Abstract
Adherence of enterohemorrhagic Escherichia coli (EHEC) to the intestinal epithelium is essential for initiation of infection. Intimin is the only factor demonstrated to play a role in intestinal colonization by EHEC O157:H7. Other attempts to identify additional adhesion factors in vitro have been unsuccessful, suggesting that expression of these factors is under tight regulation. We sought to identify genes involved in the control of adherence of EHEC O157:H7 to cultured epithelial cells. A total of 5,000 independent transposon insertion mutants were screened for their ability to adhere to HeLa cells, and 7 mutants were isolated with a markedly enhanced adherence. The mutants adhered at levels 113 to 170% that of the wild-type strain, and analysis of the protein profiles of these mutants revealed several proteins differentially expressed under in vitro culture conditions. We determined the sequence of the differentially expressed proteins and further investigated the function of OmpA, whose expression was increased in a mutant with an insertionally inactivated tcdA gene. An isogenic ompA mutant showed reduced adherence compared to the parent strain. Disruption of the ompA gene in the tdcA mutant strain abolished the hyperadherent phenotype, and anti-OmpA serum inhibited adhesion of wild-type and tdcA mutant strains to HeLa cells. Enhanced adhesion mediated by OmpA was also observed with Caco-2 cells, and anti-OmpA serum blocked adherence to HeLa cells of other EHEC O157:H7 strains. Our results indicate that multiple elements control adherence and OmpA acts as an adhesin in EHEC O157:H7.
Shiga toxin-producing strains of enterohemorrhagic Escherichia coli (EHEC) are a class of pathogenic E. coli responsible for numerous food- and water-borne outbreaks, causing a range of illnesses from nonbloody diarrhea to hemorrhagic colitis or hemolytic-uremic syndrome in humans (reviewed in references 16 and 19). Strains of EHEC O157:H7, the most common EHEC serotype in North America, colonize the intestine and produce multiple determinants, which cause the pathology associated with the disease, with Shiga toxin being a key feature of virulence. EHEC O157:H7 also harbor a large pathogenicity island, termed the locus for enterocyte effacement (LEE), which is associated with the intimate adherence to epithelial cells, initiation of host signal transduction pathways, and the formation of attaching-and-effacing intestinal lesions (reviewed in references 16 and 40).
The intestinal colonization process is incompletely understood. The LEE-encoded intimin and Tir proteins have been shown to be required for intimate adherence, with intimin being the only O157:H7 virulence determinant demonstrated to play a role in intestinal colonization in vivo (4, 14, 42). The finding that intimin-negative strains are still associated with bloody diarrhea or hemolytic-uremic syndrome (5) led several research groups to seek additional adherence factors expressed by Shiga toxin-producing E. coli strains. Several proteins have been proposed as novel adhesion factors, including Iha, implicated in adherence to HeLa cells when expressed in E. coli K-12 (35); Saa, an autoagglutinating adhesin produced by LEE-negative strains (18); and Efa-1, an adhesin from non-O157 Shiga toxin-producing E. coli serotypes necessary for in vitro adhesion to CHO cells and for colonization of the bovine intestine (17, 33). In the case of EHEC serotype O157:H7, very little is known about potential additional factors associated with adherence. Recent studies by Tatsuno et al. have identified transposon mutants of EHEC strain O157 Sakai with reduced adherence to Caco-2 cells. Such mutants mapped within LEE genes and in open reading frames (ORFs) not directly associated with adherence, which suggests that LEE genes are playing major roles in in vitro adhesion (37, 38). We recently characterized a fimbrial operon in EHEC O157:H7 that mediates adherence when introduced into a nonfimbriated E. coli K-12 strain, but EHEC strains mutated in this operon were only modestly reduced in adherence to HeLa cells (39). It is challenging to determine why, from the multiple regions in the EHEC O157:H7 chromosome with a putative role in adherence, only a limited number of genes within the LEE and other loci have been suggested to be involved in the adherence of these organisms to host cells. These data and previous reports led us to hypothesize that other unidentified factors are involved in the adhesion to epithelial cells, and their functions have been difficult to establish because their expression is tightly regulated during conditions of in vitro adherence. To address this hypothesis, we mutagenized the EHEC strain 86-24 and isolated hyperadherent mutants, which could represent relief from the putative repression of adhesion factors in vitro. Seven mutants were identified that showed an enhancement in adherence to HeLa cells, and sequence analysis of the transposon insertions revealed mutated genes associated with lipopolysaccharide (LPS) biosynthesis, amino acid metabolism, fimbrial biogenesis, and a gene of unknown function. One phenotype of a hyperadherent strain mutated in the regulatory tdcA gene was increased expression of OmpA, and we present data showing that this protein may contribute to adherence of EHEC O157:H7.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Strains were routinely grown in Luria-Bertani (LB) broth or on L agar at 37°C (11). When indicated, the bacterial strains were grown in Dulbecco minimal Eagle medium (DMEM; Gibco/Invitrogen catalog no. 11885-084) at 37°C. Antibiotics were added to media at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and streptomycin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli | ||
| 86-24 | EHEC O157:H7 strain 86-24; Smr Nalr | 36 |
| EDL933 | Prototype EHEC O157:H7 | 27 |
| 85-170 | EHEC O157:H7 (stx) | 43 |
| E2348/69 | Prototype EPEC O127:H6 | 9 |
| SM10(λpir) | thi thr leuB-tonA lacY supE recA::RP4-2-Tc::Mu-Km; Kmr | 30 |
| AGT601 | 86-24, ompA::cat; Smr Cmr | This study |
| AGT602 | P9C8F2; ompA::cat tdcA::Tnp; Smr Cmr Kmr | This study |
| Plasmids | ||
| pGEMT-Easy | Cloning vector; Apr | Promega |
| ptdcA | 2,802-bp PCR fragment containing the tdcA gene in pGEMT-Easy | This study |
| pKM201 | pMAK700 derivative with red and gam expressed from Ptac; Apr | Kenan C. Murphy |
| pKD3 | pNTS derivative containing a FRT-flanked cat gene; Apr Cmr | 3 |
| pRS551 | Protein fusion vector; Apr Kmr | 31 |
| pPOMPA | pRS551 with EcoRI/BamHI 712-bp ompA promoter region | This study |
Smr, streptomycin resistance; Cmr, chloramphenicol resistance; Apr, ampicillin resistance; Nalr, nalidixic acid resistance.
Transposon mutagenesis.
Transposon mutants of EHEC strain 86-24 were generated with the kanamycin resistance (Kmr)-encoding transposome EZ::TN <R6Kγori/KAN-2> Tnp (Epicentre catalog no. TSM08KR) by electroporation according to the manufacturer's procedures. Briefly, electrocompetent bacterial cells were transformed with 1 μl of the Tnp transposome, 1 ml of SOC medium was immediately added, and the cells were incubated at 37°C for 1 h. Transconjugants were selected on L agar containing streptomycin and kanamycin. A total of 5,000 independent transposon mutants were recovered, and groups of five colonies were pooled together and maintained in 96-well plates (Corning) containing LB broth plus 50% glycerol at −80°C.
Bacterial adhesion to epithelial cells.
For both qualitative and quantitative adhesion assays, E. coli O157:H7 and its transposon or isogenic mutant strains were evaluated for their ability to adhere to HeLa cell monolayers by our standard protocol as previously described (39). Briefly, the strains were grown in LB broth overnight at 37°C and added to tissue culture cells replenished with fresh DMEM at a concentration of 107 bacteria per well for 6 h at 37°C. For adhesion studies with Caco-2 cells, monolayers were seeded with 2 × 105 cells/well and incubated for 48 h; after the cell monolayers were washed twice with phosphate-buffered saline (PBS, pH 7.4), the infection was carried out as described above. After the incubation period, the monolayers were washed, fixed, and stained with Giemsa solution for microscopic evaluation, or bacteria were recovered with 0.1% Triton X-100 in PBS and plated on L agar plates containing the proper antibiotic for quantification.
To screen the Tnp transposome mutant library for adherence ability, each pool of five mutants was grown in LB broth overnight at 37°C. The next day, bacteria and HeLa cells were incubated for 6 h in duplicate experiments. One set of experiments was fixed and stained with Giemsa solution for microscopic evaluation. In the other set, the adherent bacterial cells were recovered and reincubated in DMEM plus antibiotics overnight at 37°C. Bacterial pools that were more adherent than the wild-type strain after visual examination were further analyzed. The following day, a new adhesion assay was performed, and the bacterial cells still showing the hyperadherent phenotype were selected and incubated overnight at 37°C for a third round of assays. After the third adhesion assay, the bacterial cells were recovered and plated on L agar containing streptomycin and kanamycin. Six independent colonies from each pool appearing to be hyperadherent were selected, and their ability to adhere to HeLa cells was reassessed. Colonies displaying the phenotype were kept at −80°C, and the genomic location of the Tnp transposome was later determined as described below. Adherence data are expressed as the percentage of the bacterial inoculum recovered from triplicate wells and are the mean of the experiments. The statistical difference was expressed as the P value as determined by a Student t test analysis.
For inhibition of bacterial adhesion to HeLa cells experiments, bacteria were grown overnight in LB medium at 37°C, and 107 CFU were mixed with a preparation of anti-OmpA-specific antibodies (serum was diluted 1:10 or 1:100 in DMEM; OmpA antiserum was kindly provided by N. V. Prasadarao). After incubation for 30 min at 37°C, the mixture was added to HeLa cell monolayers, and bacteria were allowed to adhere for 3 h at 37°C. Quantitative and qualitative determinations were performed as described above.
Determining the transposon insertion sites in hyperadherent bacterial strains.
For each hyperadherent mutant, bacterial genomic DNA was isolated by using the Easy DNA extraction kit (Invitrogen), and 1 μg of the genomic DNA was digested with EcoRV. The digested genomic DNA was self-ligated by the addition of T4 DNA ligase and incubated overnight at 16°C. The ligated products were electroporated into SM10(λpir) cells, and the rescued clones were isolated on L agar plates containing kanamycin. The self-ligated clones were extracted by using the Wizard Minipreps DNA purification system (Promega), and the DNA sequence flanking the Tnp transposome was determined at the Biopolymer Core at the University of Maryland with the <KAN-2 FP-1> forward or R6KAN-2 reverse primers included in the Tnp transposome kit.
Construction of isogenic mutants.
Disruption of the ompA gene was performed in the chromosome of EHEC strains 86-24 and P9C8F2 (Tables 1 and 2) by a marker exchange procedure described by Datsenko and Wanner (3). The primer pair 5λROMPA (5′-CGGACAACGGCATGCTGAGCCTGGGTGTTTCCTACCGTTTCGGTCCATATGAATATCCTCCTTAG-3′) and 3λROMPA (5′-AGCTGATCCAGAGCAGCCTGACCTTCCGGTTTCAGGGTTGCTTTGGTGTAGGCTGGAGCTGCTTCG-3′) was used to amplify the cat cassette from plasmid pKD3 and introduced 46 nucleotides in each side of the cassette corresponding to the sequence of the ompA gene. The purified PCR product was introduced by electroporation into strains 86-24(pKM201) and P9C8F2(pKM201), and the transformed bacterial cells were plated on L agar containing streptomycin and chloramphenicol at 37°C. Colonies resistant to chloramphenicol and streptomycin were tested for ampicillin sensitivity. The presence of the disrupted ompA gene in strains AGT601 and AGT602 was confirmed by PCR with the primers 5RVOMPA (5′-CCGATATCGGTAGAGTTAATATTGA-3′) and 3XBOMPA (5′-CCTCTAGAAAGCGGTTGGAAATGGAAG-3′).
TABLE 2.
Hyperadherent 86-24::Tnp isolates
| Tnp insertion group | Gene disrupted | Identified or proposed function |
|---|---|---|
| LPS biosynthesis (P6C6E25) | waaI | LPS biosynthesis (O-island 154) |
| Amino acid metabolism | ||
| P9C8F2 | tdcA | Activator of the tdc operon: degradation of l-threonine |
| P9C12D4 | cadA | LDC I |
| Fimbrial biogenesis | ||
| P9C8F1 | Z5221 (lpfD2) | Putative minor fimbrial subunit (O-island I54) |
| P9C8B1 | csgD | Regulator of the csg genes: curli production |
| Unknown function | ||
| P10C9E1 | yidE | Putative transport and membrane protein |
| P10C4F1 | yidE | Putative transport and membrane protein |
Preparation of whole-cell lysates.
Bacterial cultures were grown overnight at 37°C in LB medium or DMEM, divided into aliquots containing ca. 2.0 × 108 bacteria, harvested by centrifugation at 12,000 × g for 5 min at room temperature, washed in 1 ml of 1× PBS (pH 7.4), centrifuged again, resuspended in 200 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) solubilization buffer, and lysed at 100°C for 10 min. Cell debris was removed by centrifugation prior to separation of supernatant proteins by SDS-10% PAGE minigels as described by Laemmli (8). Proteins were stained with Coomassie brilliant blue.
Outer membrane preparations.
Outer membranes were prepared by the procedure of Torres and Payne (41), with the following modifications. Bacterial cells grown in DMEM were harvested in mid-log phase, pelleted by centrifugation, and resuspended in 10 mM sodium phosphate buffer (pH 7.5) before they were disrupted by using a French pressure cell at 18,000 lb/in2. The crude total membranes were collected by centrifugation at 100,000 × g, and Sarkosyl (sodium N-lauroyl sarcosine; Sigma)-insoluble outer membranes were separated from the crude total membranes by centrifugation at 100,000 × g. Outer membranes were resuspended in Laemmli solution buffer (8), and the proteins were separated by SDS-12% PAGE.
Western blot assay.
Proteins separated by SDS-10% PAGE minigels were transferred to Immobilon-P (polyvinylidene difluoride) membranes (Millipore) by using a Trans-Blot SD transfer cell (Bio-Rad) at 15 V for 22 min. Transfer of proteins was verified by staining the membrane with Ponceau S. The membrane was blocked with a PBS (pH 7.4)-0.5% Triton X-100 solution containing 5% nonfat milk. Incubations with primary (1:30,000) and secondary (1:30,000) antibodies were carried out for 1 h at room temperature. The blot was developed with enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech). The OmpA antibodies used in the present study have been purified, and their specificity was as previously reported (25).
Protein sequencing.
The proteins of interest were excised from Coomassie blue-stained SDS-polyacrylamide gels and subjected to in-gel proteolysis with trypsin. The fragments were separated by high-pressure liquid chromatography and subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass analysis at the Protein and Nucleic Acid Facility at Stanford University. A mass spectroscopy fingerprinting profile from the mass fragments generated in the MALDI-TOF analysis was obtained by using the ProFound website (http://prowl.rockefeller.edu/cgi-bin/ProFound).
Construction of the ompA-lacZ promoter fusion.
An operon fusion with lacZ was constructed by amplifying the regulatory region of the ompA operon in strain 86-24 by PCR with Pfu polymerase (Gibco-BRL) by using the primer pair 5RIPOMPA (5′-CAGAATTCGCGCTGGCAACTCTGG-3′) and 3BIPOMPA (5′-TGGGATCCCGATAGCTGTCTTTTTCATTTTT-3′). The PCR product was digested and cloned into the EcoRI and BamHI sites of plasmid pRS551, which contains a promoterless lac operon (31), to create plasmid pPOMPA.
β-Galactosidase assay.
The E. coli strains containing the ompA promoter-lacZ fusion were grown with shaking at 37°C in LB broth, diluted 1:100 in fresh DMEM or LB medium, and further grown at 37°C to mid-exponential phase (i.e., an optical density at 600 nm of ca. 0.5 to 0.6). Cultures were diluted 1:10 in Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol) and were assayed for β-galactosidase activity by using ONPG (o-nitrophenyl-β-d-galactopyranoside) as the substrate, as previously described (15).
RESULTS
Isolation of E. coli O157:H7 mutant strains hyperadherent to HeLa cells.
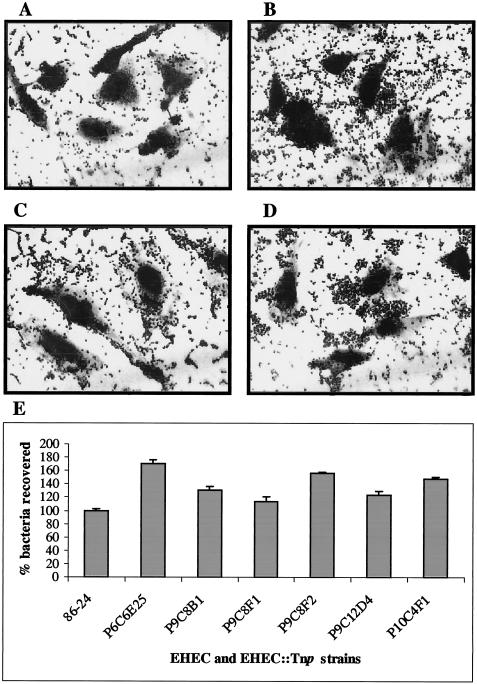
To identify bacterial factors involved in the control of adherence of EHEC O157:H7 to HeLa cells, we randomly mutagenized strain 86-24 with the EZ::TN <R6Kγori/KAN-2> Tnp transposome and collected a total of 5,000 independent insertion mutants. These Tnp mutants were used to infect HeLa cells for 3 h and, after the nonadherent bacteria were removed, the infected HeLa cells were further incubated for 3 h. The cells were either Giemsa stained for visual examination or the bacterial cells were recovered for quantification and for further enrichment as described in Materials and Methods. By this method, we were able to identify 20 Tnp mutants that showed increased adherence to HeLa cells but, after individual screening, only seven mutants reproducibly showed the hyperadherent phenotype and were selected for further characterization. When grown in DMEM these seven mutants—P6C6E25, P9C8B1, P9C8F1, P9C8F2, P9C12D4, P10C9E1, and P10C4F1—showed growth rates comparable to those of the wild-type strain (data not shown) and, in contact with HeLa cells, these Tnp mutant strains exhibited enhanced adherence ranging from 113 to 170% of the wild-type level (Fig. 1). Furthermore, adherence of the wild-type and the Tnp mutant strains to the plastic surface (24-well plates) was determined to rule out the possibility that hyperadherence is a nonspecific phenotype associated with the binding of these strains to epithelial cells. Our results indicate that the Tnp mutant strains adhere as poorly as the wild-type strain to the plastic support, and the bacteria recovered represented only 1 to 3% of the wild-type binding seen when HeLa cells are included (data not shown).
FIG. 1.
(A to D) Adhesion to HeLa cells of EHEC O157:H7 strain 86-24 (A) and its Tnp transposome mutants P10C4F1 (B), P6C6E25 (C), and P9C12D4 (D) after 6 h of incubation. (E) Percentage of EHEC O157:H7 strain 86-24 and its corresponding Tnp transposome mutants adhering to HeLa cells after 6 h of incubation. The error bars indicate the standard deviation.
Mapping the Tnp transposome insertions.
To determine the exact location of the Tnp transposome insertion, we performed genomic DNA extraction from each of the mutant strains, and the purified DNA was digested with EcoRV. Upon self-ligating and transforming the DNA fragments into an E. coli K-12 strain as described in Materials and Methods, the resulting Kmr clones were subjected to DNA sequence analysis. The specific site of the Tnp transposome insertion was determined based on basic local alignment search tool (BLAST) analysis of the flanking DNA and homologous sequences deposited in the NCBI database of EHEC strains EDL933 and O157Sakai. Based on this analysis, the mutants could be grouped into the following four functional groups: one mutant involved in LPS biosynthesis, two mutants implicated in amino acid metabolism, two mutants associated with fimbrial biogenesis, and two mutants disrupting a gene of unknown function (Table 2).
The Tnp transposome in P6C6E25 was mapped as being inserted in the waaI gene (Tnp inserted after nucleotide 281 of 1,014), a putative LPS biosynthesis enzyme, located in the chromosomal region designated O-island #145 by Perna et al. (21). Two previous studies have shown that mutants deficient in expression of the O157 polysaccharide exhibited increased adherence to HEp-2 or HeLa cells in vitro (1, 2), supporting our experimental approach to identify hyperadherent mutant strains. Due to the fact that similar mutants have been studied before, we did not further characterize mutant P6C6E25.
Two insertions were located in genes associated with amino acid biosynthesis. Mutant P9C12D4 has an insertion in cadA (Tnp inserted after nucleotide 1642 of 2,190), the structural gene encoding the lysine decarboxylase (LDC) I enzyme. Compared to the wild-type strain, P9C12D4 was unable to catabolize lysine due to a lack of LDC activity as determined by a commercial assay for LDC activity (data not shown). The second mutant of this group, P9C8F2, had an insertion in tdcA (Tnp inserted after nucleotide 466 of 938). This gene encodes the transcriptional activator of the tdc operon, which encodes a pathway for the transport and anaerobic degradation of l-threonine.
Two more Tnp transposome insertions were found within genes associated with fimbrial biogenesis. Mutant P9C8B1 was mapped to the csgD locus (Tnp inserted after nucleotide 87 of 651), which encodes a transcriptional regulator of the csg operon encoding curli. Curli fibers are thin aggregative fimbriae involved in bacterial adhesion. A property associated with the production of curli is binding to Congo red dye. When grown in Congo red indicator plates, P9C8B1 colonies remained white, indicating the inability of the strain to bind Congo red. In contrast, the wild-type strain produced red colonies (data not shown). Strain P9C8F1 had an insertion in a gene located in O-island #154. This O-island contains five ORFs putatively associated with the production of fimbriae with homology to the long polar fimbriae of EHEC O157:H7 and Salmonella enterica serovar Typhimurium (39). The Tnp transposome is located in the fourth ORF (Z5221) of this putative operon, encoding a fimbrial minor subunit with homology to the lpfD gene of EHEC O157 and serovar Typhimurium. Based on the homology, we renamed this gene lpfD2 (Tnp inserted after nucleotide 685 of 1,070).
The final insertion was mapped within the same gene in two independent Tnp transposome mutants. Both strains P10C9E1 and P10C4F1 contained insertions in the yidE gene (Tnp inserted after nucleotides 638 and 700 of 1,685) of unknown function. Homology searches indicated that this gene encodes a putative transport and membrane-associated protein, and this gene is found in other Enterobacteriaceae, such as Shigella flexneri, E. coli K-12, and S. enterica serovar Typhi.
Differential protein profiles of the different Tnp transposome mutants.
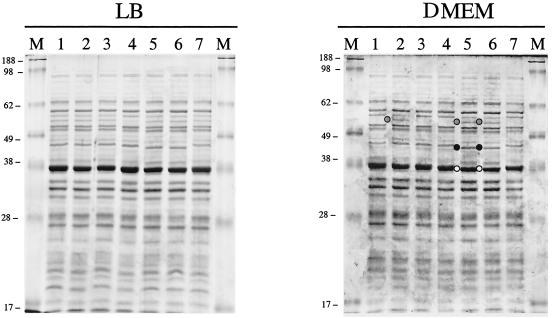
In order to start identifying those proteins that are differentially expressed in mutants associated with the hyperadherence phenotype, bacterial whole-cell lysates of the wild type and Tnp mutants were fractionated in SDS-PAGE and visualized by Coomassie brilliant blue staining. As shown in Fig. 2, whole-cell lysates obtained from bacterial strains grown in LB broth did not show any obvious difference in the protein profile when the Tnp mutants were compared to the wild-type strain. Nevertheless, when the lysates obtained from DMEM were fractionated, at least seven proteins in the Tnp mutants were differentially expressed or showed a different electrophoretic mobility compared to the wild-type strain (Fig. 2). These proteins were excised from the acrylamide gel, and their identities were revealed by MALDI-TOF MS fingerprint analysis. The results indicated that the formate acetyltransferase I enzyme was differentially expressed in the P9C12D4, P9C8F2, and P6C6E25 (cadA, tdcA, and waaI, respectively) Tnp mutants (Fig. 2; protein is identified in lanes 2, 5, and 6 by a gray circle). In addition, the tdcA and waaI Tnp mutants showed two proteins that migrated faster in the acrylamide gel compared to the wild-type strain. MALDI-TOF mass analysis revealed that the proteins were GroEL and the glutamate decarboxylase enzyme (Fig. 2; proteins are identified by black and white circles, respectively). Besides these differences, we were unable to detect other proteins whose expression was affected in DMEM in the different Tnp mutants. These data indicate that the expression of some proteins is altered upon insertion of the Tnp transposome, but it is not simple to correlate the differential protein expression in whole-cell lysates in the mutants and the hyperadherence phenotype. Therefore, we decided to further fractionate the bacterial lysates and concentrate our analysis on disrupted genes with a confirmed role as regulatory proteins.
FIG. 2.
Fractionation of whole-cell membrane lysates of EHEC O157:H7 strain 86-24 and its Tnp transposome mutants by using SDS-10% acrylamide gels and staining with Coomassie brilliant blue. Lanes: 1, EHEC 86-24; 2, P9C12D4; 3, P9C8B1; 4, P9C8F1; 5, P9C8F2; 6, P6C6E25; 7, P10C9E1; M, molecular mass markers. The proteins selected for MALDI-TOF mass spectroscopy fingerprint analyses are indicated by gray, black, and white circles. The sizes of the molecular mass markers are indicated on the left of each panel.
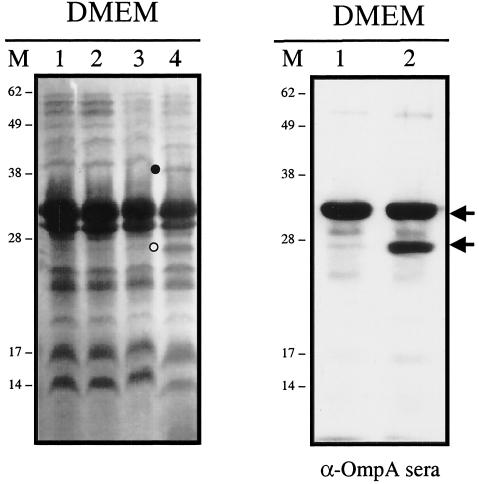
Mutant P9C8F2 differentially expresses an outer membrane protein.
Bacterial extracts of mutants P9C8F2, P9C12D4, and P9C8B, with Tnp insertions in the tdcA, cadA, and csgD genes, respectively, were further fractionated, and their proteins were separated by SDS-PAGE. The protein profiles of the inner membrane fractions of the three Tnp mutants did not show any difference compared to the wild-type strain (data not shown). In comparison, the outer membrane protein profiles of these three Tnp mutants revealed at least two proteins differentially expressed in strain P9C8F2 (Fig. 3). The first protein (Fig. 3, black circle) showed a modified electrophoretic mobility, and MALDI-TOF analysis indicated that this outer membrane protein is associated with the transport of long fatty acids. The expression of the second protein was enhanced in mutant P9C8F2 compared to the wild-type outer membrane preparation (Fig. 3, white circle). Mass spectroscopy fingerprinting analysis indicated that the protein corresponded to the outer membrane porin OmpA. In order to confirm that this protein was OmpA, the outer membrane proteins of the wild type and P9C8F2 mutant were transferred to a membrane, and a Western blot was performed with anti-OmpA antisera (kindly provided by N.V. Prasadarao). As shown in Fig. 3, the antisera specifically recognized the OmpA, thus confirming that the protein differentially expressed in P9C8F2 corresponded to OmpA. The OmpA protein appears as a doublet in the P9C8F2 mutant strain, but further analysis and construction of isogenic ompA mutants confirmed that the proteins corresponded to only one OmpA protein (see below). Furthermore, antisera raised against the OmpU protein of Vibrio cholerae and previously shown to cross-react in Western blot experiments with the OmpA protein of E. coli (32) also cross-reacted with OmpA of strains 86-24 and P9C8F2 (data not shown). These data indicated that the Tnp mutant with a disruption in the tdcA gene differentially expressed at least two proteins in its outer membrane, one of them being OmpA. Due to previous data indicating that OmpA participates in the E. coli K-1 invasive capability of brain microvascular endothelial cells (25) and that OmpU, an OmpA homologue in V. cholerae, has adhesive properties (32), we decided to further investigate whether OmpA was participating in the adherence of EHEC strain 86-24 to HeLa cells.
FIG. 3.
Outer membrane protein profiles and Western blotting with anti-OmpA serum of samples grown in DMEM and fractionated by SDS-12% PAGE. (Left) Outer membrane protein profiles. Lanes: 1, EHEC O157:H7 strain 86-24; 2, P9C12D4; 3, P9C8B1; 4, P9C8F2. (Right) Western blot analysis. Lanes: 1, EHEC 86-24; 2, P9C8F2. Lanes M, molecular mass markers in kilodaltons. The proteins selected for MALDI-TOF mass spectroscopy fingerprint analyses are indicated by black and white circles, and the OmpA protein is identified with black arrows.
Transcription of ompA is slightly stimulated in the P9C8F2 mutant strain.
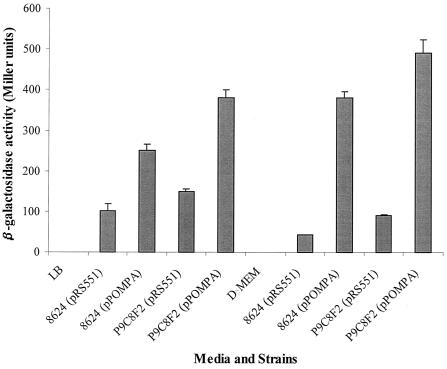
In order to determine whether the mutation of the tdcA gene in strain P9C8F2 is responsible for an increase in the expression of the OmpA protein, we generated a fusion of the ompA promoter with a reporter lacZ gene (plasmid pPOMPA) and examined the expression of this fusion during exponential growth of strains 86-24 and P9C8F2 under different medium conditions. As a control in our experiments, we used pRS551, the parent plasmid containing the promoterless lacZ gene. As shown in Fig. 4, the expression of β-galactosidase in LB broth was increased 1.5-fold in strain P9C8F2(pPOMPA) compared to 86-24(pPOMPA). To determine the effect of the medium on ompAp::lacZ expression, cultures of 86-24 and P9C8F2 containing pRS551 and pPOMPA were grown in DMEM. During exponential growth phase, β-galactosidase expression was induced only 1.3-fold in strain P9C8F2(pPOMPA) compared to 86-24(pPOMPA). These data indicate that transcription of the ompA gene was induced during these in vitro conditions and is slightly increased in the Tnp mutant.
FIG. 4.
β-Galactosidase assays of EHEC strains 86-24 and P9C8F2 carrying plasmids pRS551 (vector control) and pPOMPA (ompAp::lacZ) in LB medium and DMEM during exponential growth phase. The error bars indicate the standard deviations.
OmpA protein participates in the hyperadherence phenotype.
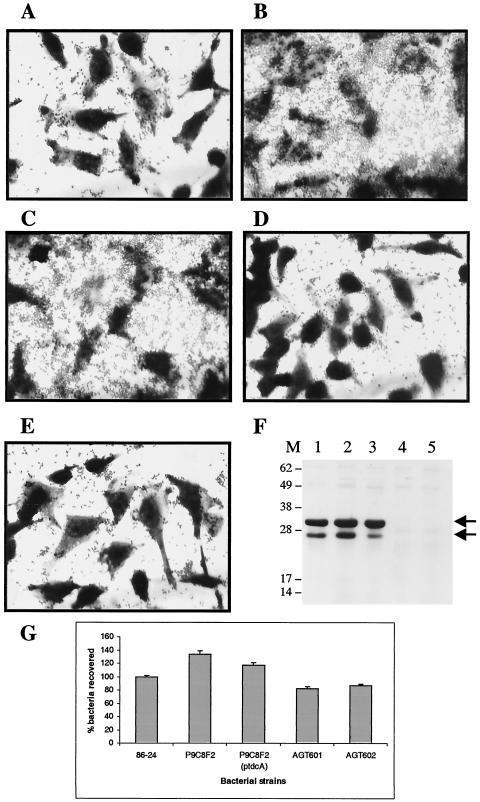
We then sought to investigate whether the hyperadherence phenotype observed in strain P9C8F2 was associated with the expression of the OmpA protein. To answer this question, we created a series of isogenic ompA mutants of strains 86-24 and P9C8F2, performed quantitative and qualitative adhesion assays, and monitored expression of the OmpA protein by Western blots with anti-OmpA antisera. We first confirmed that the mutant strain was still hyperadherent to HeLa cells and attempted to complement the mutation with the wild-type tdcA gene on a plasmid. As shown in Fig. 5, strain P9C8F2 showed the hyperadherent phenotype compared to the wild-type strain. However, the phenotype did not revert to wild-type levels upon introduction of tdcA gene on a plasmid, suggesting that the Tnp transposome is having a polar effect on genes found downstream of tdcA. We then tested whether the mutation in the ompA gene abolishes the hyperadherent phenotype. When visualized under the microscope, strain AGT602 (ompA::cat tdcA::Tnp) adhered as well as the wild-type 86-24 and did not display hyperadherence (Fig. 5). Quantification of the adherent bacteria indicated that AGT602 adhered almost as well as strain 86-24 (86.5% bacteria recovered, a 13.5% decrease compared to the wild-type strain). Furthermore, whole-cell extracts of the mutant strain did not react with the anti-OmpA sera in Western blot assays, confirming that strain AGT602 did not express the OmpA protein, a finding consistent with expression of this protein being necessary for hyper-adherence. To further investigate the role of OmpA in EHEC adherence to HeLa cells, strain AGT601 (ompA::cat) was generated and tested in a similar tissue culture cell adherence assay. As shown in Fig. 5, strain AGT601 was able to adhere to HeLa cells but to a lower extent than wild-type strain (81.9% compared to 100% bacteria recovered in strain 86-24, an 18.1% reduction). Similar to AGT602, strain AGT601 did not express the OmpA protein.
FIG. 5.
(A to E) Adhesion to HeLa cells after 6 h of incubation of EHEC strains 86-24 (A), P9C8F2 (B), P9C8F2(ptdcA) (C), AGT601 (D), and AGT602 (E). (F) Western blot with anti-OmpA serum of samples grown in DMEM and fractionated by SDS-12% PAGE. Lanes: 1, EHEC O157:H7 strain 86-24; 2, P9C8F2; 3, P9C8F2(ptdcA); 4, AGT601; 5, AGT602; M, molecular mass markers. The OmpA protein is identified with black arrows on the right. (G) Percentage of EHEC O157:H7 strain 86-24 and its corresponding Tnp transposome or isogenic mutants adherent to HeLa cells after 6 h of incubation.
We then investigated whether the antiserum raised against the OmpA protein was able to block adherence of strains 86-24 and P9C8F2 to HeLa cells. For both strains tested, adherence to HeLa cells was greatly reduced at higher concentrations of anti-OmpA serum (1:10 dilution) (P < 0.05) and was almost restored to normal levels as the serum was diluted 10-fold (1:100 dilution) (Table 3, experiment 1). It is important that the antiserum effect was more pronounced in strain P9C8F2 than in strain 86-24. Even at a 1:100 dilution of the antiserum, the mutant strain was unable to display the hyperadherence phenotype (P < 0.05). To confirm that the blocking of adherence was a specific effect due to the anti-OmpA antibodies and not to the rabbit serum, an additional experiment was performed to determine whether the sera alone were able to inhibit adherence of the two strains. As indicated in Table 3, experiment 2, rabbit serum had a slight effect in blocking the adherence of both strains at the higher concentration (1:10) that was not statistically significant (P > 0.05). At a 1:100 dilution, strains 86-24 and P9C8F2 adhere to HeLa cells as well as do strains incubated in the absence of any sera. Together, these data suggest that OmpA protein is playing a role in the adhesive properties of EHEC strain 86-24 and in P9C8F2 is responsible for hyperadherence to HeLa cells.
TABLE 3.
Adherence of EHEC strains 86-24 and P9C8F2 to HeLa cells in the presence of anti-OmpA sera
| Expt and antiserum (dilution) | % Bacteria ± SD (P)a recovered after HeLa infection with strain:
|
|
|---|---|---|
| 86-24 | P9C8F2 | |
| Expt 1 | ||
| No antiserum | 100 ± 5.6 | 131.7 ± 1.7 |
| Anti-OmpA (1:10) | 73.1 ± 16.1 (0.05) | 34.0 ± 14.3 (<0.001) |
| Anti-OmpA (1:100) | 98.6 ± 6.85 (>0.05) | 92.6 ± 8.43 (<0.05) |
| Expt 2b | ||
| No antiserum | 100 ± 7.03 | 127.2 ± 10.02 |
| Rabbit serum (1:10) | 88.8 ± 7.36 (>0.05) | 113.8 ± 6.87 (>0.05) |
| Rabbit serum (1:100) | 95.8 ± 7.66 (>0.05) | 128.4 ± 10.42 (>0.05) |
The P value was obtained by comparing the infection with no antiserum to those containing different dilutions of the antisera.
Rabbit sera were used as the control sera.
OmpA displays adhesive properties in other EHEC O157:H7 strains and participates in the binding to intestinal epithelial cells.
To determine whether the association of the OmpA protein with adherence was not just a phenotype associated with EHEC strain 86-24 used in the present study, we repeated the adherence experiments in the presence or absence of the anti-OmpA antiserum with different EHEC and EPEC strains (Table 4). The strains tested were different clinical isolates of EHEC O157:H7 strains EDL933 and 85-170, and the prototype EPEC strain E2348/69. Our results indicate that in both EHEC strains, adherence to HeLa cells was reduced when the anti-OmpA serum was present (Table 4, an approximately 25% reduction when the antiserum is present), where adhesion of the EPEC strain was not affected. The cumulative data strongly suggest that OmpA is a potential EHEC adherence factor, and the phenotype upon expression of this protein could be associated with this particular class of pathogenic E. coli strains.
TABLE 4.
Adherence of EHEC and EPEC strains to HeLa cells in the presence of anti-OmpA sera
| Antiserum (dilution) | % Bacteria ± SDa recovered after HeLa infection with strain:
|
||
|---|---|---|---|
| EHEC EDL933 (P = 0.05) | EHEC 85-170 (P = 0.05) | EPEC E2348/69 (P > 0.05) | |
| None | 100 ± 11.47 | 100 ± 14.58 | 100 ± 25.12 |
| Anti-OmpA (1:10) | 76.8 ± 16.91 | 75.6 ± 22.62 | 92.7 ± 29.28 |
The P values were obtained by comparing the infections with or without antiserum for each strain.
The adherence studies described above were performed on HeLa cells, an epithelial cell line obtained from cervix adenocarcinoma that has been used extensively in the study of adherence patterns of E. coli strains, but since EHEC O157:H7 is an intestinal pathogen, our interest was to determine whether this factor is also associated with adherence to intestinal epithelial cells. Therefore, we repeated the adherence experiments with Caco-2 cells, a colorectal adenocarcinomic cell line. We found that the percentages of bacteria recovered ± the standard deviations were as follows: 86-24, 100% ± 9.72%; P9C8F2, 121.3% ± 9.2%; AGT601, 86.5% ± 15.9%; and AGT602, 82.7% ± 16.2%. An 21.3% increase in adherence was observed with the EHEC tdcA::Tnp mutant strain compared to the wild-type strain, and a reduction in adherence was observed with the ompA mutants (the adherence of AGT601 and AGT602 was reduced 13 to 17% compared to the wild-type strain, but the adherence of AGT602 was reduced 38.6% compared to the tdcA::Tnp mutant strain). These results were comparable to the data obtained with HeLa cells, even though the differences in adhesion were not as significant as previously observed. Overall, our data indicate that OmpA is acting as an EHEC adherence factor mediating binding to different tissue culture cell lines.
DISCUSSION
Adhesion of pathogenic organisms, such as E. coli O157:H7, to host tissues is the first stage in the infectious disease process. To understand how this organism interacts with host cells may help to identify new targets for therapy or to establish the effects that adhesion has on the cell and/or tissue to which it adheres. Although a substantial amount of data has been generated in recent years regarding the interaction of E. coli O157:H7 with host cells and the role of intimin has been clearly associated with adherence in later stages during infection (4, 14, 42), it is unclear which factor(s) is involved in the initial stage of adhesion. Based on this premise, we began an investigation of additional factors potentially involved in the adhesion of E. coli O157:H7 to cultured epithelial cells. Our approach involved the isolation of hyperadherent mutants which represent relief from putative repression of adhesion factors in vitro. This “gain-of-function” phenotype is beneficial because it could lead to the identification of putative regulators and the conditions that induce their expression. Similar approaches have been used for successful identification of ClpP, a protease subunit that modulates gene expression of ail, a gene encoding the Ail adhesin of Yersinia enterocolitica (20, 22), and of CsrA, a regulator that affects glycogen biosynthesis (28). Using a similar rationale, we have found that adherence of E. coli O157:H7 to HeLa cells is affected directly or indirectly by multiple factors, such as those involved in LPS biosynthesis, amino acid metabolism, or fimbrial biogenesis. We also present evidence that OmpA has adhesive properties and is required for E. coli O157:H7 to display the hyperadherent phenotype upon contact with HeLa cells.
Previous work by Bilge et al. (1) and Cockerill et al. (2) demonstrated that E. coli O157:H7 deficient in the production of O157 LPS side chains displayed an increased binding to tissue culture cells similar to the results that we obtained in our study. Those authors concluded that the presence of the O157 polysaccharide interferes with the adherence of E. coli O157:H7 and is not required to produce the attaching-and-effacing lesion. Our Tnp transposome mutagenesis analysis confirms these studies and, in addition, we identified other mutated genes that produce a hyperadherent phenotype, which suggests a direct or indirect role of these genes in adherence. Three of these mutations resided within genes encoding transcriptional regulators or otherwise associated with the control of virulence. The mutation in the cadA gene is of particular interest because, in addition to encoding the LDC enzyme responsible for metabolizing lysine, cadA has been proposed as an antivirulence gene in enteroinvasive E. coli and Shigella spp. (12). The authors of that earlier study found that introduction of cadA in S. flexneri affects the activity of two enterotoxins produced by this organism and that the presence of the by-product cadaverine, generated from the decarboxylation of lysine, caused attenuation in S. flexneri virulence (13). It is interesting to speculate that the cadA gene in E. coli O157:H7 serves to fine-tune the virulence of this organism by controlling the expression of putative adhesion molecules only under specific environmental circumstances.
Another gene disrupted in our analysis was csgD, whose product regulates the expression of curli. The curli are highly aggregated, extracellular fibers expressed by E. coli and Salmonella spp. that are involved in bacterial adhesion to host proteins and in biofilm formation (34, 46). In strains of Shigella and enteroinvasive E. coli, the csg locus has been shown to carry insertions, deletions, or mutations, suggesting that interruption of this gene is necessary for virulence (29). In the case of E. coli O157:H7, the csgD gene product may be controlling the expression of an adhesive factor required for binding to HeLa cells but not necessary for biofilm formation, since the mutant strain containing the transposon within the csgD gene was unable to form biofilms on polyvinyl chloride plastic plates (data not shown). Moreover, it has been reported that curli fibers are infrequently expressed during in vitro growth of E. coli O157:H7 (44) and that strains containing variations at the csgD promoter region, which induced expression of curli, are associated with increased virulence in mice and increased invasion of HEp-2 cells (45). Even though EHEC is not an invasive organism, we tested EHEC strain 86-24 and Tnp transposome mutants for their ability to invade HeLa cells and did not observe any increase in the invasion ability of the mutants compared to the wild type (data not shown).
A second gene encoding a regulator identified in our study is tdcA. This activator of the tdc operon has only been implicated in the control of genes responsible for transport and anaerobic degradation of l-threonine and has not heretofore been associated with any aspect of adherence. Using this Tnp transposome mutant, we were able to identify the differentially expressed outer membrane protein A. OmpA is a heat-modifiable class of protein highly conserved among the Enterobacteriaceae, and multiple functions have been attributed to it, including serving as a mediator in F-factor-dependent conjugation, acting as a phage and colicin receptor, acting as a porin and, in combination with lipoprotein, being required for the structural integrity of the membrane and the generation of a normal cell shape (7).
The role of OmpA in bacterial virulence has been extensively studied in E. coli K-1. OmpA was originally reported as a contributor to pathogenicity and serum resistance of E. coli K-1 both in vivo and in vitro (47) and was later demonstrated to mediate E. coli invasion of brain microvascular endothelial cells (25). OmpA was first identified as a contributor to transverse the brain-blood barrier on the basis of its structural homology with Neisseria Opa proteins (25), which are involved in invasion of eukaryotic cells. Subsequent work identified a human brain microvascular endothelial cell glycoprotein (gp96) as the specific OmpA receptor and confirmed the key role of E. coli OmpA in the initial steps of the invasion process (23, 24). Although invasion is not a property of EHEC strains, it will be interesting to investigate whether the contribution to adherence of E. coli O157:H7 OmpA to the human intestinal epithelia requires a specific cell receptor and whether this ligand-receptor interaction contributes to cell signaling associated with cytoskeletal rearrangement.
Even more intriguing is the recent finding that antigen-presenting cells recognize and are activated by Klebsiella pneumoniae OmpA, triggering cytokine production by macrophages and dendritic cells and the subsequent maturation of dendritic cells and signals via Toll-like receptor 2 (6). The role of E. coli O157:H7 OmpA or other E. coli OmpA proteins in the activation of antigen-presenting cells is unknown at present, but the data obtained with K. pneumoniae OmpA suggest that the immune system has acquired the ability to recognize this new type of pathogen-associated molecular pattern (highly conserved structures on microorganisms recognized by innate immune cells).
The role of an OmpA homologue in adhesion has been suggested for V. cholerae, where OmpU has adhesive properties in vitro (32), and this is regulated by ToxR at the transcriptional level. This global regulator also modulates the expression of another outer membrane porin by inducing OmpU and repressing OmpT (10). ToxR-dependent modulation of OmpU and OmpT seems to be critical for V. cholerae bile resistance, virulence factor expression, and intestinal colonization, suggesting a relationship between porin and pathogenesis (26). Our in vitro data support the role of the E. coli O157:H7 OmpA protein as a factor that seems to be important for adherence of different EHEC O157:H7 strains to HeLa cells and which also mediates binding to Caco-2 cells. We are intrigued by these findings and are now investigating the role of this protein in intestinal colonization, as well as attempting to identify the host cell receptor interacting with this porin.
In summary, we have identified several Tnp transposome mutants within genes associated with hyperadherence to HeLa and Caco-2 cells and determined that OmpA, an outer membrane porin, has adhesive properties and is associated with the hyperadherent phenotype in one of the Tnp mutants and with the binding of EHEC O157:H7 strains to HeLa cells.
Acknowledgments
We thank Laura Quinn and Kristen Kanack for critical reading of the manuscript; William Nguyen, Jennifer Yarnall, and Susana Oaxaca-Torres for technical assistance; and Diana Gomez for maintenance of tissue culture cell lines. We also thank N. V. Prasadarao (Childrens Hospital Los Angeles) for the anti-OmpA sera.
This work was supported by grants AI41325 and DK58957 to J.B.K. and an RSUM grant from NIDDK and startup funds from the University of Texas Medical Branch to A.G.T.
Editor: A. D. O'Brien
REFERENCES
- 1.Bilge, S. S., J. C. Vary, Jr., S. F. Dowell, and P. I. Tarr. 1996. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect. Immun. 64:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cockerill, F., III, G. Beebakhee, R. Soni, and P. Sherman. 1996. Polysaccharide side chains are not required for attaching and effacing adhesion of Escherichia coli O157:H7. Infect. Immun. 64:3196-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., S. Tzipori, M. L. McKee, A. D. O'Brien, J. Alroy, and J. B. Kaper. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 92:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dytoc, M. T., A. Ismaili, D. J. Philpott, R. Soni, J. L. Brunton, and P. M. Sherman. 1994. Distinct binding properties of eae-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect. Immun. 62:3494-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeannin, P., G. Magistrelli, L. Goetsch, J. F. Haeuw, N. Thieblemont, J. Y. Bonnefoy, and Y. Delneste. 2002. Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen-presenting cells—impact on vaccine strategies. Vaccine 20:A23-A27. [DOI] [PubMed] [Google Scholar]
- 7.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Levine, M. M., E. J. Berquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Stoman, and B. Rowe. 1978. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed] [Google Scholar]
- 10.Li, C. C., J. A. Crawford, V. J. Di Rita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick, B. A., M. I. Fernandez, A. M. Siber, and A. T. Maurelli. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell Microbiol. 1:143-155. [DOI] [PubMed] [Google Scholar]
- 14.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to Hep-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 18.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69:6999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 21.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-532. [DOI] [PubMed] [Google Scholar]
- 22.Pierson, D. E. 1994. Mutations affecting lipopolysaccharide enhance ail-mediated entry of Yersinia enterocolitica into mammalian cells. J. Bacteriol. 176:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 28.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175:4744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakellaris, H., N. K. Hannink, K. Rajakumar, D. Bulach, M. Hunt, C. Sasakawa, and B. Adler. 2000. Curli loci of Shigella spp. Infect. Immun. 68:3780-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 32.Sperandio, V., J. A. Giron, W. D. Silveira, and J. B. Kaper. 1995. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 63:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens, M. P., P. M. vanDiemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukupolvi, S., R. G. Lorenz, J. I. Gordon, Z. Bian, J. D. Pfeifer, S. J. Normark, and M. Rhen. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159:344-347. [DOI] [PubMed] [Google Scholar]
- 37.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of Mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:5416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres, A. G., and J. B. Kaper. 2001. PAIs of intestinal Escherichia coli, p. 31-48. In J. Hacker and J. B. Kaper (ed.), Pathogenicity islands (PAIs) and the evolution of pathogenic microbes, vol. 1. Springer-Verlag, Berlin, Germany.
- 41.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 42.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzipori, S., H. Karch, K. I. Wachsmuth, R. M. Robins-Browne, A. D. O'Brien, H. Lior, M. L. Cohen, J. Smithers, and M. M. Levine. 1987. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect. Immun. 55:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlich, G. A., J. E. Keen, and R. O. Elder. 2002. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increase curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiser, J. N., and E. C. Gotschlich. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]