Abstract
Integrin-linked kinase (ILK) is a phosphorylated protein that regulates physiological processes that overlap with those regulated by p21-activated kinase 1 (PAK1). Here we report the possible role of ILK phosphorylation by PAK1 in ILK-mediated signaling and intracellular translocation. We found that PAK1 phosphorylates ILK at threonine-173 and serine-246 in vitro and in vivo. Depletion of PAK1 decreased the levels of endogenous ILK phosphorylation in vivo. Mutation of PAK1 phosphorylation sites on ILK to alanine reduced cell motility and cell proliferation. Biochemical fractionation, confocal microscopy, and chromatin-interaction analyses of human cells revealed that ILK localizes predominantly in the cytoplasm but also resides in the nucleus. Transfection of MCF-7 cells with point mutants ILK-T173A, ILK-S246A, or ILK-T173A; S246A (ILK-DM) altered ILK localization. Selective depletion of PAK1 dramatically increased the nuclear and focal point accumulation of ILK, further demonstrating a role for PAK1 in ILK translocation. We also identified functional nuclear localization sequence and nuclear export sequence motifs in ILK, delineated an apparently integral role for ILK in maintaining normal nuclear integrity, and established that ILK interacts with the regulatory region of the CNKSR3 gene chromatin to negatively modulate its expression. Together, these results suggest that ILK is a PAK1 substrate, undergoes phosphorylation-dependent shuttling between the cell nucleus and cytoplasm, and interacts with gene-regulatory chromatin.
Keywords: p21-activated kinase 1, signaling, nucleus
The integrin-linked kinase (ILK) is a major signaling integrator in mammalian cells. It plays critical roles in cell motility, adhesion-dependent signaling, cytoskeleton reorganization, and tumor progression and invasion (1–3). Upon activation by different stimuli, ILK phosphorylates AKT and glycogen synthase kinase 3β and participates in the related signaling cascades (1). ILK localizes to the focal adhesions by binding to the cytoplasmic tail of the β1-integrin subunit and by association with several cytoskeleton proteins, creating an ILK-based scaffold complex important for linking extracellular stimuli with cytoskeleton remodeling (1, 4). In addition to the focal adhesion points, ILK also localizes in the cytoplasm (5) and even in the nuclear compartment upon deletion of the caveolin-1-interacting site in ILK (6). Mutational analyses of the ILK kinase domain suggest that the scaffolding and enzymatic properties of ILK are intimately linked (4) and consequently that ILK's subcellular localization might depend on posttranslational modification of specific ILK residues. Nevertheless, even though ILK autophosphorylation of serine-343 within the kinase domain is considered important for ILK functions (7), regulation of ILK by direct phosphorylation has not yet been demonstrated. Moreover, even though ILK is known to be a phosphorylated protein, neither its upstream kinase, the biochemical basis for its spatiotemporal localization, nor the regulation of its nuclear localization has yet been described.
Recent studies have demonstrated that ILK-mediated regulation of cell motility and migration might be regulated by the small GTPases Rac1 and Cdc42 (reviewed in ref. 1). The serine/threonine protein kinase p21-activated kinase 1 (PAK1), a direct downstream effector of activated Rac1 and Cdc42, is an evolutionarily conserved kinase that plays clearly established roles in both the cytoplasm and the nucleus (8, 9). Upon activation by different stimuli (e.g., lipids and growth factors), PAK1 phosphorylates specific serine or threonine residues and/or interacts with its downstream targets and regulates cytoskeleton remodeling and cell migration (8, 9). In this context, some PAK1-regulated physiologic processes overlap with those of ILK-controlled cellular processes (1, 8). Collectively, these observations raise the intriguing possibility that PAK1 phosphorylation of ILK might be important in the ILK-mediated signaling network. Here we present data showing that ILK is a PAK1 substrate capable of phosphorylation-dependent nuclear-cytoplasmic shuttling and is potentially a critical protein for nuclear integrity and function.
Results
ILK Phosphorylation by PAK1.
Analysis of the ILK protein sequence revealed four putative PAK1 phosphorylation sites (i.e., K/RXXT/S) (Fig. 1A). We sought to determine whether PAK1 phosphorylates ILK. An in vitro phosphorylation assay was performed by using recombinant PAK1 as kinase and GST-tagged WT ILK (ILK-WT) as substrate. Results indicated that PAK1 phosphorylated ILK in vitro (Fig. 1B, second lane), and no detectable phosphorylation of ILK could be detected when PAK1 was not included in the assay (Fig. 1B, third lane). We next mutated the potential PAK1 phosphorylation sites in ILK-WT and used GST-ILK constructs in the in vitro PAK1 kinase assay. Mutation of either T173 to alanine (A) (T173A) or S246 to A (S246A) severely reduced PAK1-mediated ILK phosphorylation (Fig. 1C, fifth and seventh lanes). Conversely, mutation of either S228 to A (S228A) or T181 to A (T181A) only minimally affected ILK phosphorylation (Fig. 1C, fourth and sixth lanes). Because PAK1 apparently phosphorylated ILK on two different residues, we next mutated T173 and S246 to A together on ILK-WT. Interestingly, the double point mutant ILK-T173A; S246A (ILK-DM) could not be phosphorylated by PAK1 in the in vitro kinase assay (Fig. 1C, compare second and third lanes). Computer modeling of the ILK tertiary structure provided corroboratory evidence for T173 and S246 as potential ILK phosphorylation sites [supporting information (SI) Fig. 7]. Specifically, both sites reside in close proximity on the solvent-accessible molecular surface, with phosphorylation introducing a disruption of the local electrostatic surface potential.
Fig. 1.
ILK phosphorylation. (A) Potential PAK1 phosphorylation sites. (B and C) In vitro PAK1 phosphorylation of ILK (B) and ILK point mutant (C) constructs. Ponceau S staining indicates equal loading. (D) In vivo phosphorylation of ILK-WT and ILK-DM proteins in the respective MCF-7 clones. (E) Repression of PAK1 expression levels in MCF-7 cells stably expressing PAK1 shRNA. (F) Loss of in vivo phosphorylation of ILK in MCF-7 cells on depletion of PAK1 expression levels by stably overexpressing PAK1-specific shRNA. ILK bands are indicated by asterisks.
To investigate the significance of PAK1 phosphorylation of ILK in vivo, we attempted to establish stably transfected, pooled MCF-7 clones expressing V5-ILK-WT, the single mutants V5-ILK T173A and V5-ILK S246A, and ILK-DM. Stable transfection of both of the single point mutants was lethal to MCF-7 cells (three independent experiments; data not shown), but V5-ILK-WT and V5-ILK-DM pooled clones were established (SI Fig. 8A). Exponentially growing WT V5-ILK clone 21 (WT #21) and V5-ILK-DM clone 7 (DM #7) were metabolically labeled with [32P]orthophosphoric acid, and the status of ILK phosphorylation was determined by immunoprecipitation with an anti-V5 mAb (Fig. 1D). Results showed that V5-ILK was phosphorylated in vivo and that the mutation of both T173A and S246A significantly reduced ILK phosphorylation (Fig. 1D). Validation of ILK as a substrate of PAK1 was further done by studying ILK phosphorylation under conditions of depleted PAK1 levels in MCF-7 cells, which stably overexpressed PAK1-specific shRNA (Fig. 1E). In vivo labeling of ILK with [32P]orthophosphoric acid in MCF-7/PAK1 shRNA cells exhibited considerable reduction of ILK phosphorylation levels in comparison to the control cells (Fig. 1F and SI Fig. 8B, compare lanes 3′ and 4′). An asterisk indicates ILK bands in the Western blot and the autoradiogram images. Together, these findings demonstrated that ILK was phosphorylated by PAK1 on T173 and S246 and raised the possibility that PAK1 might be an upstream kinase for ILK capable of regulating ILK cellular functions.
Phosphorylation-Dependent ILK Functions.
We next evaluated whether PAK1 phosphorylation influenced ILK-mediated cell proliferation and migration. Indeed, MCF-7 ILK-WT cells showed a modestly increased proliferation (Fig. 2A) but a significantly increased migration (Fig. 2B) when compared with pcDNA vector control cells. However, both of these endpoints were reduced in MCF-7 ILK-DM-expressing cells (Fig. 2 A and B). These findings support a potential functional role for PAK1-mediated phosphorylation in ILK-regulated cellular processes.
Fig. 2.
Role of PAK1 phosphorylation sites on ILK cellular activities and localization. (A and B) Cell growth curve (A) and Boyden chamber assays (B) in exponentially growing (10% serum) MCF-7/pcDNA, ILK-WT, and ILK-DM clones. Data are the means ± SD of three independent experiments. ∗, differences from MCF-7/pcDNA cells; #, differences from ILK-WT #21 cells. Analyses included using the Student t test for overall significant differences within groups (P < 0.05). (C) MCF-7 cells transiently transfected with ILK-expressing vectors as labeled were fixed and stained for V5 tag. (Scale bars: 20 μm.) (D) Western blot analysis of exponentially growing Hec1A and MCF-7 cells after subcellular fractionation. Blots show representative results of three different experiments. ∗ indicates ILK in the nuclear fraction.
Cellular activities of ILK appear to depend on ILK subcellular localization, and ILK has been reported to localize in different intracellular compartments (1, 6). We next used laser scanning confocal microscopy to evaluate the potential effect of mutating PAK1 phosphorylation sites on ILK intracellular localization. Transient transfection of V5-ILK constructs demonstrated that ILK-WT localized primarily in the cytoplasm, but a small pool of ILK also localized in the nucleus (Fig. 2C). Transfection with the point mutants ILK-T173A, ILK-S246A, or ILK-DM altered the pattern of ILK localization (Fig. 2C). Surprisingly, in addition to localizing in the cytoplasm, ILK-T173A showed strong nuclear localization in 100% of transfected cells (Fig. 2C Upper Right). In contrast, ILK-S246A accumulated variably at cell focal adhesion points and in the nucleus (88% and 38% of transfected cells, respectively) (Fig. 2C Lower Left). Nuclear accumulation of ILK-DM was seen in 100% of the transfected cells, whereas localization to the focal adhesion points was observed in 67% of the transfected cells (Fig. 2C Lower Right). To further corroborate this evidence for the nuclear localization of ILK, we subjected MCF-7 and Hec1A cells to biochemical fractionation. As expected, endogenous ILK was detected mainly in the cytoplasmic fractions of both cell lines; however, ILK was also found in the nucleoplasmic fractions of both MCF-7 and Hec1A cells and in the nuclear matrix fraction of Hec1A cells (Fig. 2D). The quality of the cell fractionation was confirmed through analysis of protein markers of chromatin [poly(ADP-ribose)polymerase (PARP)], cytoplasm (vinculin), and nuclear matrix (lamin B) fractions. These results confirmed that, in addition to residing in the cytoplasm, ILK is a nuclear protein.
Regulation of ILK Subcellular Localization.
The changes in ILK localization with phosphorylation site mutations suggested that PAK1-dependent ILK phosphorylation might impact ILK's intracellular localization. For active nuclear-cytoplasmic shuttling, proteins predominantly require nuclear localization signals (NLS) and nuclear export signals (NES), which mediate the association with cargo proteins (i.e., importins and exportins). Protein phosphorylation can also influence intercompartmental protein shuttling (10). To evaluate the role of ILK NLS (363KKpedtnRR371) in ILK nuclear localization we created an ILK NLS mutant by directly mutating lysine (K) 363 to A (K363A) in V5-ILK-WT. Confocal microscopy analyses showed that the ILK NLS mutant remained almost completely cytoplasmic in 83% of transfected MCF-7 cells (Fig. 3A, compare with ILK-WT in Fig. 2C). Interestingly, 12% of ILK NLS mutant-expressing cells were binucleate (Fig. 3A Center); 42% showed gross alterations in nuclear shape, nuclear DNA distribution, or both; and 12% exhibited large DNA-containing cytoplasmic vacuoles (Fig. 3A Right). These findings suggested that the ILK NLS modulates the nuclear importation of ILK and that precluding ILK nuclear entry may adversely affect normal cell function.
Fig. 3.
ILK NLS and NES. MCF-7 cells transiently transfected with the V5-ILK NLS mutant (K363A) (A) or the V5-ILK NES mutant (I400A) (B) expressing vectors as labeled were fixed and stained for V5 tag (red), the F-actin-binding protein phalloidin (green), and DNA (blue). (Scale bars: 20 μm.) Transfected cells frequently displayed abnormalities (arrows).
To locate a possible NES in ILK, we used NetNES 1.1 software (www.cbs.dtu.dk/services/NetNES) to analyze the ILK protein sequence (11). We identified a putative NES in the ILK protein sequence consisting of a single amino acid (i.e., I400) (Fig. 3B and SI Fig. 9). Although NESs are generally leucine (L)/isoleucine (I)-rich, no specific consensus NESs have been established (11), and other investigators have reported that a single amino acid in the proper context can function as a NES (12). When MCF-7 cells were transfected with V5-ILK with I400 mutated to A (I400A), the expressed protein accumulated in the nucleus in 87% of transfected MCF-7 cells (Fig. 3B). These results demonstrate dramatically increased nuclear retention and at least a partial role for I400 as a NES. However, expression of ILK-I400A also altered cell morphology, causing DNA condensation, nuclear honeycombing, and marginalization of nuclear DNA in 28% of transfected cells (Fig. 3B Left and Right, white arrows); formation of binucleate cells (6%, Fig. 3 Center); altered nuclear shapes, including irregular membrane shape and nuclear blebbing or bulging (34%); and formation of large DNA-containing vacuoles (18%, Fig. 3B, red arrows) and micronuclei (14% of cells). Together, these data suggested a critical role for regulated ILK subcellular localization in the maintenance of both nuclear and cell homeostasis.
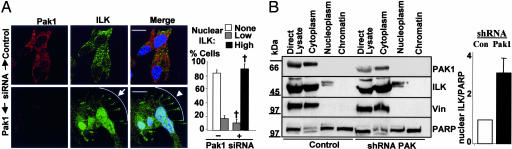
ILK is phosphorylated by PAK1, and, because mutation of PAK1 phosphorylation sites on ILK altered ILK subcellular localization (Figs. 1 and 2), we hypothesized that curtailing PAK1 kinase activity might affect ILK subcellular localization. Currently there is no commercially available, specific PAK1 chemical inhibitor; therefore, we examined the effects of PAK1-specific siRNA on ILK subcellular distribution. In scrambled oligonucleotide-transfected MCF-7 cells (control), ILK localized mainly in the cytoplasm, with minimal colocalization of endogenous ILK and PAK1 (Fig. 4A Upper). In PAK1 siRNA-transfected MCF-7 cells, PAK1 expression levels (as indicated by red staining) were effectively reduced (Fig. 4A Lower Left). Cells lacking PAK1 showed a dramatic redistribution of endogenous ILK from primarily cytoplasmic to marked accumulation in focal adhesions (Fig. 4A, arrow) and the nucleus (Fig. 4A). To support these observations, we stably transfected MCF-7 cells with a vector encoding short hairpin RNA (shRNA) against PAK1. Exponentially growing cells were then biochemically fractionated. PAK1 expression in the stable clones was effectively reduced by ≈50% when compared with that in vector control MCF-7 cells (Fig. 4B, top blot). ILK was detected in both cytoplasmic and nucleoplasmic fractions of both control and PAK1 shRNA-expressing clones. However, the reduction in cellular PAK1 levels induced an increase in the amount of ILK in the nucleoplasm fraction (Fig. 4B), suggesting that PAK1 may be required for ILK nuclear export. Accordingly, quantitation of the ratio of cytoplasmic ILK and vinculin to estimate the relative levels of cytoplasmic ILK in control and PAK1 shRNA clones showed a measurable decrease of cytoplasmic ILK in PAK1 shRNA clones by 7.2 ± 1% (n = 2). This also reflected an increased tendency of ILK to localize to the nucleus in cells with depleted PAK1 levels, thus supporting the involvement of phosphorylation by PAK1 in modulating subcellular localization of ILK.
Fig. 4.
Role of PAK1 in ILK nuclear localization. (A) MCF-7 cells were transfected with either scrambled oligonucleotides or PAK1 siRNA. After 48 h, cells were fixed and stained for endogenous ILK (green), PAK1 (red), and DNA (blue). (Scale bars: 20 μm.) (B) Western blot analysis of exponentially growing control vector and PAK1 shRNA stably transfected cells that were subjected to subcellular fractionation. Vin, vinculin. Blots show representative results of two different experiments. The graph shows densitometric analyses.
To delineate this potential role for PAK1 in the nuclear export of ILK, we next studied the cellular distribution of ILK in MCF-7 cells treated with EGF, a known PAK1 activator (9), or with leptomycin B, a selective inhibitor of nuclear export (13). Both EGF and leptomycin B induced nuclear accumulation of ILK (Fig. 5A). This nuclear accumulation of ILK was confirmed by confocal microscopy (Fig. 5B), with the combination of EGF and leptomycin B resulting in even greater nuclear ILK retention (Fig. 5B Lower Right). ILK distribution was scored in 200 cells under each condition, and results are presented in SI Table 1. Thus, the flux of ILK between the cytoplasmic and nuclear pools may be regulated not only by ILK nuclear import and export signals but also by ILK phosphorylation.
Fig. 5.
Role of PAK1 in ILK nuclear export. (A) MCF-7 cells were serum-starved for 48 h and treated with EGF (100 ng/ml) or leptomycin B (2 ng/ml) for 1 h, and lysates from different subcellular fractions were immunoblotted with the indicated antibodies. (B) Immunofluorescent staining of MCF-7 cells. Cells were treated as described above and immunostained for the endogenous ILK (green), F-actin-binding protein phalloidin (red), and DNA (blue). (Scale bars: 20 μm.) The results shown in B are quantified in SI Table 1.
Potential Nuclear Functions of ILK.
Regulated nuclear import and export of ILK suggest a functional role in the nucleus for this traditionally cytoplasmic protein. Because interfering with normal ILK localization altered the nuclear phenotype (Fig. 3) and because lamins are critical proteins for nuclear integrity and function (14, 15), we evaluated whether stably transfected ILK-WT #21and DM #7 clones showed changes in the expression or distribution of nuclear lamins. Lamin B distribution was minimally affected in WT or DM clones (data not shown). Conversely, lamin A/C expression was reduced in ILK-DM #7 (Fig. 6A Right) and showed atypical distribution patterns. These alterations included decreased nucleoplasm staining, areas of nuclear membrane thickening and disruption of contiguous nuclear membrane staining, cytoplasmic perinuclear accumulation, and polarized nuclear lamin distribution (Fig. 6A and SI Table 2). Altered distribution was seen in 26% of the WT #21 clones and 80% of the DM #7 clones.
Fig. 6.
ILK nuclear functions. (A) pcDNA/MCF-7, ILK-WT #21, and ILK-DM #7 clones were fixed and stained for lamin A/C and DNA. The results represented here are quantified in SI Table 2. Nuclear abnormalities were scored in at least 200 cells. (B) Schematic representation of the ILK-chromatin target region for the CNKSR3 gene. (C Upper) ChIP with V5 antibody validated the association of ILK-WT and ILK-DM with the CKNSR3 regulatory chromatin. (C Lower) ChIP with ILK antibody shows similar chromatin association of ILK in HEC1A cells. (D) Association of ILK-WT and ILK-DM with chromatin region 500 bp proximal to the CNKSR3 transcription start site. (E) Luciferase assay in MCF-7 cells shows the possible transcription-repressing functions of the ILK target chromatin. (F and G) Repressive effects of ILK-WT and ILK-DM overexpression on PGL3-ILK chromatin luciferase in the respective MCF-7 clones (H). Silencing of ILK expression in MCF-7 cell results in the relieving of repression on PGL3-ILK chromatin luciferase. Western blotting shows efficient silencing of ILK by siRNA (H Inset). (I) Luciferase assay in MCF-7 cells shows the effect of ILK-WT, ILK-DM, and ILK-NLS mutant proteins on PGL3-ILK target chromatin luciferase. (J) RT-PCR analysis of CNKSR3 expression in MCF-7 pcDNA, ILK-WT, and ILK-DM clones shows distinct repression of the gene by ILK. Quantitation from two different experiments is provided below. Data are the means ± SD of three independent experiments. ∗, Significantly different from values for the respective controls (P < 0.05, Student's t test for differences within groups).
Because extranuclear proteins can also function as transcription cofactors when localized in the nucleus (16, 17), we explored the possibility that ILK might interact with specific gene promoter chromatins. A genome-wide double ChIP assay was performed by using V5-ILK-WT #21 cells and a specific V5 antibody employing the method described in detail elsewhere (18). These experiments revealed one candidate chromatin target of ILK. This 336-bp chromatin fragment was from a possible regulatory sequence located on chromosome 6 and 110.2 kb upstream of the CNKSR3 gene. To validate this finding, the ChIP assay was repeated several times in ILK-WT #21 and also in Hec1A cells, using the V5-specific antibody and an ILK-specific antibody, respectively (Fig. 6C). We also designed ChIP primers for the 500-bp region immediately upstream of the CNKSR3 transcriptional start site and tested for ILK interaction in this traditional gene-regulatory region. Results indicated that, indeed, ILK was also associated with this region of regulatory chromatin (Fig. 6D). ILK-DM, which exhibited nuclear accumulation, was also capable of associating with these regions of CNKSR3 chromatin (Fig. 6 C Upper and D). These results confirmed the association of ILK with the candidate region of chromatin in both cell lines. The ILK pulled-down 336-bp fragment was then cloned into a PGL3 basic vector devoid of an intrinsic mammalian promoter or enhancer sequence upstream of a luciferase reporter gene. Transfection of MCF-7 cells with the PGL3-ILK chromatin target reporter showed that introducing the ILK chromatin target significantly reduced the basal level of PGL3 reporter activity (Fig. 6E). Luciferase assay conducted in the ILK-WT clone showed the inhibitory effect of ILK overexpression on this chromatin region (Fig. 6F), and this was more pronounced in the ILK-DM clone (Fig. 6G). Furthermore, the PGL3-ILK target reporter activity was increased in MCF-7 cells cotransfected with ILK-specific siRNA (Fig. 6H), indicating a relieving of repression by silencing ILK expression. In MCF-7 cells, transient overexpression of ILK-WT repressed ILK-chromatin target luciferase activity, and the repression was more prominent by ILK-DM (Fig. 6I). There was no repression by ILK-NLS mutant, which reflected its inability to localize to the nucleus (Fig. 6I). RT-PCR analyses of CNKSR3 mRNA levels showed down-regulation in both ILK-WT and -DM stable clones (Fig. 6J), confirming the inhibitory effect of ILK on CNKSR3 gene transcription. These data indicated that ILK associated with chromatin and may function as a transcriptional repressor of CNKSR3 gene. Thus, regulation of transcription by associating with specific target sequences may be an additional function of nuclear ILK.
Discussion
On the basis of commonly driven cellular outcomes (8) and the presence of PAK1 phosphorylation sites on ILK, we identified PAK1 as a physiological upstream kinase for ILK. We identified two solvent-exposed ILK residues (i.e., T173 and S246) as the sites of PAK1 phosphorylation on ILK. Mutation of the two PAK1 phosphorylation sites inhibited cell growth and migration. Analysis of the role of phosphorylation in ILK subcellular distribution led to several key observations. First, we observed that a pool of ILK localizes in the nucleus and that ILK's cellular localization is altered when ILK is phosphorylated. Second, we identified a functional NLS in ILK whose mutation abrogates ILK's nuclear localization. Third, we established that ILK localization is also regulated by an NES that we identified as I400. Fourth, we implicated PAK1 activity in ILK nuclear export by observing that ILK accumulates in the nucleus when PAK1 protein levels are reduced. Although treatment of cells with EGF, a potent PAK1 activator, increased nuclear ILK, our data indicate a potential role for PAK1 in ILK nuclear export, and other EGF-induced pathways may also trigger ILK nuclear localization.
ILK seems to be important for the maintenance of nuclear integrity because disruption of normal ILK subcellular localization patterns led to several nuclear aberrations, including altered morphology and abnormal lamin A/C distribution. Some of these nuclear alterations were reminiscent of those seen in diseases of compromised nuclear integrity, such as laminopathy leading to muscular dystrophy (19), to premature aging syndromes (20, 21), and to aging of the kidney (22). However, there are currently no literature reports connecting ILK and specific laminopathies, and this may warrant further exploration.
Our data also indicate another potential role for ILK in the nucleus. We found that ILK associates with target chromatin and acts as a gene repressor. This finding is not without precedent, because nonconventional, location-specific functions have been for both cytoplasmic (16, 17) and nuclear (23) proteins. Functional plasticity of proteins overcomes the restraints of subcellular localization and expands the repertoire of cytoplasmic proteins found in the nuclear compartment. In addition, in light of the nuclear localization of EGF receptor (16), myosin VI (17), actin (24), and ILK (present study), it is tempting to speculate that a network of cytoskeleton proteins may also extend functionally into the nucleus. Nonetheless, our findings raise as many questions as they answer. For example, identify the cellular compartment in which ILK becomes phosphorylated by PAK1. Considering that PAK1 localizes in the nucleus upon EGF treatment (25), this phosphorylation may occur in the nucleus to regulate ILK nuclear export. In this respect, the regulation of ILK's association with specific exportins (10) might be an additional level of regulation for ILK nuclear/cytoplasmic shuttling. It is also possible that phosphorylation might affect the ability of ILK to interact with and regulate its cytoskeleton-binding partners. Nevertheless, our present findings warrant the conclusion that PAK1-mediated ILK phosphorylation controls ILK-regulated cellular processes and that ILK is a nuclear protein with nuclear functions.
Methods
Cell Culture and Reagents.
MCF-7 and NIH 3T3 were purchased from American Type Culture Collection (Gaithersburg, MD). Details of antibodies and other reagents are provided in SI Methods.
Phosphorylation Studies.
In vitro phosphorylation was done by incubating GST-tagged ILK or ILK point mutants with GST-PAK1 enzyme and [γ-32P]ATP. In vivo phosphorylation was done by treating the cells with [32P]orthophosphoric acid followed by immunoprecipitation of ILK and separation by SDS/PAGE. Details are provided in SI Methods.
Immunofluorescent Labeling and Confocal Microscopy.
The cellular localization of proteins of interest was accomplished by indirect immunofluorescence as previously described (26) and as detailed in SI Methods.
ChIP Assay.
A genome-wide ChIP screening for ILK-chromatin targets was done by conducting the ChIP analysis in MCF-7 overexpressing V5-tagged ILK. The analysis was done by following the procedure explained in detail elsewhere (18) and detailed in SI Methods.
Supplementary Material
Acknowledgments
We thank J. Mascarenhas (University of Texas M. D. Anderson Cancer Center) for ILK mutagenesis and S. Dedhar (University of British Columbia, Vancouver, BC, Canada) for ILK construct. This work was supported by National Institutes of Health Grants CA109379 and CA90970 (to R.K.).
Abbreviations
- ILK
integrin-linked kinase
- PAK1
p21-activated kinase 1
- NES
nuclear export signal
- NLS
nuclear localization signal
- shRNA
short hairpin RNA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701999104/DC1.
References
- 1.Hannigan G, Troussard AA, Dedhar S. Nat Rev Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- 2.Esfandiarei M, Suarez A, Amaral A, Si X, Rahmani M, Dedhar S. Circ Res. 2006;99:354–361. doi: 10.1161/01.RES.0000237022.72726.01. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Yurecko RS, Dedhar S, Cleary PP. Cell Microbiol. 2006;8:257–266. doi: 10.1111/j.1462-5822.2005.00618.x. [DOI] [PubMed] [Google Scholar]
- 4.Legate KR, Montanez E, Kudlacek O, Fassler R. Nat Rev Mol Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 5.Driver GA, Veale RB. Cancer Cell Int. 2006;27:6–12. doi: 10.1186/1475-2867-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun J, Hyun S, Kwon T, Lee EJ, Hong SK, Kang SS. Cell Signal. 2005;17:751–760. doi: 10.1016/j.cellsig.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Persad S, Attwell S, Gray V, Mawji N, Deng JT, Leung D, Yan J, Sanghera J, Walsh MP, Dedhar S. J Biol Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- 8.Kumar R, Gururaj AE, Barnes CJ. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 9.Bokoch GM. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 10.Cyert MS. J Biol Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 11.Chevalier SA, Meertens L, Calattini S, Gessain A, Kiemer L, Mahieux R. Retrovirology. 2005;2:70–81. doi: 10.1186/1742-4690-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taagepera S, McDonald D, Loeb JE, Whitaker LL, McElroy AK, Wang JY, Hope TJ. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob KN, Garg A. Mol Genet Metab. 2006;87:289–302. doi: 10.1016/j.ymgme.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 16.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 17.Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, Bussi M, Biffo S. Mol Cell. 2006;23:749–755. doi: 10.1016/j.molcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Mazumdar A, Wang R-A, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ. Nat Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 21.Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Proc Natl Acad Sci USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Chen X, Xie Y, Shi S, Feng Z, Fu B, Zhang X, Cai G, Wu C, Wu D, Gu YJ. Gerontol A Biol Sci Med Sci. 2004;59:984–996. doi: 10.1093/gerona/59.10.b984. [DOI] [PubMed] [Google Scholar]
- 23.Marino M, Ascenzi P, Acconcia F. Steroids. 2006;71:298–303. doi: 10.1016/j.steroids.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Miralles F, Visa N. Curr Opin Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Singh RR, Song C, Yang Z, Kumar R. J Biol Chem. 2005;280:18130–18137. doi: 10.1074/jbc.M412607200. [DOI] [PubMed] [Google Scholar]
- 26.Kumar R, Wang RA, Mazumdar A, Talukder AH, Mandal M, Yang Z, Bagheri-Yarmand R, Sahin A, Hortobagyi G, Adam L, et al. Nature. 2002;418:654–657. doi: 10.1038/nature00889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.