Abstract
Although cardiac hypertrophy has been the subject of intensive investigation, regression of hypertrophy has been significantly less studied, precluding large-scale analysis of the relationship between these processes. In the present study, using pharmacological models of cardiac hypertrophy in mice, expression profiling was performed with fragments of more than 4,000 genes to characterize and contrast expression changes during induction and regression of hypertrophy. Administration of angiotensin II and isoproterenol by osmotic minipump produced increases in heart weight (15 and 45%, respectively) that returned to preinduction size after drug withdrawal. From multiple expression analyses of left ventricular RNA isolated at daily time-points during cardiac hypertrophy and regression, we identified sets of genes whose expression was altered at specific stages of this process. While confirming the participation of 25 genes or pathways previously shown to be altered by hypertrophy, a larger set of 30 genes was identified whose expression had not previously been associated with cardiac hypertrophy or regression. Of the 55 genes that showed reproducible changes during the time course of induction and regression, 32 genes were altered only during induction, and 8 were altered only during regression. This study identified both known and novel genes whose expression is affected at different stages of cardiac hypertrophy and regression and demonstrates that cardiac remodeling during regression utilizes a set of genes that are distinct from those used during induction of hypertrophy.
The heart increases its muscle mass in response to increased wall stress resulting from physiologic or pathologic states. The most characteristic cellular feature of this process is the enlargement of existing myocytes caused by the accumulation of sarcomeric proteins and reorganization of myofibrillar structures. Although this response is initially compensatory, it may progress to a pathologic state. As a result, cardiac hypertrophy is a predictor of cardiac morbidity and mortality, independent of hypertension or other risk factors (1, 2).
Analysis of gene expression during the induction of cardiac hypertrophy has revealed a specific transcriptional pattern associated with this process. Early mediators of the hypertrophic transcriptional program include the immediate-early genes (e.g., c-fos, c-myc, c-jun, and Egr1), followed by a cascade of mitogen-activated protein kinases (3–7). These changes contribute to substantial alterations in the expression and organization of sarcomeric and structural proteins (3, 8).
In contrast to the numerous studies examining the response to induction of hypertrophy, surprisingly few studies have examined genes whose expression is specifically altered during recovery from cardiac hypertrophy. The few studies that have examined gene expression changes during regression have largely focused on a small number of reporter genes such as myosin heavy chain (9, 10) and have not sought to identify new genes expressed specifically during this process. Because of the paucity of information concerning genes with altered expression during regression of hypertrophy, it has not been determined the extent to which cardiac remodeling during regression involves a simple reversal of the induction program, or activation of a separate transcriptional program. Knowledge of genes whose expression is altered specifically during regression might suggest novel strategies to limit hypertrophy and its progression to heart failure.
Large-scale efforts to identify the complete set of genes expressed by a variety of organisms, including humans and mice, have recently been coupled with the development of hardware for assessing the expression of many genes in parallel. Although genome-wide expression profiling has largely been applied to the analysis of unicellular organisms, tissue culture systems, and tumors (11–16), its application to tissues of whole animals offers an approach for examining gene expression in response to complex physiologic processes in vivo.
In the present study, we applied microarray expression profiling to identify genes altered during induction and regression of cardiac hypertrophy. To minimize the inherent variability of gene expression in vivo, an inbred mouse line was used. Pharmacologic inducers of hypertrophy were chosen so that induction and regression of hypertrophy could be precisely controlled, and two drugs were used to minimize drug-specific effects. Genes, both known and novel, were identified in these studies whose expression was altered specifically during induction and/or regression of hypertrophy.
Materials and Methods
Induction and Regression of Hypertrophy.
Male, 6-week-old FVB mice were treated with isoproterenol (ISO) (15 mg/kg/day × 7 days), angiotensin II (AII) (200 ng/kg/min × 14 days), or vehicle by osmotic minipump (Alza), implanted during Avertin anesthesia. Doses of these agents were chosen to mimic previous reports (17, 18). At the onset of treatment, drug-treated mice were paired with vehicle-treated mice, were matched for body weight, and were housed together throughout the experiment. After induction, the pumps were removed. ISO-treated animals were followed for up to 7 days, and AII animals were followed for up to 14 days after pump removal. Mice were killed at daily time points during induction and regression for both drugs. Whole heart and body weights were measured. The atria and right ventricular free wall were dissected away from the left ventricle, and samples were frozen within 3 min. The left ventricle was used for expression analysis. All procedures were approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
Histologic Analysis.
Parallel series of mice were treated with ISO, AII, or their respective vehicles, and hearts were prepared for histologic analysis. Animals were killed by cervical dislocation, and hearts were removed and fixed for 48 h in 4% paraformaldehyde before embedding in paraffin. Ten-micrometer sections were cut in the coronal plane, were rehydrated, and were double-stained with FITC-conjugated wheat germ agglutinin and 4′,6-diamidino-2-phenylindole (Molecular Probes). Myocyte size was quantified from photomicrographs of the left ventricular free wall by using imagepro software (http://www.mediacy.com). Other sections were stained with sirius red for analysis of collagen content.
Microarray Preparation.
Microarrays were prepared, hybridized, and scanned by using the protocols of http://cmgm.stanford.edu/pbrown (13, 19). The library used contained over 3,000 IMAGE consortium clones from Research Genetics, plus an additional 900 transcription factors, for a total of approximately 4,000 probes (46). A complete clone list is available at http://rubin.lbl.gov. The PCR amplified cDNA inserts were spotted on poly-l-lysine-coated microscope slides by capillary action using stainless steel pins. All clones named in the text or figures were sequence-verified.
RNA Extraction, Hybridization, and Quantification.
Total RNA was extracted from tissue by using the TRIzol reagent (GIBCO/BRL) or the RNeasy Midi Kit (Qiagen, Chatsworth, CA). For each time point, RNA was pooled from two drug or four vehicle-treated mice and 40 μg of total RNA labeled by reverse transcription using Superscript II (GIBCO/BRL), oligo(dT) primer, and Cy5-dUTP or Cy3-dUTP (Amersham) for experimental and control samples, respectively. Experimental and control cDNAs were hybridized to microarrays at 65°C overnight and were imaged by using a dual-laser scanner. Expression ratios were calculated by using the scanalyze software package (20). Ratios had a mean close to 1 and a SD of 0.4 for each array, which is comparable to the variation seen in other array studies (15, 19). Preliminary experiments were performed comparing hybridizations in which the same RNA sample was labeled with both dyes with hybridizations in which different control RNA samples were compared. These experiments suggest that one-third of the variation seen during experimental comparisons is attributable to variation intrinsic to the arrays and two-thirds is attributable to biologic variation between animals.
To allow comparisons between arrays, expression ratios were expressed as the number of SD from the mean. To correct for any gene-, sequence-, or intensity-specific artifacts, a set of six arrays comparing untreated hearts was used to calibrate each gene as described (19). The median expression ratio for each gene over the six control arrays was subtracted from each subsequent hypertrophy array.
Time Course of Gene Expression.
A set of approximately 4,000 cDNA clones was analyzed over a time course of cardiac hypertrophy induction. Array hybridizations were performed at daily intervals during the 7-day induction and regression of hypertrophy by ISO, and the 14-day induction and regression by AII. The use of a time course maximized our ability to compare two different drug models of hypertrophy that operated over different time frames and allowed us to identify genes altered in both models without having to predict the specific day of action. Adjacent time points also served as de facto replicates.
Data Extraction.
Those spots that were in the top 50% of total intensity for each array were analyzed. Empirically, these clones represent the subset of clones with detectable expression in heart tissue. To ensure sufficient data for individual analysis of each gene, only those genes with data in the top 50% of intensity for more than half of the time points were analyzed. This yielded a set of 1,310 genes for the 53 hypertrophy arrays.
Confirmation of Expression Level.
To validate expression ratios obtained by microarray, expression levels for atrial natriuretic factor were measured by quantitative reverse transcription (RT)–PCR using the 7700 Sequence Detector (PE Biosystems) and were compared with microarray results at several time points. Additional candidates were selected for post hoc confirmation at time points that led to their identification by microarray.
Results
Induction and Regression of Cardiac Hypertrophy.
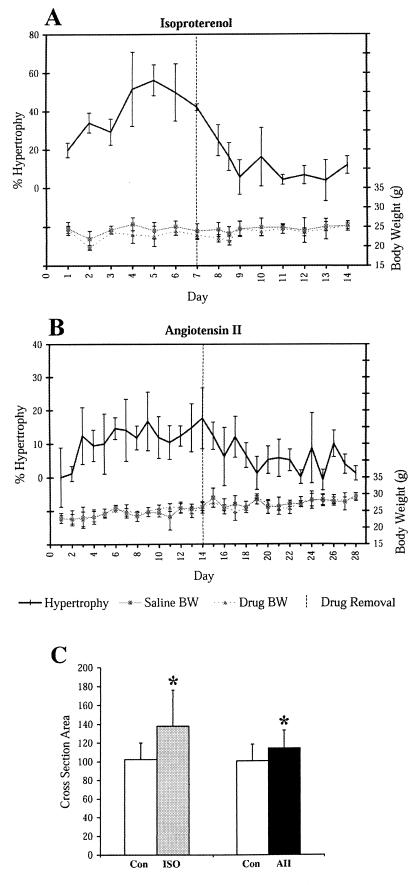
Isoproterenol (ISO) treatment induced an increase in heart weight of more than 45% (Fig. 1A) whereas angiotensin II (AII) treatment induced a 20% increase (Fig. 1B). During regression, heart weight returned to normal for both drugs, consistent with previous observations (21). Body weights for ISO- and AII-treated mice were not significantly different from vehicle-treated mice throughout induction and regression.
Figure 1.
Induction and regression of hypertrophy. Shown is induction and regression of hypertrophy induced by ISO (A) and AII (B). Percent hypertrophy is calculated as the heart/body weight of the drug-treated mice, divided by the heart/body weight of the matched, vehicle-treated controls. Lower curves show body weights on scale to the right. Drug was removed after 7 (A) or 14 (B) days of induction (dashed vertical line). (C) Relative myocyte cross-sectional area was calculated from photomicrographs of AII and ISO treated hearts. Bars represent means of 50 cells +/− SD. *, P < 0.0005.
It was expected that cardiac hypertrophy would be accompanied by an increase in cardiac myocyte size. To confirm myocyte hypertrophy in our models, we examined sections of the left ventricular free wall after 14 days of AII or 7 days of ISO treatment. Care was taken to examine sections that contained predominantly myocytes with central nuclei, located at midventricular level. Measurement of myocyte cross-sectional area (Fig. 1C) showed that ISO increased myocyte size by 35% and AII by 13%. Collagen staining did not show hypertrophy-associated fibrosis in either model (data not shown).
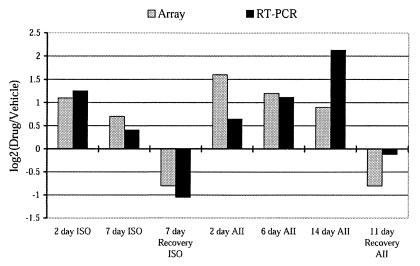
As a further validation of our models and as a validation of expression ratios determined by microarray, expression levels for atrial natriuretic factor, a robust marker of cardiac hypertrophy (22, 23), were measured by quantitative RT-PCR and were correlated with ratios determined by microarray (Fig. 2). As expected, atrial natriuretic factor expression ratios showed comparable increases during induction of hypertrophy, and decreases during regression of hypertrophy, when measured by RT-PCR and microarray. This demonstrates the ability of our microarray to detect modest changes in expression.
Figure 2.
Expression of atrial natriuretic factor during induction and regression. The log2 of the expression ratio (Drug/Vehicle) as measured by cDNA microarray and quantitative RT-PCR is given for several time points during induction and regression of hypertrophy.
Detection of Hypertrophy-Specific Changes.
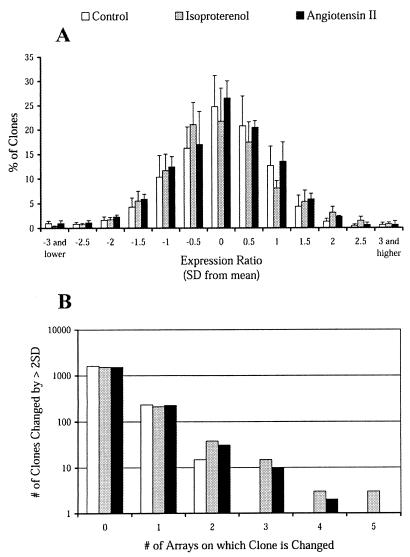
Because expression profiling has not previously been reported for whole cardiac tissue, the magnitude of normal variation in gene expression in the hearts of inbred FVB mice was determined. This was essential for assessing the threshold and reproducibility of “real” changes in expression during induction and regression of hypertrophy. When left ventricular RNA from pairs of saline-treated mice was hybridized to the microarray, detectable expression was demonstrated for approximately 50% of clones (1,860). Expression ratios (expressed as log2 values) were normally distributed and centered around a ratio of 1 (log2 ratio = 0) (Fig. 3A). As expected from the normal distribution, 5% of expressed genes had ratios lying more than 2 standard deviations (SD) from the mean for any given array. If the genes lying beyond 2 SD in this comparison represent normal stochastic variation in expression, the identity of clones present in the tails of the frequency distribution should vary randomly. To test this, 10 independent control RNA samples were hybridized to five arrays. Of the 1,860 expressed genes, 249 showed ratios more than 2 SD from the mean on one array, 15 showed consistent changes on two of the five arrays, and no gene showed a ratio more than 2 SD from the mean on more than two arrays (Fig. 3B). These values were not different from the numbers predicted by random variation. These results suggest that the expression ratio in untreated inbred mice follows a random distribution around 1, and that any systematic bias is small.
Figure 3.
Distribution of gene expression ratios for treated and untreated mice. The distribution of expression ratios for saline- and isoproterenol-treated mice is shown. A shows the mean number of genes, expressed as a percentage of the total number of expressed genes, with a given expression ratio (n = 5). Expression ratios are expressed as the number of standard deviations from the mean. B shows the number of genes that exhibited expression changes greater than 2 SD for zero, one, two, three, four, and five of 5 arrays tested, for vehicle vs. vehicle, ISO vs. vehicle, and AII vs. vehicle.
To determine the threshold for changes in gene expression that are attributable to hypertrophic stimuli, the distribution of expression ratios was determined for five ISO- vs. vehicle-treated ventricles (Fig. 3). As before, expression ratios were normally distributed with 5% of genes having ratios more than 2 SD from the mean. If, unlike the control arrays, some of the genes in ISO-treated animals exhibit real changes, these genes should be found reproducibly in the tails of the frequency distribution. When five arrays pairing RNA from ISO and saline-treated mice were examined, 270 clones were changed by more than 2 SD on one or more arrays, 59 on two or more arrays, 21 on three or more, 6 on four or more, and 3 on all five arrays (P < 0.001). Similar results were found for AII arrays (Fig. 3B). This suggests that reproducible hypertrophy-specific changes in gene expression can be identified through expression profiling. These results provided a basis for identifying real changes during the time course of hypertrophy induction and regression.
Genes with Altered Expression During Induction of Hypertrophy.
Two approaches were used to identify genes affected during hypertrophy. The first approach maximized the detection of genes with brief expression changes. Those clones showing expression changes greater than 2 SD from the mean (≈1.8-fold) were identified on each array. To separate true responses from experimental noise, a clone was required to demonstrate a change in expression ratio of more than 2 SD for two or more arrays during hypertrophy induction by ISO and two or more arrays during AII induction (P < 0.001). This algorithm provided a stringent filter against statistical noise because fewer than two genes would be expected to satisfy these criteria by chance. In addition, the requirement that clones show similar expression changes for both ISO and AII enriches for hypertrophy-specific effects at the expense of drug-specific effects.
The second approach maximized the detection of genes that show smaller, but more prolonged changes. Such genes may not be detected by the previous analysis, yet still contribute greatly to the phenotype. The average expression ratio for each gene over the induction time points was calculated after combining the data for ISO and AII. The average expression ratio over the regression time points was then subtracted. The mean difference was 0 and the standard deviation for this difference was calculated, assuming a normal distribution. Those genes with expression changes that differed from 0 by more than 3 SD (P < 0.01) were identified. If more than 90% of the change in mean expression ratio for a particular clone occurred during one phase of the experiment, that clone was designated as induction or regression-specific. The remaining genes were considered to be biphasic.
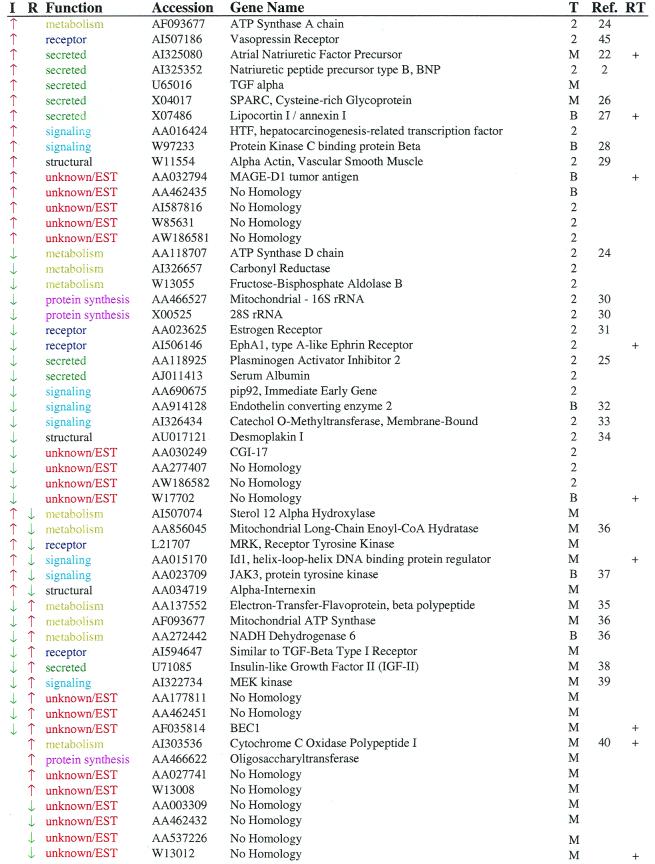
Thirty-two genes show changes in expression during only the induction phase of the time course (Table 1) and were evenly divided between those showing increases and those showing decreases in expression. These genes participate in a variety of cellular processes. For example, seven genes encode secreted factors, including three genes, atrial natriuretic factor, BNP, and PAI-2, previously implicated in hypertrophy. Similarly, four genes encode proteins participating in signaling pathways, and three genes, ECE-2, COMT, and PKC-bp, participate in pathways known to be important in hypertrophy. Two structural genes were identified, including a clone for α-smooth muscle actin. This clone is highly similar to α-skeletal muscle actin, a gene known to be up-regulated during hypertrophy (41), as well as α-cardiac actin. Our microarray may not distinguish these isoforms, so we cannot say with certainly which actin isoform is actually changing. This is a fundamental problem of any sequence-based approach, and it should be considered when selecting genes for follow-up analysis.
Table 1.
Genes that respond to angiotensin II and isoproterenol
Arrows indicate the direction of expression change for induction (I) and regression (R). Probable function and GenBank accession numbers are listed for each gene. Genes were identified based on tests for changes of two standard deviations on two arrays (2), based on mean levels of expression (M) or both (B). Where available, references are given linking the gene to cardiac hypertrophy. Genes indicated by “+” were confirmed by quantitative RT-PCR.
As expected, several genes participating in energy metabolism were identified. The one transcription factor identified, hepatocarcinoma-related transcription factor, had not been shown previously to be expressed in the heart. Finally, nine genes show no sequence similarity to any known gene. One-third of the 23 named genes altered during induction had not been linked previously to the hypertrophic process.
Changes in Gene Expression During Regression of Hypertrophy.
Given that few studies have investigated the regulation of gene expression during the regression of cardiac hypertrophy, gene expression was analyzed during regression in the same manner as had been done during induction of hypertrophy. Expression levels for eight genes were specifically altered during regression of hypertrophy. Four showed increased expression, and four showed decreased expression. A surprisingly large percentage of the clones identified during regression represent novel genes (six of eight). This may reflect the limited extent to which this process has been explored.
Genes Altered During Both Induction and Regression.
An additional 15 genes showed significant, reciprocal changes in expression during induction and regression such that the change could not be attributed more to one phase than the other. As noted for the genes above, these genes are spread over several functional classes. Eight of the fifteen were identified in previous studies of hypertrophy (Table 1) whereas three show no similarity to known genes. Included in this group are IGF-II, an orphan tyrosine kinase receptor MRK that is implicated in cardiac development, two kinases (JAK3 and MEK kinase), and a negative regulator of basis helix-loop-helix transcription factors (Id1) that has been implicated in cardiac myocyte apoptosis (42).
Post Hoc Confirmation of Expression Changes.
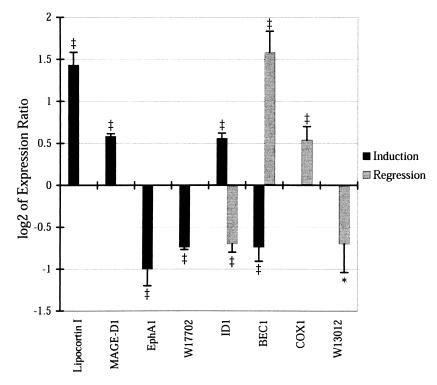
We chose eight genes from the induction, regression, and biphasic groups for confirmation by RT-PCR (Fig. 4). Four of the genes were up-regulated and four were down-regulated. Expression changes tended to be smaller when measured by this independent technique, but, in each case, the changes were in the same direction by array and RT-PCR and each expression change achieved statistical significance.
Figure 4.
Confirmation of expression levels by quantitative RT-PCR. The log2 of the expression ratio (Drug/Vehicle) as measured by quantitative RT-PCR is given for several genes at the time points during induction and/or regression that lead to their inclusion in Table 1. P values indicate probability that log2 ratio is different from 0. *, P < 0.05; ‡, P < 0.001.
Discussion
This study illustrates the utility of genome-wide screens of whole cardiac tissue as a tool for the discovery of new genes altered during cardiac hypertrophy and regression. The success of expression profiling of pharmacological models of hypertrophy is supported by the identification in our study of 25 hypertrophy-associated genes previously identified through analysis of cellular and physiologic models. This suggests that substantial similarity exists between the transcriptional programs activated by pharmacological and physiological models of hypertrophy. Indeed, 4 of 15 new hypertrophy genes identified in a recent genomic screen of pressure overload hypertrophy (43) were also identified in our screen.
Our analysis identified a large set of 40 genes whose expression changes were limited to the induction or regression phase and a smaller group of 15 genes whose expression changes appeared to be biphasic. The 32 genes specifically altered during induction of hypertrophy represent a spectrum of functional classes including secreted factors, receptors, intracellular signaling molecules, proteins involved in intermediary metabolism, structural proteins, and protein synthetic genes. Because the hypertrophic program is well known to cause alterations in many aspects of cell physiology, it is not surprising that genes were identified in each of these broad functional classes.
An important finding of this study was the identification of a distinct set of genes specifically altered during the regression of hypertrophy. This supports the concept that regression involves a separate transcriptional program from that operating during induction of hypertrophy. In contrast to the induction-specific genes, most of the regression-specific genes have no homology to genes of known function, in humans or other organisms. These novel genes would not likely have been identified through biochemical analysis of known pathways of hypertrophy induction, and their characterization may provide new insights into cardiac remodeling.
In addition to the induction- or regression-specific genes, a third set of genes was identified whose expression is differentially regulated between induction and regression but not obviously classified as induction- or regression-specific. This group includes genes in a variety of functional classes and relatively few genes of unknown function. Because the expression of this group of genes is not confined to induction or regression, they may be altered as a secondary response to hypertrophy rather than as primary regulators of the process. This is suggested by the proportionally larger number of genes involved in energy metabolism included in this set.
Our study identified 30 genes not previously associated with hypertrophy, and over half of these genes are completely novel. This was accomplished with a microarray comprised of less than 3% of the mouse genome. A screen employing all expressed sequences of the mouse would likely identify several hundred candidate genes, a significant fraction of which are predicted to be novel. Although a large list of candidates may be an important substrate for sequence polymorphism-based screens of populations in the search for disease modifiers, such a set is too large for in-depth biological study of each candidate gene. Additional methods for stratifying the list of candidate genes clearly will be needed to identify a subset of genes to target for further biological testing.
Several of the specific experimental hurdles imposed by expression profiling of whole tissues are illustrated in these studies. The fundamental challenge is to separate real changes in gene expression from normal variation, which in turn critically depends on the magnitude of normal variation in expression levels and the magnitude of changes observed during experimental manipulation. Because whole tissues are made up of many cell types, variation in expression level of some genes may be difficult to detect. Nonetheless, in our analysis, we were able to identify several hypertrophy genes expressed primarily by nonmyocytes (e.g., ECE-2, SPARC). In addition, gene expression appears to be more tightly controlled in vivo than it is in vitro. In our models, expression ratios rarely exceeded three-fold (3 SD; Fig. 3). This is consistent with observations of the skeletal muscle response to aging and caloric deprivation (44). Our study maximized the detection of real changes by making multiple comparisons between drug-treated and control animals on a defined genetic background, using two different models of hypertrophy and constraining our candidate list to those genes with the highest statistical likelihood of change. Although many known regulators of hypertrophy were not identified in our screen, this was expected because our experimental design and statistical analyses were specifically conceived to minimize false positive results.
It is striking that the majority of the genes identified during different stages of hypertrophy have not been implicated previously in cardiac hypertrophy. This is particularly true of the regression phase. Because regression of hypertrophy is less well studied and appears to involve a transcriptional program that is separate from induction, study of regression is uniquely suited to analysis by expression profiling. The genes identified in these studies may provide insight into basic pathophysiological mechanisms in cardiac remodeling, as well as suggesting potential leads for therapeutic intervention.
Acknowledgments
We thank David Kingsley and Greg Barsh for their gift of expressed sequence tag clones. This work was supported by National Institutes of Health Grant HL 63897-01 and Department of Energy Contract DE-AC0376SF00098 (Lawrence Berkeley National Laboratory) to E.M.R. C.J.F. is an Alexander Hollaender Distinguished Postdoctoral Fellow of the U.S. Department of Energy, Office of Biological and Environmental Research. J.B. is an Established Investigator of the American Heart Association.
Abbreviations
- ISO
isoproterenol
- AII
angiotensin II
- RT
reverse transcription
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. in Table 1).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100127897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100127897
References
- 1.Agabiti-Rosei E, Muiesan M L. Adv Exp Med Biol. 1997;432:199–205. doi: 10.1007/978-1-4615-5385-4_22. [DOI] [PubMed] [Google Scholar]
- 2.Schmieder A. J Am Med Assoc. 1996;275:1507–1513. [Google Scholar]
- 3.Chien K R, Knowlton K U, Zhu H, Chien S. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 4.Zimmer H G. J Mol Med. 1997;75:849–859. doi: 10.1007/s001090050176. [DOI] [PubMed] [Google Scholar]
- 5.Lijnen P, Petrov V. Methods Find Exp Clin Pharmacol. 1999;21:363–374. doi: 10.1358/mf.1999.21.5.541915. [DOI] [PubMed] [Google Scholar]
- 6.Robbins R J, Swain J L. Am J Physiol. 1992;262:H590–J597. doi: 10.1152/ajpheart.1992.262.2.H590. [DOI] [PubMed] [Google Scholar]
- 7.Sugden P H, Clerk A. Circ Res. 1998;83:345–352. doi: 10.1161/01.res.83.4.345. [DOI] [PubMed] [Google Scholar]
- 8.Schaub M C, Hefti M A, Harder B A, Eppenberger H M. J Mol Med. 1997;75:901–920. doi: 10.1007/s001090050182. [DOI] [PubMed] [Google Scholar]
- 9.Gupta M, Gupta M P. Mol Cell Biochem. 1997;176:273–279. [PubMed] [Google Scholar]
- 10.Cutilletta A F. Eur Heart J. 1984;5, Suppl. F:193–197. doi: 10.1093/eurheartj/5.suppl_f.193. [DOI] [PubMed] [Google Scholar]
- 11.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 14.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perou C M, Jeffrey S S, van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C, et al. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soonpaa M H, Field L J. Am J Physiol. 1994;266:H1439–H1445. doi: 10.1152/ajpheart.1994.266.4.H1439. [DOI] [PubMed] [Google Scholar]
- 18.Harada K, Komuro I, Shiojima I, Hayashi D, Kudoh S, Mizuno T, Kijima K, Matsubara H, Sigaya T, Murakami K, Yazaki Y. Circulation. 1998;97:1952–1959. doi: 10.1161/01.cir.97.19.1952. [DOI] [PubMed] [Google Scholar]
- 19.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 21.Saadane N, Alpert L, Chalifour L E. Br J Pharmacol. 1999;127:1165–1176. doi: 10.1038/sj.bjp.0702676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenzie J C, Tanaka I, Inagami T, Misono K S, Klein R M. Proc Soc Exp Biol Med. 1986;181:459–463. doi: 10.3181/00379727-181-rc3. [DOI] [PubMed] [Google Scholar]
- 23.Tsunoda K, Hodsman G P, Sumithran E, Johnston C I. Circ Res. 1986;59:256–261. doi: 10.1161/01.res.59.3.256. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien T X, Schuyler G T, Rackley M S, Thompson J T. J Mol Cell Cardiol. 1999;31:167–178. doi: 10.1006/jmcc.1998.0852. [DOI] [PubMed] [Google Scholar]
- 25.Feener E P, Northrup J M, Aiello L P, King G L. J Clin Invest. 1995;95:1353–1362. doi: 10.1172/JCI117786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelmann G L, Campbell S E, Rakusan K. Mol Cell Biochem. 1996;163–164:47–56. doi: 10.1007/BF00408640. [DOI] [PubMed] [Google Scholar]
- 27.Jans S W, de Jong Y F, Reutelingsperger C P, van der Vusse G J, van Bilsen M. Mol Cell Biochem. 1998;178:229–236. doi: 10.1023/a:1006803900554. [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki T, Komuro I, Zou Y, Yazaki Y. Hypertens Res. 1999;22:113–119. doi: 10.1291/hypres.22.113. [DOI] [PubMed] [Google Scholar]
- 29.Nemenoff R A. Front Biosci. 1998;3:D194–D207. doi: 10.2741/a274. [DOI] [PubMed] [Google Scholar]
- 30.Meerson F Z, Iavich M P. Kardiologiia. 1983;23:5–11. [PubMed] [Google Scholar]
- 31.Pelzer T, Shamim A, Wolfges S, Schumann M, Neyses L. Adv Exp Med Biol. 1997;432:83–89. doi: 10.1007/978-1-4615-5385-4_9. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Hoshi H, Sasaki H, Mitsui Y. J Cardiovasc Pharmacol. 1991;17, Suppl. 7:S182–S186. doi: 10.1097/00005344-199100177-00052. [DOI] [PubMed] [Google Scholar]
- 33.Krakoff L R, Buccino R A, Spann J F, Jr, De Champlain J. Am J Physiol. 1968;215:549–552. doi: 10.1152/ajplegacy.1968.215.3.549. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Gerdes A M. J Mol Cell Cardiol. 1999;31:333–343. doi: 10.1006/jmcc.1998.0886. [DOI] [PubMed] [Google Scholar]
- 35.Barger P M, Kelly D P. Am J Med Sci. 1999;318:36–42. doi: 10.1097/00000441-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Thurich T, Bereiter-Hahn J, Schneider M, Zimmer G. Arzneimittelforschung. 1999;49:212–220. doi: 10.1055/s-0031-1300404. [DOI] [PubMed] [Google Scholar]
- 37.Kunisada K, Tone E, Fujio Y, Matsui H, Yamauchi-Takihara K, Kishimoto T. Circulation. 1998;98:346–352. doi: 10.1161/01.cir.98.4.346. [DOI] [PubMed] [Google Scholar]
- 38.Vetter U, Kupferschmid C, Lang D, Pentz S. Basic Res Cardiol. 1988;83:647–654. doi: 10.1007/BF01906959. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez M T, Sah V P, Zhao X L, Hunter J J, Chien K R, Brown J H. J Biol Chem. 1997;272:14057–14061. doi: 10.1074/jbc.272.22.14057. [DOI] [PubMed] [Google Scholar]
- 40.Xia Y, Buja L M, Scarpulla R C, McMillin J B. Proc Natl Acad Sci USA. 1997;94:11399–11404. doi: 10.1073/pnas.94.21.11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson P C, Long C S, Waspe L E, Henrich C J, Ordahl C P. J Mol Cell Cardiol. 1989;21, Suppl. 5:79–89. doi: 10.1016/0022-2828(89)90774-8. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka K, Pracyk J B, Takeda K, Yu Z X, Ferrans V J, Deshpande S S, Ozaki M, Hwang P M, Lowenstein C J, Irani K, Finkel T. J Biol Chem. 1998;273:25922–25928. doi: 10.1074/jbc.273.40.25922. [DOI] [PubMed] [Google Scholar]
- 43.Shimkets R A, Lowe D G, Tai J T, Sehl P, Jin H, Yang R, Predki P F, Rothberg B E, Murtha M T, Roth M E, et al. Nat Biotechnol. 1999;17:798–803. doi: 10.1038/11743. [DOI] [PubMed] [Google Scholar]
- 44.Lee C K, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 45.Muders F, Kromer E P, Bahner U, Elsner D, Ackermann B, Schunkert H, Palkovits M, Riegger G A. Cardiovasc Res. 1995;3:416–421. [PubMed] [Google Scholar]
- 46.Lennon G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]