Abstract
Helicobacter pylori encodes three histidine kinases and five response regulators belonging to the family of two-component regulatory systems which are involved in transcriptional control. Here we demonstrate that isogenic mutants of H. pylori P76 with deletions of the response regulator open reading frame (ORF) HP1365 and ORFs HP244, HP165, and HP1364 encoding histidine kinases are unable to colonize the stomachs of BALB/c mice, suggesting an essential role of these systems in the regulation of important virulence properties of H. pylori. Furthermore, we demonstrate that the genes under the control of the PHP1408 and PHP119 promoters which are regulated by the two-component system HP166-HP165 are not essential for single mutant colonization of mice but are required under competitive colonization conditions.
Helicobacter pylori is the causative agent of chronic type B gastritis and peptic ulcer disease in humans and is also considered a risk factor for the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (14, 21). A number of bacterial factors contributing to the pathogenicity of H. pylori have already been identified (2, 5, 15, 22, 27). By using different animal models of infection it has been demonstrated that some of these factors are essential for the successful colonization of the host organism, such as urease (9, 30), the urea channel protein UreI (25), flagella and chemotaxis proteins (10, 12), superoxide dismutase (23), and the iron storage protein Pfr (31). Furthermore, a comprehensive systematic screen using signature tag mutagenesis recently identified a set of 47 H. pylori genes which proved to be essential for colonization in the H. pylori gerbil model (16). The expression of virulence traits that enable a pathogen to colonize the host and to cope with the hostile environment within the host has to be tightly regulated to ensure the production of the required virulence factor at the right time point of infection. Two-component systems are signal transduction systems which are able to perceive environmental stimuli and to trigger an adequate cellular response. Consequently, they are frequently involved in the coordinate regulation of virulence-associated genes. Usually they are composed of a histidine kinase which monitors environmental signals and a cognate response regulator whose function is modulated by a phosphotransfer reaction from its cognate histidine kinase. Analysis of the genome sequence revealed that H. pylori contains only three histidine kinases and five response regulators involved in transcriptional regulation (1, 29). Interestingly, it turned out that two response regulator genes (open reading frames [ORFs] HP166 and HP1043) are essential for cell viability, while deletion of the response regulator ORF HP1021 resulted in a pronounced growth defect of the bacteria grown under in vitro culture conditions (4, 18). The aim of the present study was to analyze whether the H. pylori two-component genes HP244, HP165, and HP1364, encoding histidine kinases, and the response regulator ORF HP1365 are required for colonization in a mouse model of infection.
Construction and characterization of isogenic mutants of H. pylori P76 with deletions in the two-component ORFs.
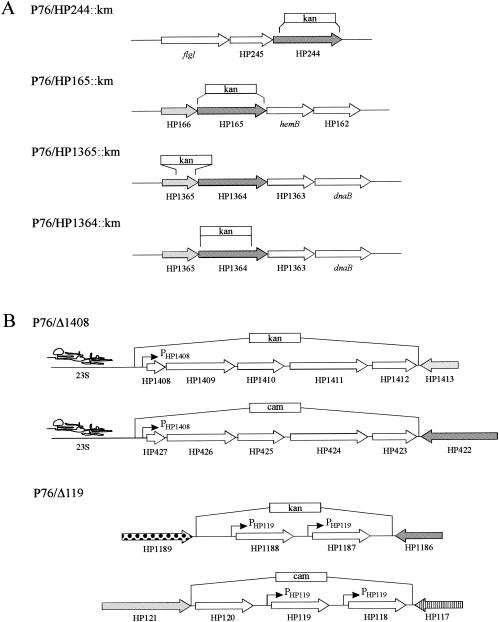
Derivatives of the mouse-adapted H. pylori strain P76 (13), carrying deletions in the two-component ORFs HP244, HP165, HP1364, and HP1365, were constructed by allelic exchange mutagenesis (3) (Fig. 1). Therefore, the suicide plasmids pSL-244::km, pSL-165::km, pSL-1364::km, and pSL-1365::km, which have been described previously (4), were used for transformation of H. pylori strain P76 (20), carrying streptomycin resistance for optimal quantitative reisolation from the infected mouse stomach by streptomycin selection. The correct replacement of the two-component ORFs by the kanamycin resistance cassette was confirmed by PCR experiments performed on chromosomal DNA of the mutants with oligonucleotide primer pairs flanking the insertion site (data not shown). As the H. pylori strain J99 contains an additional copy of the response regulator ORF HP1365 (JHP1283, which is located adjacent to the histidine kinase ORF JHP1282, and JHP1443) (1), the mutant P76/HP1365::km was checked carefully for the presence of a second HP1365 response regulator gene by PCR analysis of its chromosomal DNA with primer pairs derived from this ORF. No PCR fragment of the length to be expected if the fragment was derived from a wild-type copy of ORF HP1365 was obtained, demonstrating that H. pylori P76, similar to strain 26695 (29), harbors a single response regulator, ORF HP1365. A deletion mutant of ORF HP1021 could be obtained by transformation of strain P76 with plasmid pSL-1021::km (4). However, similar to the mutant of strain G27 this strain was severely hampered in its growth on blood agar plates and was therefore not considered for the infection of mice.
FIG. 1.
Construction of H. pylori P76 mutants used in this study. (A) For the construction of the mutants P76/HP244::km, P76/HP165::km, P76/HP1365::km, and P76/HP1364::km the parts of the two-component ORFs indicated by brackets were replaced with a kanamycin resistance cassette via allelic exchange mutagenesis. Histidine kinase and response regulator ORFs are indicated by shading in dark and light gray, respectively. (B) The target genes of the HP166-HP165 two-component system, which are under the control of the PHP1408 and PHP119 promoters, respectively, are located at two different loci in the genome of H. pylori 26695. These loci show a high degree of variability between the two sequenced H. pylori strains 26695 and J99 (1, 29), and their precise structure in H. pylori P76 is not known. In P76/Δ1408 the regions between the 23S ribosomal DNA and ORFs HP1413 and HP422, encoding target genes under the control of the PHP1408 promoter, were replaced by kanamycin and chloramphenicol resistance cassettes. Similarly, in P76/Δ119 the regions between ORFs HP1189 and HP1186 and ORFs HP121 and HP117, encoding target genes under the control of the PHP119 promoter, were replaced by kanamycin and chloramphenicol cassettes. See reference 8 for details.
Both urease activity and motility are prerequisites for colonization. Therefore, mutants P76/HP244::km, P76/HP165::km, P76/HP1364::km, and P76/HP1365::km were analyzed for these phenotypes by measuring ammonia production of bacterial lysates from urea hydrolysis with the Berthelot reaction (6) and by stabbing bacteria into semisolid agar and scoring for the presence of motility rings (17). All mutant strains showed urease activities similar to that of the wild-type P76 (Table 1), indicating that the two-component systems are not involved in the basal expression of urease under in vitro culture conditions. This is in agreement with the results of a recent analysis of the PureA promoter, demonstrating that deletion of sequences located upstream of the −35 region does not affect transcription from the PureA promoter under these conditions (7). While strains P76/HP165::km, P76/HP1364::km, and P76/HP1365::km were fully motile, mutant P76/HP244::km showed no swarming on semisolid agar (data not shown). This observation is due to a nonflagellated phenotype of this deletion mutant, because HP244 is the cognate histidine kinase phosphorylating the NtrC-like response regulator HP703, which activates the transcription of several σ54-dependent flagellar basal body and hook genes (4, 26).
TABLE 1.
Urease activities of the P76 wild type and isogenic two-component mutants grown on blood agar platesa
| Strain | Urease activity (μmol/mg/min) |
|---|---|
| P76 | 12.0 (3.1) |
| P76/HP1365::km | 10.9 (3.2) |
| P76/HP1364::km | 11.9 (3.6) |
| P76/HP165::km | 18.8 (9.8) |
| P76/HP244::km | 14.8 (5.2) |
| P76/Δ1408 | 13.0 (2.0) |
| P76/Δ119 | 13.4 (2.9) |
The results shown are the averages of four independent experiments. Standard deviations are given in brackets. As determined by Student's t test differences in urease activity between H. pylori P76 and the mutants are not statistically significant.
Isogenic two-component mutants of H. pylori P76 are unable to infect mice.
To investigate whether deletion of the two-component genes affects the ability of H. pylori P76 to colonize the mouse stomach, mice were infected with the P76 wild-type strain and its mutant derivatives P76/HP244::km, P76/HP165::km, P76/HP1364::km, and P76/HP1365::km. Female 6- to 8-week-old BALB/c mice (RCC, Itingen, Switzerland) were inoculated three times intragastrically with doses of 1.0 × 109 CFU of H. pylori P76 or its mutant derivatives at 2-day intervals. This experiment was performed two times independently with five mice per group. For the infection strain P76 and the mutants were grown on GC agar plates (Difco) supplemented with horse serum (8%), vancomycin (10 mg/liter), trimethoprim (5 mg/liter), nystatin (1 mg/liter), and 250 mg of streptomycin/liter (serum plates with streptomycin) for 2 days. The bacteria were harvested and suspended in brucella broth, and the final concentration was adjusted to 5 × 109 cells/ml. Aliquots of 200 μl (109 bacteria) were given orogastrically to mice. Four weeks after the third challenge with H. pylori mice were sacrificed by CO2 asphyxiation, and the stomachs were excised, weighed, and opened along the great curvature. For assessment of H. pylori colonization by culture stomachs were cut and homogenized in 2 ml of brucella broth by a hand homogenizer (Fisher Scientific, Schwerte, Germany) and 1/200 serial dilutions were spread over the surface of serum plates with streptomycin. Plating of bacteria was performed in duplicate. The plates were incubated for 5 to 7 days under standard conditions, and colonies were counted.
The wild-type P76 strain was recovered from all animals, and in two independent experiments the medians for the bacterial load were 1.48 × 107 and 1.58 × 107 CFU per g of stomach tissue. None of the P76 mutants with deletions of the two-component genes could be cultured from the stomachs of mice, suggesting that these regulatory genes are necessary for the successful colonization of the host by controlling the expression of bacterial factors required at some stage of the infection process. We cannot completely rule out polar effects of the insertion of the kanamycin cassette on the genes located downstream of ORFs HP1365-HP1364 and HP166-HP165 (Fig. 1). However, since these downstream genes include orthologs of dnaB (HP1362) and hemB (HP163), whose gene products are involved in DNA replication and heme biosynthesis, respectively, we would have expected an effect of a strongly polar mutation on the in vitro growth of the mutants, which is not observed.
It has been demonstrated that the two-component systems are not involved in the expression of some of the well-known factors which have proved to be essential for host colonization, i.e., urease, the iron storage protein Pfr (S. Bereswill and D. Beier, unpublished observation), and—with the exception of the HP244-HP703 system—motility and chemotaxis. In the case of the mutant P76/HP244::km the colonization defect was expected and, as mentioned above, is probably due to the aflagellated phenotype of this strain. From the observation that histidine kinase HP1364 and response regulator HP1365 are encoded by adjacent genes and that these genes are cotranscribed from a promoter located upstream of ORF HP1365 (P. Dietz and D. Beier, unpublished observations), it seems likely that these proteins are cognate phosphorylation partners; however, this has not been proven experimentally. So far, the target genes of the HP1365-HP1364 two-component system are completely unknown. As both the sensor protein HP1364 and the cognate response regulator HP1365 are necessary for the infection of mice, it can be concluded that transcription of target genes involved in the colonization process requires phosphorylation of the response regulator HP1365. Response regulator HP166 is essential for cell viability, while the cognate histidine kinase HP165 is easily dispensable for the growth of H. pylori under in vitro culture conditions (4, 18). Therefore, it was suggested that the nonphosphorylated regulator protein HP166 is required for the transcription of target genes which are essential for cell viability, while additional nonessential target genes are regulated by the phosphorylated response regulator in the presence of appropriate environmental stimuli, leading to the activation of the cognate histidine kinase HP165 (8). The expression and proper regulation of some of these nonessential target genes seem to be necessary for colonization, as suggested by the observation that histidine kinase HP165 is required for establishing stable infection of mice.
Role of a subset of the target genes of the two-component system HP166-HP165 for colonization.
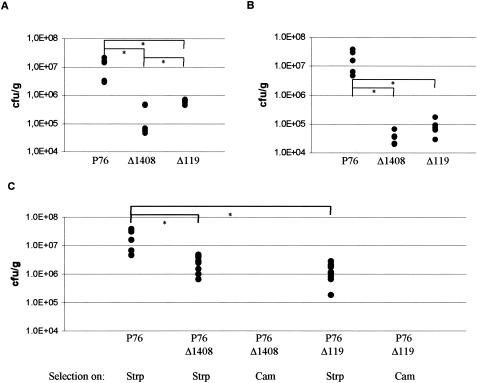
Recently, the identification of nonessential target genes of the two-component system HP166-HP165, whose transcription is activated by the phosphorylated response regulator HP166, was reported (8). Genes under the control of the PHP119 promoter constitute a gene family encoding H. pylori specific proteins of unknown function. In strain 26695 this gene family comprises five members (HP1188, HP1187, HP118, HP119, and HP120), which are clustered at two different loci in the genome (29). The target genes under the control of the PHP1408 promoter are also H. pylori specific and form an operon which is present in two identical copies in the genome of H. pylori 26695 (HP1408-HP1412 and HP427-HP423) (29). To investigate whether the PHP119- and PHP1408-dependent target genes contribute to host colonization, mutants P76/Δ119 and P76/Δ1408 were used to infect mice. P76/Δ119 and P76/Δ1408 are derivatives of H. pylori P76 in which the two chromosomal loci comprising the target genes under the control of the PHP119 and PHP1408 promoters, respectively, were replaced by kanamycin and chloramphenicol resistance cassettes and have been described earlier (Fig. 1) (8). Again, in two independent experiments five mice per mutant were inoculated with three consecutive doses of 1.0 × 109 CFU of H. pylori. Both mutants were competent for the colonization of the mouse stomach, yielding similar bacterial loads (P76/Δ1408, 7.0 × 104 and 3.6 × 104 CFU/g of stomach tissue; P76/Δ119, 6.1 × 105 and 7.4 × 104 CFU/g of stomach tissue; Fig. 2). Although, compared to the P76 wild type, both mutants showed a significantly reduced ability to infect mice, these data demonstrate that ORF HP119 and its paralogs, as well as ORFs HP1408-HP1412, are dispensable for host colonization and, therefore, that additional target genes regulated by the phosphorylated response regulator HP166 must be responsible for the colonization defect of the histidine kinase mutant P76/HP165::km. A recent transcriptome analysis of a deletion mutant of the histidine kinase ORF HP165 identified several such target genes, representing mainly H. pylori specific genes of unknown function (11). When mice (10 animals per group) were inoculated with a 10:90 mixture of wild-type P76 and the mutant P76/Δ119 or P76/Δ1408, respectively, only wild-type bacteria could be recovered from the animals 4 weeks after the last challenge with the mixed inoculum (Fig. 2), as investigated by plating of stomach homogenates in parallel on serum plates containing streptomycin (250 mg/liter) or both streptomycin and chloramphenicol (4 mg/liter). Therefore, although not essential for colonization per se, the genes under the control of the PHP119 and PHP1408 promoters provide the bacteria with a clear advantage for survival under competitive conditions within the host organism.
FIG. 2.
Colonization of BALB/c mice with wild-type H. pylori P76 and isogenic deletion mutants of target genes of the two-component system HP166-HP165 (P76/Δ1408 and P76/Δ119, respectively). The colonization abilities of wild-type and mutant P76 are shown as CFU per gram of stomach tissue for single mice. The experiments lasted for 4 weeks each. In two independent experiments (A and B) a significantly reduced colonization ability of P76/Δ1408 and P76/Δ119 mutants compared to that of P76 was observed (n = 5). In a third experiment (C) a 10/90 mixed infection with P76 (streptomycin resistant) and P76/Δ1408 or P76/Δ119 (both of which are streptomycin and chloramphenicol resistant) was performed (n = 10 for mixed infection and n = 5 for P76 single infection). Neither P76/Δ1408 nor P76/Δ119 could be recovered from mixed infections with P76 as indicated by the lack of growth on serum plates supplemented with chloramphenicol. Groups presented in panels B and C were evaluated in the same experiment, and the wild-type control experiments were therefore performed with the same animals. *, P < 0.05.
Taken together our data suggest that besides regulating the expression of genes essential for bacterial viability the two-component systems of H. pylori are also involved in host adaptation and survival in vivo. Similarly, a deletion analysis of the 13 histidine kinase and 14 response regulator genes predicted by the genome sequence of Streptococcus pneumoniae revealed that inactivation of seven two-component systems and one orphan response regulator caused attenuation in a mouse respiratory tract infection model, while in vitro growth of the mutants under nutrient-rich conditions was not affected (28). It is becoming increasingly evident that successful infection of a host organism requires not only the presence of specific virulence factors interacting in a sophisticated way with host tissues and cells but also a tight regulation of metabolic processes adapted to the particular niche that a pathogen is encountering. This is also reflected by the vast array of metabolic and general stress response genes identified in screens for virulence-attenuated pathogens by using signature-tagged mutagenesis (19, 24). Therefore, the H. pylori two-component systems HP1365-HP1364 and HP166-HP165 might be involved either in the regulation of H. pylori-specific virulence traits or in maintaining bacterial fitness by allowing the adaptation of metabolic processes in response to frequently changing environmental conditions within the human stomach. The comprehensive identification of the target genes of these two-component systems by transcriptome analysis will be needed to clarify this question.
Acknowledgments
We thank R. Gross for critically reading the manuscript.
This work was supported by grants Be1543/2-1, Be1543/2-3, and HA2697/6-1 from the Deutsche Forschungsgemeinschaft.
Editor: A. D. O'Brien
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E., Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 3.Beier, D., G. Spohn, R. Rappuoli, and V. Scarlato. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, B. J., N. de Vries, S. G. Rijpkema, A. H. M. van Vliet, and C. W. Penn. 2002. Transcriptional and mutational analysis of the Helicobacter pylori urease promoter. FEMS Microbiol. Lett. 213:27-32. [DOI] [PubMed] [Google Scholar]
- 8.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Duarte, O. G., B. Lucas, Z.-X. Yan, K. Panthel, R. Haas, and T. F. Meyer. 1998. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella typhimurium producing urease subunits A and B. Vaccine 16:460-471. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin, C. S. 1997. Helicobacter pylori gastritis, peptic ulcer, and gastric cancer: clinical and molecular aspects. Clin. Infect. Dis. 25:1017-1019. [DOI] [PubMed] [Google Scholar]
- 15.Ilver, D., A. Arnqvist, J. Ögren, I.-M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 16.Kavermann, H., P. B. Burns, K. Angermüller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 18.McDaniel, T. K., K. C. DeWalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15-23. [DOI] [PubMed] [Google Scholar]
- 19.Mecsas, J. 2002. Use of signature-tagged mutagenesis in pathogenesis studies. Curr. Opin. Microbiol. 5:33-37. [DOI] [PubMed] [Google Scholar]
- 20.Panthel, K., W. Jechlinger, A. Matis, M. Rohde, M. Szostak, W. Lubitz, and R. Haas. 2003. Generation of Helicobacter pylori ghosts by PhiX protein E-mediated inactivation and their evaluation as vaccine candidates. Infect. Immun. 71:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 22.Satin, B., G. Del Giudice, V. Della Bianca, S. Dusi, C. Laudanna, F. Tonello, D. Kelleher, R., R. Rappuoli, C. Montecucco, and F. Rossi. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shea, J. A., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 25.Skouloubris, S., J.-M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 29.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, M. Hayes, W. S. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 30.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stähler, J. Guhl, E. Bisse, A. H. M. van Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. J. Bacteriol. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]