01-94060 and a grant from the Keck Foundation. G.P.G. is supported by an NIH Medical Scientist Training Program grant and a Department of Defense Breast Cancer Research Program predoctoral award. D.X.N. is a postdoctoral fellow of the Damon Runyon Cancer Research Foundation. J.M. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- LMS
lung metastasis gene-expression signature
- SAM
significance analysis of microarray
- FDR
false discovery rate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701138104/DC1.
References
- 1.Giordano SH, Buzdar AU, Smith TL, Kau SW, Yang Y, Hortobagyi GN. Cancer. 2004;100:44–52. doi: 10.1002/cncr.11859. [DOI] [PubMed] [Google Scholar]
- 2.Heimann R, Hellman S. J Clin Oncol. 2000;18:591–599. doi: 10.1200/JCO.2000.18.3.591. [DOI] [PubMed] [Google Scholar]
- 3.Koscielny S, Tubiana M, Le MG, Valleron AJ, Mouriesse H, Contesso G, Sarrazin D. Br J Cancer. 1984;49:709–715. doi: 10.1038/bjc.1984.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fidler IJ. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 5.Chambers AF, Groom AC, MacDonald IC. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 6.Bernards R, Weinberg RA. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy S, Ross KN, Lander ES, Golub TR. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 8.Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA. J Clin Oncol. 2006;24:2261–2267. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 9.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 10.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 12.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 13.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 14.Massagué J. N Engl J Med. 2007;356:294. [Google Scholar]
- 15.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massagué J. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 17.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 19.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishwaran H, Kogalur UB. R News. 2007 in press. [Google Scholar]
- 21.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pittman J, Huang E, Dressman H, Horng CF, Cheng SH, Tsou MH, Chen CM, Bild A, Iversen ES, Huang AT, et al. Proc Natl Acad Sci USA. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta GP, Nguyen DX, Chiang A, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massagué J. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 26.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.