Abstract
We report the complete nucleotide sequence and genetic organization of the Vat-encoding pathogenicity island (PAI) of avian pathogenic Escherichia coli strain Ec222. The 22,139-bp PAI is situated adjacent to the 3′ terminus of the thrW tRNA gene, has a G+C content of 41.2%, and includes a bacteriophage SfII integrase gene, mobile genetic elements, two open reading frames with products exhibiting sequence similarity to known proteins, and several other open reading frames of unknown function. The PAI encodes an autotransporter protein, Vat (vacuolating autotransporter toxin), which induces the formation of intracellular vacuoles resulting in cytotoxic effects similar to those caused by the VacA toxin from Helicobacter pylori. The predicted 148.3-kDa protein product possesses the three domains that are typical of serine protease autotransporters of Enterobacteriaceae: an N-terminal signal sequence of 55 amino acids, a 111.8-kDa passenger domain containing a modified serine protease site (ATSGSG), and a C-terminal outer membrane translocator of 30.5 kDa. Vat has 75% protein homology with the hemagglutinin Tsh, an autotransporter of avian pathogenic E. coli. A vat deletion mutant of Ec222 showed no virulence in respiratory and cellulitis infection models of disease in broiler chickens. We conclude that the newly described PAI and Vat may be involved in the pathogenicity of avian septicemic E. coli strain Ec222 and other avian pathogenic E. coli strains.
Avian pathogenic Escherichia coli induces extraintestinal infections which may cause respiratory disease, septicemia, swollen head syndrome, cellulitis, or combinations of these syndromes (16, 17). Respiratory infections due to E. coli occur primarily in young broilers whose respiratory tract is damaged by infectious agents or noxious stimuli (17). As a sequel to the respiratory infection, the E. coli may invade the bloodstream and cause septicemia. Avian pathogenic E. coli strains commonly belong to certain O serogroups, particularly O1, O2, O35, and O78 (9, 17, 43). However, several other O serogroups and nontypeable E. coli strains are also avian pathogenic, albeit at lower frequencies.
Properties associated with virulence of avian pathogenic E. coli include production of F1 (type 1) and P fimbrial adhesins that mediate adherence to tissues (10, 13, 38), secretion of aerobactin for scavenging iron, resistance to the bactericidal activity of serum, the formation of certain polysaccharide capsules, the production of colicin V (16, 32, 34, 49), and the synthesis of temperature-sensitive hemagglutinin (Tsh) (39). These virulence factors are protein structures located at the cell surface, proteins that are secreted to the bacterial cell surface, or proteins that are released into the external environment. Autotransporters are one category of secreted proteins implicated in virulence of avian pathogenic E. coli and certain other types of pathogenic E. coli. The autotransporter proteins constitute a distinct family of proteins secreted from gram-negative bacteria that represent bacterial virulence factors, such as adhesins, proteases, and toxins (14, 23). The autotransporters have an overall unifying structure comprising (i) an N-terminal signal sequence, (ii) the secreted mature protein (or passenger domain), and (iii) a C-terminal domain, which forms a pore in the outer membrane through which the passenger domain passes to the cell surface (23). Members of this family include virulence factors of important human pathogens, such as the immunoglobulin A1 proteases from Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae, the VacA cytotoxin from Helicobacter pylori (23), Tsh from avian pathogenic E. coli (39), E. coli secreted protein C (EspC) from enteropathogenic E. coli (45), E. coli serine protease P (EspP) from enterohemorrhagic E. coli (5), Sat from uropathogenic E. coli (19), and PssA from Shiga toxin-producing E. coli (11).
It was recently reported that avian pathogenic E. coli produces a cytotoxin that is similar to the H. pylori VacA cytotoxin (40). The cytotoxic substances in supernatants of both avian pathogenic E. coli and H. pylori cause similar morphological changes in target cells and are similar with respect to heat lability and susceptibility to proteolytic enzymes. The similarities in vacuolation of cells by the avian E. coli culture supernatants and the VacA toxin from H. pylori were remarkable (40). The fact that the H. pylori cytotoxin is considered to be a virulence factor suggested that the E. coli cytotoxin may also contribute to virulence.
Virulence genes of bacterial pathogens may be carried on plasmids, bacteriophages, or the chromosome and tend to be clustered in the genome. Pathogenicity islands (PAIs) are regions on the genomes of certain pathogenic bacteria that include virulence genes and are absent from related nonpathogenic strains (22). The acquisition of PAIs has been proposed as a major mechanism in pathogen evolution (22). PAIs have been identified in several bacterial species, including uropathogenic E. coli (46), enteropathogenic E. coli (29), H. pylori (6), Vibrio cholerae (4), and Shigella spp. (2, 31). The genes for several autotransporter proteins have been identified in PAIs. The she PAI of Shigella flexneri 2a encodes two autotransporter proteins, namely, Pic, a protease with mucinase and hemagglutinin activities, and SigA, a cytopathic protease that contributes to intestinal fluid accumulation (2, 24). The espC PAI of enteropathogenic E. coli encodes EspC, a member of the autotransporter family with enterotoxic activity (30).
In the present study, we report the cloning and sequence determination of the structural gene encoding the vacuolating autotransporter (Vat) cytotoxin from an avian pathogenic E. coli strain and a preliminary evaluation of the role of Vat in virulence. In addition, we describe the complete sequence of a novel PAI in which the vat gene is located.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strain Ec222 (serogroup O78) was isolated from a septicemic chicken and has been shown to produce vacuolating cytotoxicity (40). E. coli XL10-Gold (Stratagene, La Jolla, Calif.) was used as the recipient strain for genetic manipulations. Strains were grown in Luria-Bertani (LB) broth or agar. Plasmid vectors pUC18 (Fermentas, Burlington, Ontario), pBluescript II SK(+) (Stratagene), and pOK12 (47) were used as cloning vectors and for plasmid library construction. Suicide vector pRE107 (oriT oriV sacB Apr) and E. coli SM10(λpir) (thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr) were used for allelic exchange mutagenesis. When antibiotic selection was necessary, the growth medium was supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml). All strains were stored at −70°C in LB broth containing 15% glycerol.
Plasmid library construction.
All genetic manipulations were performed by standard methods as described by Sambrook et al. (41). Plasmid DNA was purified by using a plasmid mini kit (Qiagen Inc., Santa Clarita, Calif.). Purification of DNA fragments and extraction from agarose gel slices were performed with Geneclean (Geneclean III Kit; BIO101 Inc., La Jolla, Calif.). DNA ligations and restriction endonuclease digestions were performed with enzymes supplied by New England Biolabs (Beverley, Mass.) or Amersham Pharmacia Biotech (Piscataway, N.J.). DNA transformations were performed by using the heat shock method described by Nishimura et al. (33).
Cloning strategy. (i) Recombinant DNA technique.
Initially, a plasmid library was constructed by ligating Sau3A-digested E. coli Ec222 genomic DNA into BamHI-digested pBluescript II SK(+). The ligation mixture was used to transform E. coli XL-I Blue (Stratagene) competent cells. The transformants were screened for vacuolating cytotoxin activity on chicken embryo fibroblasts (CEFs) as previously described (40).
(ii) Chromosome walking.
For the initial sequence information, we cloned and sequenced the PCR product obtained with E. coli Ec222 as the template and primers based on the C-terminal translocator domain which is highly conserved among autotransporters. The information obtained was used to (i) select specific restriction enzymes that were used for digestion of the chromosomal DNA, (ii) design specific primers, and (iii) generate probes which were used in Southern blotting or colony blotting to select for cloning DNA fragments of convenient sizes adjacent to the previously sequenced areas and to identify positive subclones. The entire PAI sequence was obtained by chromosome walking by using sequence information obtained from each round of sequencing.
Preparation of DIG-labeled probes.
DNA probes for detection of all steps of chromosome walking were labeled by PCR amplification in the presence of digoxigenin-11-dUTP (DIG; Boehringer Mannheim) according to the manufacturer's recommendation. DNA probes were amplified from E. coli strain Ec222.
Southern and colony blot hybridizations.
The preparation of genomic DNA of E. coli Ec222 and DNA hybridizations were described previously (35). The genomic DNA of E. coli Ec222 was extracted with the Qiagen genomic tip and associated reagents (Qiagen Inc.). In Southern blots and colony hybridizations, the nylon membranes were prehybridized for at least 4 h at 42°C in hybridization solution without labeled probe and then hybridized separately at 42°C with specific DNA probes for 16 h. The membranes were washed at 68°C under high-stringency conditions. DNA hybridizations and detection were performed by using the DIG labeling and detection system according to the manufacturer's recommendation (DIG system user's guide for filter hybridization, Boehringer Mannheim GmbH, Mannheim, Germany).
Cytotoxin production.
Cytotoxicity assays were used to confirm toxin production by the recombinant E. coli XL10/pVP1685 (vat). CEF cells were grown at 37°C in Eagle's minimal essential medium with 10% fetal calf serum (Sigma-Aldrich Co., St. Louis, Mo.) as described by Salvadori et al. (40). Transformants were cultivated overnight in LB broth with ampicillin at 37°C, with shaking at 150 rpm. Culture supernatants were assayed in twofold serial dilutions. The plates were incubated at 37°C in humidified 5% CO2. Morphological changes were observed over a 6-h period, and the cells were stained with neutral red as described by Cover et al. (8).
Plasmid and strain construction.
DNA fragments of E. coli Ec222 detected by Southern blot were cloned, and then inserts in plasmids pVP16 (Apr) and pVP85 (Apr) were sequenced. Sequence analysis showed that these two plasmids contained inserts which likely included the entire sequence of a Tsh-related protein. A clone (pVP16-85) that was expected to express the Vat protein was constructed by ligating together fragments from pVP16 (AccI-KpnI) and pVP85 (AccI-EcoRV) into the KpnI-EcoRV site of pOK12. pVP16-85 contained the complete vat coding region, native promoter, and downstream termination motif. Vat production was determined by cytotoxicity assays on CEF cells.
Construction of a vat deletion mutant of E. coli strain Ec222 was performed by suicide vector mutagenesis as described by Gunzer et al. (18). We generated PCR fragments of 1,021 and 970 bp from E. coli Ec222 by using Advantage HF-2 polymerase (Clontech, Palo Alto, Calif.) and the primer pairs Vat74up-Vat74down and Vat73up-Vat73down, respectively (Table 1). The two amplified DNA fragments were ligated at their XbaI restriction sites, generating a vat gene with an internal 1,161-bp deletion (corresponding to amino acid positions 389 to 767) (see Fig. 2), which was cloned into the SalI-SacI site of suicide vector pRE107 to generate plasmid pREΔvat (Apr). The vat mutant strain (Ec222VPΔ2) was generated by allelic exchange of the vat gene in pREΔvat by using ampicillin and sucrose selection as previously described (18). PCR amplification with the primers Vat74up and Vat73down was performed to confirm that the deletion in the vat gene had occurred, and the isogenic pair, Ec222 and Ec222VPΔ2, were tested for cytotoxicity on CEF cells.
TABLE 1.
Oligonucleotide primers used in allelic exchange experiment
| Primer | 5′-3′ Sequencea |
|---|---|
| Vat74up | GC-GTCGAC-CTTACCTCTCTGGCACT (SalI) |
| Vat74down | CG-TCTAGA-GTCAGTGAACCGGCACC (XbaI) |
| Vat73up | GC-TCTAGA-TCTTCAACGGCACCGTC (XbaI) |
| Vat73down | CG-GAGCTC-GATGTGCATGAATGTGG (SacI) |
Recognition sites for restriction endonucleases are underlined, and restriction endonucleases are indicated. The reference for all primers is this study.
FIG. 2.
Structure of the Vat precursor protein (not drawn to scale). The protein consists of 1,377 amino acids, with a signal sequence (SS) of 5.7 kDa, the Vat cytotoxin of 111.8 kDa, and a C-terminal outer membrane translocator of 30.5 kDa. A, signal sequence cleavage site; B, two cysteine residues; C, atypical serine protease motif; D, region that is deleted in vat mutant strain Ec222VPΔ2; E, outer membrane translocator cleavage site.
DNA sequencing and sequence analysis.
Nucleotide sequencing was performed at the Guelph Molecular Supercentre with the ABI Prism fluorescent Big Dye Terminator system by using universal vector primers or synthetic oligonucleotides designed on the basis of preceding sequences. The DNA sequences in the PAI and the sequences of the corresponding translated proteins were compared with sequences in the GenBank genetic sequence database by using the BLAST programs accessed from the National Center for Biotechnology Information. Comparisons of multiple sequence alignments were done using the program Vector NTI (InforMax, Inc., Bethesda, Md.).
Infection studies.
The virulence of E. coli Ec222 and its vat deletion mutant Ec222VPΔ2 were tested in experimental models of respiratory-septicemic disease and cellulitis. The respiratory-septicemic disease model consisted of aerosol administration of the challenge E. coli 4 days after intranasal administration of the Massachusetts strain of infectious bronchitis virus to 17-day-old broiler chicks (36). Two groups of 10 chickens received wild-type E. coli Ec222, and two groups of 10 received E. coli Ec222VPΔ2. Six days after the E. coli challenge, chickens that survived were euthanized and gross lesions of airsacculitis, pericarditis, and perihepatitis were scored on the basis of severity. Absence of lesions was scored as 0.01, airsacculitis was scored as 1 to 3, and pericarditis and perihepatitis were each scored as 1 or 2. Mean total lesion scores were determined for each group, and differences were compared by the two-tailed t test, with P of <0.05 considered significant. Air sacs, pericardial fluid, and livers were also cultured by direct plating of swabs and by immersion of the swabs in Trypticase soy broth (Difco, Detroit, Mich.).
The cellulitis model of disease in 3-week-old broiler chicks involved disinfecting the skin of each chick with 70% ethanol, making a 1-cm scratch with a sterile 22-gauge needle on the skin of the left side of the caudal abdomen, and then swabbing the scratched area with a cotton swab dipped in an overnight culture of the challenge organism (37). One group of 12 chickens was challenged with wild-type E. coli Ec222, and a second group of 12 chickens was challenged with E. coli Ec222VPΔ2. Ten days after infection, the chickens were euthanized and the lesions were scored as mild (score of 3), moderate (score of 5), or severe (score of 7). Absence of a lesion was scored as 0.01.
Nucleotide sequence accession number.
The sequence reported here has been assigned GenBank accession no. AY151282.
RESULTS
The cloning of fragments of E. coli Ec222 DNA into high-copy-number vectors yielded two colonies whose culture supernatants possessed vacuolating activity on CEF cells. However, the positive clones rapidly lost the ability to express vacuolating cytotoxicity. This loss of biological activity was accompanied by loss of some or all of the insert DNA in the recombinant plasmids. This problem was not solved by use of a low-copy-number vector for cloning. We therefore modified our strategy. We designed primers based on gene sequences corresponding to the highly conserved C termini of E. coli autotransporters (Tsh, SepA, EspP, Pet, and Sat) and made a probe called C-AT. A Southern blot of digested genomic DNA from strain Ec222 was hybridized with the C-AT probe, and a single band of 2,360 bp was detected. The 2,360-bp fragment was cloned, subsequent recombinants were screened with the C-AT probe, and a positive clone was sequenced. The entire PAI sequence was obtained by chromosome walking by using sequence information obtained from each round of sequencing.
Sequence analysis of VAT-PAI in E. coli Ec222. (i) Size and site.
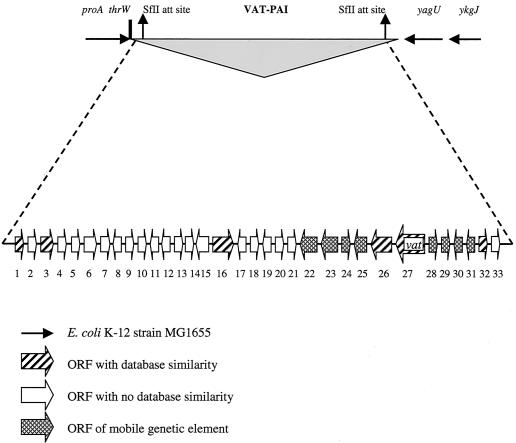
The assembly of the sequences inserted into different plasmid vectors revealed a PAI which we have designated VAT-PAI. The complete VAT-PAI is 22,139 bp in size and exhibits several features which are typical of PAIs. The chromosomal integration site of VAT-PAI is the thrW tRNA gene at one end and the yagU gene at the other, representing loci that are 40 kb apart from each other in the E. coli K-12 MG1655 chromosome.
(ii) Structure of VAT-PAI.
Analysis of the unique DNA sequences from this PAI revealed the presence of genes encoding proteins with potential virulence roles and open reading frames (ORFs) with unknown functions. The PAI includes 33 ORFs that encode putative proteins which are >60 deduced amino acids long and are numbered sequentially (Table 2; Fig. 1). Nineteen of the ORFs and their products showed either no similarity or no significant homology to any protein or nucleotide sequence in the database. Significant homology was defined as greater than 30% identity over at least 50% of the query sequence. There are eight ORFs that are related to mobile genetic elements, such as bacteriophage integrase genes and insertion sequences (IS), and six ORFs that encode putative proteins including the vacuolating autotranporter toxin and the PapX protein (Table 2; Fig. 1).
TABLE 2.
ORFs and mobile genetic elements within the VAT-PAI
| ORF or feature | G + C content (%) | No. of aaa encoded | Positionb (bp) | Gene or protein with similar sequence | % ntc (protein) identity | Accession no. |
|---|---|---|---|---|---|---|
| 1-641 | proA (E. coliK-12 MG1655) | 95 | AE000132 | |||
| 642-717 | thrW (E. coliK-12 MG1655) | 100 | AE000132 | |||
| att | 699-720 | Bacteriophage SflI attachment site (S. flexneri) | 100 | AF021347 | ||
| 1 | 39.3 | 149 | 1818-2267 | Phosphoserine aminotransferase (Spinacea oleracea) | (24) | D84061 |
| 2 | 40.4 | 60 | 3736-3918 | No significant homology | ||
| 3 | 33.8 | 208 | 4165-4791 | Putative protein (A. thaliana) | (24) | AL049730 |
| 4 | 33.3 | 89 | 4801-5070 | No significant homology | ||
| 5 | 39.1 | 68 | 5342-5548 | No significant homology | ||
| 6 | 36.9 | 130 | 5727-6119 | No significant homology | ||
| 7 | 46.9 | 117 | 7362-7715 | No significant homology | ||
| 8 | 47.1 | 96 | 7708-7418 | No similarity | ||
| 9 | 46.5 | 164 | 7732-8226 | No significant homology | ||
| 10 | 48.2 | 169 | 8252-8761 | No significant homology | ||
| 11 | 47.7 | 173 | 8319-7798 | No similarity | ||
| 12 | 48.8 | 206 | 8725-8105 | No similarity | ||
| 13 | 36.9 | 65 | 8926-9123 | No significant homology | ||
| 14 | 44.5 | 190 | 9158-9730 | No similarity | ||
| 15 | 43.9 | 84 | 9654-9400 | No similarity | ||
| 16 | 44 | 424 | 9715-10989 | gp22 Mycobacteriophage Bxb1 | (30) | AF271693 |
| 17 | 43.6 | 90 | 10424-10152 | No significant homology | ||
| 18 | 41 | 225 | 11037-11714 | No significant homology | ||
| 19 | 46.4 | 60 | 11146-10964 | No significant homology | ||
| 20 | 41.4 | 61 | 11164-11349 | No significant homology | ||
| 21 | 48.3 | 68 | 12099-12305 | No significant homology | ||
| 22 | 44.2 | 159 | 12275-11796 | Putative integrase (E. coli O157:H7 EDL933) | (33) | AE005658 |
| 23 | 43.2 | 290 | 13131-12259 | Integrase (X. axonopodis 306) | (27) | AE011739 |
| 24 | 46 | 112 | 13389-13727 | Integrase (E. coli) | (49) | AJ278144 |
| 25 | 41 | 130 | 13831-13439 | Integrase (E. coli) | (50) | AJ278144 |
| 26 | 30.5 | 174 | 14685-14161 | PapX regulatory protein (E. coli) | (48) | P42193 |
| 27d | 48.4 | 1377 | 18944-14811 | Tsh and Hbp (E. coli) | 80 (75) | AF218073 |
| AJ223631 | ||||||
| 28 | 48.3 | 71 | 19534-19749 | iso-IS1 insA (S. flexneri 2a) | 93 (89) | AF386526.1 |
| 29 | 52.8 | 64 | 19831-20025 | iso-IS1 insB (S. flexneri 2a) | 94 (88) | AF386526.1 |
| 30 | 52.3 | 64 | 20012-19818 | iso-IS1 insB (S. flexneri 2a) | 94 (88) | AF386526.1 |
| 31 | 51.1 | 74 | 20206-19982 | iso-IS1 insB (S. flexneri 2a) | 93 (65) | AF386526.1 |
| att | 20421-20571 | Bacteriophage SflI attachment site (S. flexneri) | 91 | AF021347 | ||
| 32 | 56.7 | 22 | 20679-20745 | Unknown function (pathogenic poultry E. coli) | 86 | AF524932.1 |
| 33 | 41.1 | 63 | 20709-20900 | No similarity | ||
| 191 | 21045-21620 | Protein YagU (E. coliK-12 MG1655) | 91 | AE000136 | ||
| 76 | 22138-21908 | Ferredoxin (E. coliK-12 MG1655) | AE000136 |
aa, amino acids.
Data for E. coli K-12 are in bold.
nt, nucleotide sequence; percent identity with the translated amino acid sequence is shown in parentheses.
ORF 27 encodes the Vat protein.
FIG. 1.
Genetic map of the VAT-PAI with 22,139 bp of unique DNA downstream of the tRNA gene thrW and upstream of the yagU gene of E. coli K-12 MG1655. Numbers indicate ORFs described in Table 2. ORFs have not been drawn to scale.
In the VAT-PAI, the tRNA gene thrW is followed by a fragment of the sequence for the bacteriophage SfII attachment site, followed by inserted DNA that contains 16 ORFs encoding proteins with >60 deduced amino acids (Table 2). Among these ORFs, which are numbered sequentially from 1 to 16, only three (ORFs 1, 3, and 16) showed similarities to sequences in the database. ORFs 1 and 3 encode proteins with low homology (24% identity) to a phosphoserine aminotransferase of Spinacea oleracea and to a hypothetical protein of Arabidopsis thaliana, respectively. ORF 16 encodes a fragment similar to gp22 of mycobacteriophage Bxb1 and is followed by sequences encoding products with homology to integrases found in E. coli MG1655, E. coli O157:H7 EDL 933, and Xanthomonas axonopodis strain 306. The terminal 18 nucleotides of the thrW gene are repeated between ORFs 23 and 24 (bp 13217 to 13234). In the adjacent region, ORF 26 encodes a putative protein similar to the 17-kDa transcriptional regulator PapX (43% homology) and ORF 27 encodes a protein 75% similar to Tsh or Hbp of E. coli. ORF 27 is followed by IS elements that are homologous to iso-IS1 of S. flexneri 2a. At the right junction site of VAT-PAI, there is an ORF that encodes the integrase of bacteriophage SfII, the remaining sequence of the attachment site of bacteriophage SfII, and the upstream region of the interrupted gene yagU. If one includes ORFs that encode proteins of <60 amino acids, then ORF 32 (22 amino acids) is identified. This ORF is similar to a region of unknown function in pathogenic poultry E. coli. The DNA inserted from thrW to yagU contains approximately 6.5 kb of fragments without any nucleotide or translated amino acid sequence homology.
The average G+C content of the VAT-PAI is 41.2%, which is lower than the average for the E. coli genome (51%). Interestingly, the G+C content has high variability across the VAT-PAI (Table 2).
(iii) Sequence analysis of the vat toxin gene.
The largest ORF (27) in the VAT-PAI consists of 4,134 nucleotides and was designated the vat gene. This gene encodes a putative protein with 1,377 amino acids and a deduced molecular mass of 148.3 kDa. The G+C content of the vat gene was determined to be 48.4%, slightly lower than the average in E. coli. Two cysteines are present in the mature cytotoxin (positions 239 and 251) (Fig. 2). A predicted Shine-Dalgarno site, with sequence GGAA, was identified 13 bp upstream of the proposed start codon. A possible rho-independent stem-loop transcriptional termination signal was also identified 6 bp downstream of the TGA termination codon of the vat gene. Regions flanking the vat gene show the presence of IS-like elements and phage-related integrase genes (Fig. 1).
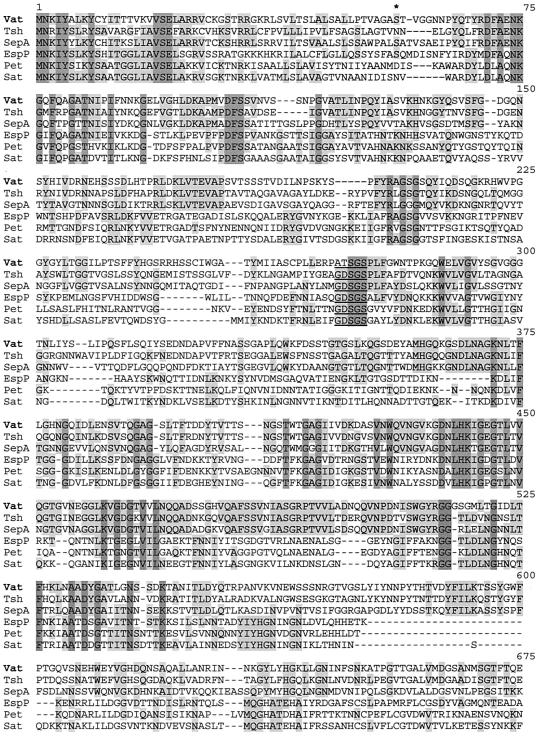
Comparison of the amino acid sequence translated from vat with sequences in GenBank revealed that the Vat sequence was similar to sequences of a family of serine protease autotransporters of Enterobacteriaceae (SPATEs), including Tsh (78%), Pic (47%), SepA (45%), EspP (36%), Sat (36%), and Pet (35%) (Fig. 3). As expected, a high degree of homology was found among the C-terminal domains, with homologies that ranged from 60 to 98% identity. Homologies among the N-terminal regions were less and varied from 42 to 60% identity. The alignments between Vat and some SPATEs showed homologous areas and also the presence of large gaps (Fig. 3). A serine protease motif (GDSGSP) has been found at the same position in several SPATEs, but at the corresponding site in Vat, the sequence was ATSGSP. Computer analysis of the deduced amino acid sequence of Vat indicated the characteristics of a signal sequence followed by a signal peptidase cleavage site between residues 55 and 56 (AGA-ST). By comparison with other autotransporters, the cleavage site of the β domain was predicted to lie between residues N1100 and N1101 (Fig. 2).
FIG. 3.
Alignment of the sequence of the predicted Vat protein (accesion number AY151282) with those of its closest homologues, Tsh (AJ223631), SepA (Z48219), EspP (X97542), Pet (AF056581), and Sat (AF289092). Residues identical in all six sequences are shaded in dark grey and those residues conserved in at least three sequences are shaded in light grey. The asterisk at residue 56 indicates the first amino acid of the mature Vat protein. The serine protease motif is underlined.
To confirm that the vat ORF encoded a secreted protein, supernatants of overnight cultures of recombinant E. coli XL10/pVP16-85 (vat), the control strain (vector only), and wild-type Ec222 were tested on CEF cells for the presence of vacuolating cytotoxic effects. Supernatants from the recombinant and wild-type strains showed the characteristic vacuolation on CEF cells, while supernatant from the control bacteria with vector only had no effect.
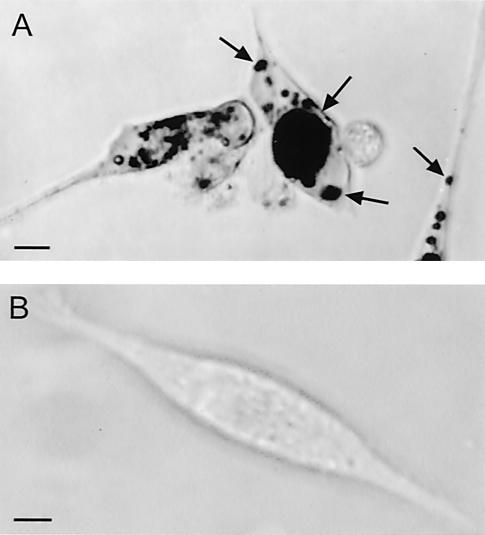
Tests of culture supernatants of the isogenic E. coli pair Ec222 and Ec222VPΔ2 with the cytotoxicity assay showed that the wild-type Ec222 strain possessed vacuolating cytotoxic activity whereas Ec222VPΔ2 lacked this activity. Cytotoxicity was seen as vacuolation which was detected by 2 h, reached maximal activity at 6 h, and was still observable at 24 h. The vacuoles were observed in unstained cells and after staining with neutral red. Figure 4A shows the appearance of vacuoles in CEF cells stained with neutral red after exposure to a 1/16 dilution of supernatant of an overnight LB culture of E. coli Ec222. A similar preparation from the vat deletion mutant Ec222VPΔ2 had no effect on the cells (Fig. 4B).
FIG. 4.
Vacuolation of CEF cells caused by culture supernatant of E. coli Ec222. (A) Vacuoles (arrows) were evident after 6 h of exposure of CEF cells to a 1/16 dilution of the filter-sterilized supernatant of an overnight culture of wild-type E. coli Ec222. The cells were stained with neutral red, which accumulated in the vacuoles. (B) Normal appearance of CEF cells 6 h after treatment as described for panel A except that the E. coli strain was Ec222ΔVP, a vat deletion mutant derivative of Ec222. Bars, 10 μm.
Infection studies.
The results of infection of chickens with the wild type and the vat deletion mutant of E. coli Ec222 (Ec222VPΔ2) are summarized in Table 3. For the aerosol infection, the results for both groups were pooled as there was no significant difference in the results for the groups which received the same treatments. Among the 20 chickens challenged with E. coli Ec222, nine chickens had severe lesions (scores of 5 to 7), three had moderate lesions (scores of 2 to 3), three had mild lesions (score of 1), and four had no lesions. Among the 20 chickens challenged with E. coli Ec222VPΔ2, two had mild lesions of airsacculitis and 18 had no lesions. In the cellulitis infection study, all 12 chickens which received wild-type E. coli Ec222 developed cellulitis; one chicken had a severe lesion, 10 had moderate lesions, and one had a mild lesion. All but one of these chickens showed signs of septicemia (perihepatitis and pericarditis), and four of them died. None of the chickens infected with E. coli Ec222VPΔ2 developed cellulitis. The challenge E. coli was recovered from the cellulitis lesions. In both models of infection, the difference in virulence levels of the wild type and the vat deletion mutant of Ec222 was highly significant (P < 0.0001).
TABLE 3.
Mortality, lesion scores, and recovery of challenge E. coli from infected chickens
| Challenge E. coli | Mortality (%) | Mean lesion score ± SEM | Recovery of E. coli
|
|
|---|---|---|---|---|
| Directa | Totalb | |||
| Aerosol challenge | ||||
| Wild-type Ec222 | 15 | 3.65 ± 0.65c | 43/60 | 56/60 |
| Ec222 vat deletion mutant | 0 | 0.11 ± 0.07 | 9/60 | 32/60 |
| Cellulitis challenge | ||||
| Wild-type Ec222 | 33 | 5.0 ± 0.25c | 12/12 | NDd |
| Ec222 vat deletion mutant | 0 | 0.01 ± 0 | 0/12 | ND |
Number recovered by direct plating of swabs on MacConkey agar/total.
Number recovered by culture of swabs in Trypticase soy broth followed by subculture on MacConkey agar/total.
Significantly higher than that for lesions caused by infection with for Ec222 vat mutant. P = <0.0001. Maximum score was 7.0. Normal was scored as 0.01.
ND, not done.
DISCUSSION
We describe for the first time a PAI in avian E. coli that encodes an autotransporter protein. We designated the gene for the autotransporter protein vat, for vacuolating autotransporter toxin, and the PAI was designated VAT-PAI.
PAIs are clusters of genes that include virulence genes, are present in the genomes of pathogenic strains, and are absent from nonpathogenic strains. PAIs occupy large genomic regions (>10 kb) often associated with tRNA genes which act as integration sites for foreign DNA, reflecting horizontal gene transfer. These islands have G+C contents that differ from those of the core genomic DNA and often carry genes encoding mobility factors such as integrases and transposases and IS elements (21). All these characteristics are shared by the VAT-PAI.
The VAT-PAI is inserted at a chromosomal site adjacent to a tRNA-thrW gene, a site that is preferentially occupied in uropathogenic E. coli by PAI III536 (12) and in S. flexneri by a PAI that encodes the O antigen (1). The VAT-PAI is located between the thrW and yagU genes and interrupts the attachment site of bacteriophage SfII. Additionally, integrase genes of E. coli were found throughout the PAI, a feature which is also characteristic of PAIs. Insertion of the VAT-PAI is associated with absence of a large fragment of E. coli K-12 DNA at thrW. The integration of other PAIs into thrW also appears to have caused deletions (1, 12). In E. coli K-12 MG1655, the region from thrW to yagU covers approximately 40 kb and contains several IS elements, which may lead to high recombination in this region and could explain the loss of a large fragment. Maurelli et al. suggest that the strategy of “black hole” formation or deletion of large regions could precede additions of PAIs, making the recipient background more favorable for expression of virulence genes (28). The VAT-PAI, with approximately 22 kb, has a G+C content of 41.2%, which is lower than the 51% present in the E. coli K-12 genome, suggesting that it may have been acquired by horizontal transfer. The variability across the PAI may represent insertion of DNA from a number of sources over time. In particular, the G+C content of the portion of the PAI inserted between thrW and the E. coli integrase gene (bp 718 to 13439) is 43.3%, whereas the G+C content of the remainder of the PAI is 37.9%.
Another common characteristic of PAIs is the presence of unidentified ORFs. On the VAT-PAI, there are 19 ORFs encoding proteins of >60 deduced amino acids with no homology to known DNA or protein sequences. It is not known at this time whether these ORFs contribute to gene expression or regulation in the VAT-PAI.
In the VAT-PAI, two ORFs, numbered 26 and 27, are possibly related to virulence. ORF 26 shows no homology to known DNA sequences in GenBank, but it encodes a putative protein that has 43% homology with regulatory protein PapX (accession number P42193) or Mar (accession number P27245) (7). papX was first identified as a gene at the end of the pap operon which encoded a 17-kDa protein that was not required for assembly of P fimbriae in uropathogenic E. coli strain J96 (27). mrpJ occupies a similar location in the mrp fimbrial operon of Proteus mirabilis, and although PapX and the MrpJ protein of P. mirabilis have only 26 to 46% amino acid sequence identity in three domains, these two proteins are functional homologs (26). Both proteins acted as transcription regulators to repress flagella synthesis and motility in P. mirabilis when the fimbrial operon was turned on. The coordination of motility and fimbrial expression was shown to contribute to virulence of P. mirabilis in a mouse model of urinary tract infection (26).
ORF 27 is the largest ORF, with 4,134 nucleotides, and encodes a putative protein with 1,377 amino acids and a deduced molecular mass of 148.3 kDa. The product of this gene induces the formation of intracellular vacuoles in cell culture and shows 75% homology with the hemagglutinin Tsh of avian pathogenic E. coli (accession number AF218073) and the hemoglobin protease (Hbp) from a human pathogenic E. coli strain (accession number AJ223631), which are both members of the autotransporter family of proteins. In a previous study, we showed that a putative toxin of avian pathogenic E. coli Ec222 exhibited vacuolating cytotoxic effects on primary cultures of CEF (40). The present study shows that the gene encoding the vacuolating toxin has high homology with genes encoding members of the autotransporter protein family. The autotransporter family of proteins comprises a rapidly growing number of virulence determinants of gram-negative bacteria. In the autotransporters, an N-terminal amino acid leader sequence directs secretion via the sec pathway and is cleaved at the inner membrane by a signal peptidase. Once in the periplasm, the C-terminal portion of the protein then forms a β-barrel structure which inserts into the outer membrane to form a pore for the passage of the passenger domain to the bacterial cell surface. The passenger domain may then be released into the extracellular milieu or remain associated with the bacterial cell surface (23).
Many of its structural features indicate that Vat is a new member of the subfamily of autotransporters termed SPATEs. This hypothesis is supported by our analysis of the vat gene structure and its predicted protein. Vat shows a signal sequence in the N-terminal sequence and a signal peptidase cleavage site between residues 55 and 56 (AGA-ST). Analysis of the C terminus of Vat suggested the presence of 14 amphipathic β-strands, typical for members of the autotransporter family, and the amino acid motif YSF, which is necessary for correct localization of autotransporters to the outer membrane (25). However, members of the SPATE subfamily show the presence of a conserved serine protease motif (GDSGSPL), whereas in Vat there is a modification in the first two amino acids (ATSGSPL). It has been shown that the serine protease motif is important for phenotypic functions such as cytotoxic and proteolytic activities (20). Some authors predict that casein hydrolytic activity is dependent on the conserved serine protease active-site motif (2, 3). The difference in this motif may be the reason that Vat was negative in tests for proteolytic activity on casein-based substrate (data not shown). It is possible that related SPATE proteins may have been adapted to fulfill specific roles in pathogenesis in their host organisms since the precise role of SPATE proteins in pathogenesis has not been determined yet. Dutta et al. suggest that SPATE proteins are derived from a common ancestor and have been modified in each pathogenic species to permit adaptation to specific niches (14).
Several SPATE proteins have been described for E. coli and Shigella species. These include Tsh of avian pathogenic E. coli, with hemagglutinin activity (39); EspP of Shiga toxin-producing E. coli that produces cytopathic effects on epithelial cells (5); Pet, an enterotoxin of enteroaggregative E. coli (15); Sat of uropathogenic E. coli, which exhibits a cytopathic and vacuolating activity (20); and SepA of S. flexneri that produces cytopathic effects (3). An important role for Vat in pathogenesis may lie in its vacuolating activity. The similarities in vacuolation of cells by the avian E. coli culture supernatants and the VacA toxin from Helicobacter pylori are remarkable. Similarly, the VacA precursor contains three functional domains: a 33-amino-acid N-terminal signal sequence, the 87-kDa cytotoxin domain, and the C-terminal 50-kDa domain. Interestingly, there are no significant similarities between nucleotide sequences of vat and vacA or between protein sequences of Vat and VacA. However, among the SPATE proteins, two cysteine residues are found only in VacA (42) and Vat and are located in the passenger domain 11 and 12 amino acids apart, respectively. This characteristic may be important for the mechanism of action of these proteins.
A vacuolating cytopathic effect similar to that demonstrated for Vat and VacA has also been described for Sat, a SPATE protein of uropathogenic E. coli (20). Guyer et al. demonstrated that Sat causes vacuolation and glomerular damage in experimental urinary tract infection in mice and contributes to the pathogenesis of urinary tract infection, thereby demonstrating its important role in the pathogenicity of uropathogenic E. coli (20). Interestingly, although Vat and Sat elicit vacuolation on cell lines, a comparison of SPATE protein sequences (Fig. 3) shows that Vat is more closely related to Tsh (78%) than it is to Sat, with which it shares only 36% similarity. It would be interesting to conduct comparative studies on the mechanisms of vacuole formation by SPATE proteins.
During the revision of the manuscript, the sequence of the genome of uropathogenic E. coli CFT073 was published (48), and section 2 of the complete genome (accession no. AE016756) contained a region (bp 669932 to 78853) with 98% nucleotide sequence homology with the sequence of the VAT-PAI between bp 13217 and 22130. The region in strain CFT073 contains the intact thrW gene followed by a region homologous with papX and then a gene for a hemoglobin protease. The homology with the VAT-PAI begins with the terminal 19 bp of the CFT073 thrW gene.
It appears that the vat gene is required for virulence of E. coli strain Ec222 as its inactivation resulted in an E. coli mutant that was nonpathogenic in both the respiratory-septicemic and the cellulitis models of disease. The wild-type organism was exceptionally invasive, and lesions of pericarditis and/or perihepatitis were observed in over 50% of the chickens which received an aerosol challenge and in 92% of the chickens which received the challenge via scratched skin. We would like to have complemented the mutation and demonstrated a restoration of virulence, but the instability of the cloned vat gene, even in a low-copy-number vector, has so far prevented the creation of the complemented mutant. Nonetheless, this preliminary examination of the role of Vat in virulence suggests that it is a major virulence factor.
According to the present data and the similarity between Vat and SPATE, we predict that the Vat structure consists of a signal sequence of 5.7 kDa, the Vat cytotoxin of 111.8 kDa, and a C-terminal outer membrane translocator of 30.5 kDa, following the model of secretion described for autotransporters. We hypothesize that other closely related members of this class of autotransporters may also produce vacuolating effects.
The expression of many bacterial virulence proteins, including the autotransporters Tsh (44), EspP (5), and SepA (3), is thermoregulated. We found that Vat was detectable in the culture supernatant of E. coli Ec222 grown at 37°C but not in that of E. coli grown at 21 or 42°C (data not shown).
In conclusion, we have characterized the novel gene vat, which is part of a PAI newly identified in an avian pathogenic E. coli strain. We provide experimental evidence that vat is required for virulence of septicemic E. coli Ec222. Nucleotide sequence homologies and analysis of the protein encoded by vat indicate that it is a member of the SPATE family of autotransporter proteins. The protein encoded by vat was shown to possess a vacuolating cytotoxic activity on CEF cells. Further investigation of the VAT-PAI and Vat protein may reveal that the PAI carries additional virulence determinants and may identify the specific role of Vat in the virulence of avian E. coli.
Acknowledgments
The Poultry Industry Council, Guelph, Ontario, Canada, is gratefully acknowledged for funding this work (grant no. 115-01).
We appreciate the valuable assistance provided by S. Kariyawasam and H. Ghunaim, whose help was critical in conducting the infection studies.
Editor: V. J. DiRita
REFERENCES
- 1.Adhikari, P., G. Allison, B. Whittle, and N. K. Verma. 1999. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexneri chromosome. J. Bacteriol. 181:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hasani, K., I. R. Henderson, H. Sakellaris, K. Rajakumar, T. Grant, J. P. Nataro, R. Robins-Browne, and B. Adler. 2000. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect. Immun. 68:2457-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjelloun-Touimi, Z., M. S. Tahar, C. Montecucco, P. J. Sansonetti, and C. Parsot. 1998. SepA, the 110 kDa protein secreted by Shigella flexneri: two-domain structure and proteolytic activity. Microbiology 144:1815-1822. [DOI] [PubMed] [Google Scholar]
- 4.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 6.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover, T. L., W. Puryear, G. I. Perez-Perez, and M. J. Blaser. 1991. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect. Immun. 59:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 10.Dho-Moulin, M., J. F. van den Bosch, J. P. Girardeau, A. Bree, T. Barat, and J. P. Lafont. 1990. Surface antigens from Escherichia coli O2 and O78 strains of avian origin. Infect. Immun. 58:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djafari, S., F. Ebel, C. Deibel, S. Kramer, M. Hudel, and T. Chakraborty. 1997. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol. Microbiol. 25:771-784. [DOI] [PubMed] [Google Scholar]
- 12.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Funfstuck, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dozois, C. M., J. M. Fairbrother, J. Harel, and M. Bosse. 1992. pap- and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect. Immun. 60:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta, P. R., R. Cappello, F. Navarro-Garcia, and J. P. Nataro. 2002. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect. Immun. 70:7105-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eslava, C., F. Navarro-Garcia, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomis, S. M., C. Riddell, A. A. Potter, and B. J. Allan. 2001. Phenotypic and genotypic characterization of virulence factors of Escherichia coli isolated from broiler chickens with simultaneous occurrence of cellulitis and other colibacillosis lesions. Can. J. Vet. Res. 65:1-6. [PMC free article] [PubMed] [Google Scholar]
- 17.Gross, W. B. 1994. Diseases due to Escherichia coli in poultry, p. 237-259. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 18.Gunzer, F., U. Bohn, S. Fuchs, I. Muhldorfer, J. Hacker, S. Tzipori, and A. Donohue-Rolfe. 1998. Construction and characterization of an isogenic slt-ii deletion mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 66:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. Mobley. 2000. Identification of Sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-66. [DOI] [PubMed] [Google Scholar]
- 20.Guyer, D. M., S. Radulovic, F. E. Jones, and H. L. Mobley. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect. Immun. 70:4539-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 20:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marklund, B. I., J. M. Tennent, E. Garcia, A. Hamers, M. Baga, F. Lindberg, W. Gaastra, and S. Normark. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol. Microbiol. 6:2225-2242. [DOI] [PubMed] [Google Scholar]
- 28.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 30.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 32.Ngeleka, M., J. K. P. Kwaga, D. G. White, T. S. Whittam, C. Riddell, R. Goodhope, A. A. Potter, and B. Allan. 1996. Escherichia coli cellulitis in broiler chickens: clonal relationships among strains and analysis of virulence-associated factors of isolates from diseased birds. Infect. Immun. 64:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura, A., M. Morita, Y. Nishimura, and Y. Sugino. 1990. A rapid and highly efficient method for preparation of competent Escherichia coli cells. Nucleic Acids Res. 18:6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parreira, V. R., C. W. Arns, and T. Yano. 1998. Virulence factors of avian Escherichia coli associated with swollen head syndrome. Avian Pathol. 27:148-154. [DOI] [PubMed] [Google Scholar]
- 35.Parreira, V. R., and C. L. Gyles. 2002. Shiga toxin genes in avian Escherichia coli. Vet. Microbiol. 87:341-352. [DOI] [PubMed] [Google Scholar]
- 36.Peighambari, S. M., R. J. Julian, and C. L. Gyles. 2000. Experimental Escherichia coli respiratory infection in broilers. Avian Dis. 44:759-769. [PubMed] [Google Scholar]
- 37.Peighambari, S. M., R. J. Julian, J. P. Vaillancourt, and C. L. Gyles. 1995. Escherichia coli cellulitis: experimental infections in broiler chickens. Avian Dis. 39:125-134. [PubMed] [Google Scholar]
- 38.Pourbakhsh, S. A., M. Dho-Moulin, A. Bree, C. Desautels, B. Martineau-Doize, and J. M. Fairbrother. 1997. Localization of the in vivo expression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb. Pathog. 22:331-341. [DOI] [PubMed] [Google Scholar]
- 39.Provence, D. L., and R. Curtiss III. 1994. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salvadori, M. R., T. Yano, H. E. Carvalho, V. R. Parreira, and C. L. Gyles. 2001. Vacuolating cytotoxin produced by avian pathogenic Escherichia coli. Avian Dis. 45:43-51. [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schmitt, W., and R. Haas. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12:307-319. [DOI] [PubMed] [Google Scholar]
- 43.Sojka, W. J., and R. B. A. Carnaghan. 1961. Escherichia coli infection in poultry. Res. Vet. Sci. 2: 340-352. [Google Scholar]
- 44.Stathopoulos, C., D. L. Provence, and R. Curtiss III. 1999. Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect. Immun. 67:772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein, M., B. Kenny, M. A. Stein, and B. B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 48.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wooley, R. E., K. R. Spears, J. Brown, L. K. Nolan, and O. J. Fletcher. 1992. Relationship of complement resistance and selected virulence factors in pathogenic avian Escherichia coli. Avian Dis. 36:679-684. [PubMed] [Google Scholar]