Abstract
Replication and transcription activator (RTA) (also referred to as ORF50), an immediate-early gene product of Kaposi's sarcoma-associated herpesvirus (KSHV)/(human herpesvirus 8), plays a critical role in balancing the viral life cycle between latency and lytic replication. RTA has been shown to act as a strong transcription activator for several downstream genes of KSHV. Direct binding of RTA to DNA is thought to be one of the important mechanisms for transactivation of target genes, while indirect mechanisms are also implicated in RTA transactivation of certain selected genes. This study demonstrated direct binding of the DNA-binding domain of RTA (Rdbd) to a Kaposin (Kpsn) promoter sequence, which is highly homologous to the RTA-responsive element (RRE) of the PAN promoter. We undertook a comparative study of the RREs of PAN RNA, ORF57, vIL-6, and Kpsn to understand how RTA regulates gene expression during lytic replication. Comparing RNA abundance and transcription initiation rates of these RTA target genes in virus-infected cells suggested that the transcription initiation rate of the promoters is a major determinant of viral gene expression, rather than stability of the transcripts. RTA-mediated transactivation of reporters containing each RRE showed that their promoter strengths in a transient-transfection system were comparable to their transcription rates during reactivation. Moreover, our electrophoretic mobility shift assays of each RRE demonstrated that the highly purified Rdbd protein directly bound to the RREs. Based on these results, we conclude that direct binding of RTA to these target sequences contributes to their gene expression to various extents during the lytic life cycle of KSHV.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is an etiological agent of all clinical forms of Kaposi's sarcoma (KS), including AIDS-associated KS, classic KS, endemic forms of KS, and renal transplant-related KS (16, 20, 31, 35). KSHV is also associated with two AIDS-related lymphoproliferative diseases, primary effusion lymphoma (PEL) (2) and multicentric Castleman's disease (45). Like other herpesviruses, KSHV exhibits two distinct phases of its life cycle, latency and lytic replication. Although latent infection by KSHV has been proposed to be critical for tumorigenesis, it is clear that lytic replication is also important in KSHV pathogenesis. Most tumor cells in KS and PEL are latently infected with KSHV; however, the virus undergoes lytic replication in a small number of cells in these tumor lesions (11, 40, 46, 47, 50, 60). Upon reactivation of KSHV from latency to lytic replication, the infected cells express viral chemokines and proinflammatory cytokines, such as viral macrophage inflammatory proteins and viral interleukin-6 (vIL-6) (34, 46, 50). These lytic gene products may play a key role in disease progression in an autocrine or paracrine fashion (1, 12, 32). In addition, antibodies against lytic proteins are significantly elevated in the sera of patients developing KS (54). Treatment with ganciclovir, a drug that inhibits lytic replication of herpesviruses, significantly reduced the incidence of KS development in AIDS patients at high risk for KS (30). Therefore, it is important to study the molecular mechanisms of reactivation for a better understanding of KSHV pathogenesis.
KSHV replication and transcription activator (RTA) (also referred to as ORF50) is an immediate-early gene product encoded primarily by open reading frame 50 (ORF50), which is well conserved among all gammaherpesviruses (29, 52, 55, 56). Ectopic expression of RTA is sufficient to disrupt latency and activate lytic replication to completion in latently KSHV-infected PEL cell lines (13, 28, 49). A dominant-negative mutant of RTA with a 160-amino-acid deletion at the C terminus inhibited the ability of RTA to reactivate KSHV from latency (27). Thus, the expression of KSHV RTA is sufficient and necessary for viral reactivation, indicating a central role of RTA in the switch of the viral life cycle from latency to lytic replication. In Epstein-Barr virus (EBV) (a gamma-1 herpesvirus), ZEBRA (also referred to as BZLF1, Zta, or Z) plays a key role in disruption of viral latency in synergy with RTA (5, 6, 24, 37, 58).
The RTA homologues from gammaherpesviruses function as a transcriptional activator, and RTA autostimulates its own expression (9, 13, 38). The Oct-1, Sp1, and Sp3 cognate sites appear to be involved in RTA auto-activation (4, 42, 59). KSHV RTA also interacts with cellular factors, such as the cyclic AMP-responsive element binding protein-binding protein (CBP), histone deacetylase-1, RBP-Jκ, and MGC2663 (named KSHV RTA binding protein) (14, 23, 53). EBV RTA has been shown to interact with CBP and the retinoblastoma protein (51, 57). KSHV RTA transactivates the expression of several downstream genes, including polyadenylated nuclear (PAN) RNA, Kaposin (K12), ORF57, K-bZIP (K8, the ZEBRA homologue of KSHV), K5, K9, ORF6 (single-stranded DNA binding protein), ORF59 (DNA polymerase-associated processivity factor), TK, K14 (vOX-2), viral G-protein coupled-receptor, and viral interleukin-6 (vIL-6) (4, 8, 10, 15, 19, 27, 43, 59). EBV RTA alone can activate a subset of lytic promoters that do not require ZEBRA, and vice versa (39). In other cases, RTA activates downstream genes in synergy with ZEBRA. However, the molecular mechanism by which RTA activates target genes during lytic replication has not yet been elucidated. To date, both direct and indirect mechanisms have been suggested for RTA transactivation (7, 26, 42, 43).
We previously showed a very strong binding affinity of RTA to the RTA-responsive element (RRE) of the PAN promoter (pPAN RRE; 5′-GCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGGAAC-3′) (44). A sequence search of the KSHV genome revealed a sequence remarkably similar to that of pPAN RRE in the Kaposin promoter. This sequence of the Kaposin (Kpsn) promoter (5′-CCCGGGAAATGGGTGGCTAACCCCTACATAAGCAGTTTG-3′) contains a 25-bp sequences (shown in italics) with 16-bp and additional 5-bp matches with pPAN RRE (shown in bold type). Other research groups have shown that the RTA-responsive sequence of ORF57 (pORF57 RRE; 5′-GCAAGTGTAACAATAATGTTCCCACGGCCC-3′) confers direct binding affinity for RTA (26). A highly homologous sequence of the pORF57 RRE was also found in the K-bZIP promoters (5′-TATTTGTGAAACAATAATGATTAAAGGGG-3′) and has been shown to have a binding affinity to RTA (10, 26). However, a recent study by Liang et al. showed that a cellular transcriptional factor, RBP-Jκ, plays an important role in recruitment of RTA to the ORF57 promoter (23), suggesting a complicated mode of gene regulation by RTA. The RREs of the ORF57 and KbZIP promoters do not appear to have any significant sequence homology with those of the PAN and Kpsn promoters. Another RRE was recently identified which has been shown to regulate vIL-6 gene expression (pvIL-6RRE; 5′-GTGGTTCTAAGTCGCACGTTAGAAACCCCGCCCCCTGGTGCTCACTTT-3′) and to confer binding affinity for RTA (8). Again, there is no obvious homology in the RRE of the vIL-6 promoter to other known RREs. It will be intriguing to investigate how RTA can regu-late target gene expression during lytic replication, utilizing diverse sets of responsive sequences.
We undertook a comparative study of a group of viral early genes that are strongly activated by RTA. We measured steady-state RNA levels and transcription initiation rates in virus-infected cells, promoter activities of reporter plasmids in a transient-transfection system, and in vitro RTA-DNA binding affinities. The results from these analyses suggest that direct DNA binding of RTA make a significant contribution to the expression of these target genes during lytic replication.
MATERIALS AND METHODS
Cell culture.
All cells were cultured at 37°C in the presence of 5% CO2. 293T (a human embryonic kidney cell line transformed with the E1 region of adenovirus and the simian virus 40 [SV40] T antigen) and 293 cells were cultured in Dulbecco's modified Eagle's medium (Cellgro) containing 10% fetal bovine serum (FBS) and antibiotics (50 U of penicillin/ml and 50 μg of streptomycin/ml). The KS-1 cell line was derived from primary effusion lymphoma (PEL) patient at the University of California at Los Angeles (UCLA), which is the same type of lymphoma as the BC-3 cell line, infected with KSHV. KS-1 cells were maintained in RPMI 1640 medium supplemented with 10% FBS and antibiotics. BJAB or DG75, a human B-cell line not infected by any herpesvirus, was grown in RPMI 1640 containing 10% FBS and antibiotics.
Plasmid construction.
To make the reporter construct pE4T/Luc, the adenovirus E4 minimal TATA box region was PCR amplified from plasmid Z6E4Tluc (kindly provided by M. Carey, UCLA), using a pair of primers, E4T-F/BglII (5′-tctggaagatctgactctagaggatcc-3′) and E4T-R/HindIII (5′-atacccaagcttacaccactcgacacg-3′), and Pfuturbo DNA polymerase (Stratagene, La Jolla, Calif.). In all sequences, the underlined nucleotides represent restriction enzyme sites. The PCR product was cut with BglII and HindIII and cloned into the pGL3-basic plasmid (Promega, Madison, Wis.) which had been cut with the same enzymes. The luciferase reporter construct pE4T/RREs contains a single copy of the RREs in the promoters of PAN RNA, Kaposin (Kpsn), ORF57, and vIL-6 in a forward orientation. Each RRE sequence is shown in Fig. 6A. We used the double-stranded oligonucleotide pan1 (5′-cgggatccGCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGGAACAgatcttcg-3′) containing a region from nucleotide (nt) −78 to −37 in the PAN promoter sequence (nt 28588 to 28629 in the KSHV genome) to construct pE4T/PAN. The uppercase letters indicate viral sequences. The double-stranded oligonucleotide pan1 was cut with BamHI and BglII and cloned into pE4T/Luc, which had been cut with BglII. The double-stranded oligonucleotide Kpsn (5′-cgggatccCCCGGGAAATGGGTGGCTAACCCCTACATAAGCAGTTTGagatcttcg-3′; nt 118863 to 118825) was used to generate constructs for pE4T/Kpsn, K-ORF572 (5′-cgggatccGCAAGTGTAACAATAATGTTCCCACGGCCCagatcttcg-3′; nt 81900 to 81929) for pE4T/ORF57, and K2p25 (5′-cgggatccGTGGTTCTAAGTCGCACGTTAGAAACCCCGCCCCCTGGTGCTCACTTTagatcttcg-3′; nt 18337 to 18290) for pE4T/vIL-6. In addition, each RRE was also cloned into the pGL3 promoter plasmid (Promega) containing the SV40 promoter in a similar manner, resulting in pSV40/PAN, pSV40/Kpsn, pSV40/ORF57, and pSV40/vIL-6 in a reverse orient-ation. The copy numbers and the orientations of the inserts were confirmed by sequencing analysis.
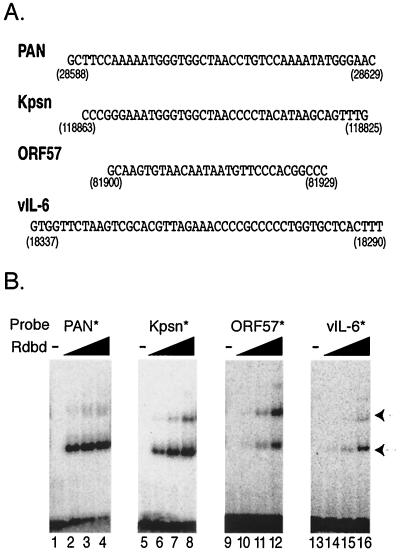
FIG. 6.
Rdbd binds with various affinities to the RREs of PAN, Kpsn, ORF57, and vIL-6 promoters. (A) Sequences of the RREs from the promoters of PAN, Kpsn, ORF57, and vIL-6 of KSHV. Numbers indicate the locations of each RRE in the KSHV genome. (B) Dose-dependent binding of Rdbd to the RREs. Each probe contained an RRE with common flanking sequences at both ends. Increasing amounts of Rdbd (0, 50, 150, and 500 ng) were incubated with end-labeled probes, PAN* (lanes 1 to 4), Kpsn* (lanes 5 to 8), ORF57* (lanes 9 to 12), and vIL-6* (lanes 13 to 16), respectively. Arrows indicate Rdbd-specific binding.
For nuclear run-on assays, the sequences of PAN RNA, Kaposin, and ORF57 were PCR amplified from BC-1 genomic DNA, using a set of primers specific for each gene. pan10/KpnI and pan2/EcoRI were used for pPAN, containing the region from nt 28659 to 29803 of the KSHV genome, KaposinF (5′-ccggaattcACGGAGGACGGATCTCTTGG-3′) and KaposinR (5′-cgcggatcctctagaGAACATGTGGCACACGCTGC-3′) for pKpsn (nt 118741 to 117454), and kORF57F (5′-gccgagaattcATGGTACAAGCAATGATAGAC-3′) and kORF57R (5′-cgcggatcctctagaGCCATGGGGTTTGGCAATCCTTAAG-3′) for pORF57 (nt 82069 to 83564). The PCR product for PAN was cloned into the pcDNA3 plasmid. A TA cloning kit (Invitrogen) was used to clone pKpsn and pORF57 in the pCRII vector provided by the manufacturer.
Slot blot analysis of RNA.
Total RNA was extracted from KS-1 cells uninduced or induced with either chemicals or RTA transfection, using TriReagent (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer's instructions. Five micrograms of RNA was denatured with 7.4% formaldehyde and 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min at 65°C and applied to nylon membranes using the slot blot apparatus. Various amounts of DNA containing the gene of interest without any vector sequences were also denatured in 0.2 N NaOH and then neutralized with an equal volume of 10 M ammonium acetate. The denatured DNA mixtures were dotted onto the same nylon membrane to serve as a standard. DNA fragments only containing a gene of interest were labeled, using a random priming method. The membrane was prehybridized and then hybridized with labeled probes, as previously described (36). The membranes were exposed to a phosphoimage screen, and quantitative analysis of signals was performed using a STORM imaging system (Molecular Dynamics, Sunnyvale, Calif.).
Nuclear run-on assays.
Nuclei from 4 × 107 KS-1 cells untreated or treated with 3 mM sodium butyrate for 18 h were isolated as previously described (25). Briefly, cells were washed twice with cold 1× phosphate-buffered saline (PBS) and resuspended in 8 ml of cold lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2). The cells were then centrifuged, resuspended in 4 ml of cold lysis buffer with 0.5% NP-40, and incubated on ice for 20 min. The lysates were passed 10 times through a 25G5/8 needle and incubated on ice for another 10 min. The nuclei pellet was resuspended in 210 μl of nuclear storage buffer (50 mM Tris-HCl [pH 8.0], 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA) and stored at −80°C. Nuclear run-on assays were performed as previously described (25). Each reaction consisted of 210 μl of nuclei and 60 μl of 5× nuclear run-on buffer (5× consisting of 25 mM Tris-HCl [pH 8.0], 12.5 mM MgCl2, 750 mM KCl, 1.25 mM ATP, GTP, and CTP). [α-32P]-UTP (30 μl; 3,000 Ci/mM) was then added, and the nuclear suspension was incubated at 30°C for 30 min, after which DNaseI (345 U; GibCO-BRL, Grand Island, N.Y.) in 10 mM CaCl2 was added. After 5 min at 30°C, the reaction was stopped with 1× SET (1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 10 mM Tris-HCl [pH 7.4]), and proteinase K was added to a concentration of 200 μg/ml. After 45 min of incubation at 37°C, the solution was extracted with an equal volume of a mixture of phenol and chloroform, and the interphase was again extracted with 100 μl of 1× SET. Ammonium acetate (10 M) was added to the combined aqueous phases (original plus reextraction) to a final concentration of 2.3 M, an equal volume of isopropanol was added, and nucleic acid was precipitated (−70°C for 15 min). The precipitate was centrifuged, and the pellet was resuspended in 100 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Unincorporated nucleotides were removed using a G-25 spin column. NaOH was added to the elute at a final concentration 0.2 N and incubated for 10 min on ice. HEPES was added to a concentration of 0.24 M, and then 2.5 vol of ethanol was added to precipitate labeled RNA overnight at −20°C. The pellet was resuspended in hybridization buffer (10 mM TES [pH 7.4], 0.2% SDS, 10 mM EDTA, 0.3 M NaCl, 1× Denhardt's solution, yeast tRNA [250 μg/ml]). Nylon membranes containing denatured plasmid DNAs were prepared using a slot blot apparatus (GibCO-BRL). Plasmids used for the templates were pPAN (a 1.1-kb fragment of the PAN RNA sequence in pcDNA3), pKpsn (a 1.3-kb fragment of the Kpsn ORF and GC-rich upstream direct repeats in pCRII), pORF57 (a 1.5-kb fragment of the ORF57 coding sequence in pCRII), pvIL-6 (a 0.6-kb fragment of vIL-6 ORF in pcDNA3.1), and pGAPDH (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]; a 0.6-kb fragment of the 3′-end of GAPDH in pCRII). After overnight prehybridization, the membranes were hybridized with the nuclear run-on products (107 cpm) for 36 h at 65°C. The radioactivity of labeled products was measured using a scintillation counter, LS1800 (Beckman Coulter, Fullerton, Calif.). After hybridization, the membranes were washed twice with 0.1% SDS and 2× SSC for 30 min and then washed twice with 0.1% SDS and 0.1× SSC for 30 min at 65°C. The membranes were exposed to a phosphoimage screen, and quantitative analysis of signals was performed using a STORM imaging system (Molecular Dynamics).
Transfections.
For the luciferase reporter assays, 2.5 × 105 293T cells were transfected with 100 ng of pcDNA3 or pcDNA3/RTA and 20 ng of a reporter plasmid into 12-well plates, using a calcium-phosphate transfection method. Each transfection for reporter assays was performed in duplicate and contained 10 ng of pRLSV40 (Promega) as a control for transfection efficiencies, as well as 270 ng of pcDNA3. At 24 h posttransfection, cells were washed with 1× PBS and subjected to reporter assays. BJAB or KS-1 cells (5 × 105) were transfected with 250 ng of pcDNA3 or pcDNA3/RTA and 50 ng of a reporter plasmid into 24-well plates, using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. In addition, each transfection contained 10 ng of pRLSV40 and 700 ng of pcDNA3. At 24 h posttransfection, cells were harvested, washed with 1× PBS, and subjected to reporter assays.
Dual luciferase assays.
The dual luciferase reporter assay system (Promega) was used to test promoter activity. Transfected 293T cells in 12-well plates were washed with 1× PBS and incubated with 250 μl of 1× passive lysis buffer provided by the manufacturer. Transfected BJAB or KS-1 cells were resuspended in 100 μl of 1× passive lysis buffer after being washed with 1× PBS. Lysates were frozen, thawed once, and centrifuged at top speed in a microcentrifuge for 5 min. Lysates were assayed using a Luminometer Lmax (Molecular Devices, Sunnyvale, Calif.). The reporter assays were carried out according to the manufacturer's protocol for the dual luciferase reporter assay system (Promega). The Renilla luciferase activities from the pRLSV40 plasmid were not affected in cotransfection of RTA in the cell lines tested, including 293, 293T, BJAB, and KS-1 cells, and were therefore, used as an internal control to normalize transfection efficiency in different transfection samples.
EMSAs.
The recombinant Rdbd protein (amino acids 1 to 320) tagged with a FLAG peptide at the N terminus and six histidine residues at the C terminus was expressed in bacteria and purified as previously described (44). A set of double-stranded oligonucleotides, including pan1 (pPAN RRE), MJmulti (a mutant version of pPAN RRE), Kpsn (pKpsn RRE), Kpsn/TG (a mutant version of pKpsn RRE), K-ORF572 (pORF57 RRE), and K2p25 (pvIL-6 RRE) were used for electrophoretic mobility shift assays (EMSAs), (see Fig. 1 and 6 for sequences). Two pairs of unrelated sequences, NS1 (5′-cgagatcggggtgaggcatgggggatcccg-3′) and NS2 (5′-cgggatccgagatcgaagtgaggcatgggggatgagatcttcg-3′), were synthesized and serve as negative controls for EMSA. All double-stranded oligonucleotides were end labeled with [γ-32P]ATP followed by fill-in reaction, and EMSAs were performed as previously described (43, 44). For competition assays, various amounts of cold competitors (indicated in Fig. 7) were incubated with the purified Rdbd protein 30 min prior to the addition of end-labeled probes. Supershift assays were performed using a monoclonal antibody (2.2 μg) against a FLAG peptide (Sigma) and polyclonal rabbit sera against RTA (1 μl). Normal rabbit sera (1 μl) as well as polyclonal rabbit sera against an irrelevant protein (1 μl) were also used as a negative control for supershift assays.
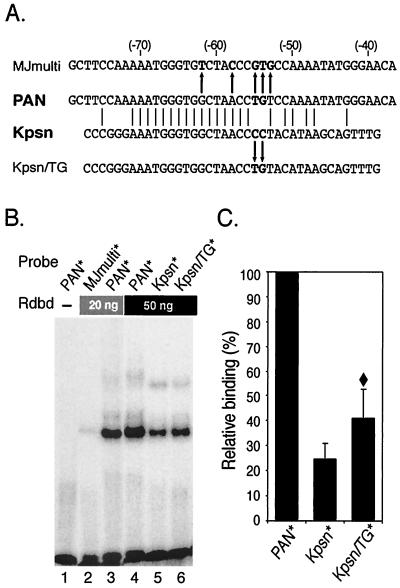
FIG. 1.
RTA DNA-binding domain protein (Rdbd) binds a Kpsn promoter sequence which is highly homologous to the RRE of the PAN promoter. (A) Comparison of the pPAN RRE and the homologous Kpsn promoter sequence. The pPAN RRE (PAN) shares significant homology to the Kpsn promoter sequence, and matched sequences are aligned. MJmulti is a mutant version of pPAN RRE with 5 nt mismatched (44), as indicated with arrows. For Kpsn/TG, a mutation of Kpsn (CC→TG) was introduced to further liken Kpsn to pPAN RRE. (B) EMSA of PAN*, MJmulti*, Kpsn*, and Kpsn/TG*. End-labeled probes were incubated with 0, 20, or 50 ng of Rdbd, as indicated. Rdbd was expressed in bacteria with a FLAG peptide at the N terminus and 6× histidine residues at the C terminus and purified using a Ni+-nitrilotriacetic acid column. (C) Quantitative analysis of Rdbd binding. Rdbd binding affinities to Kpsn* and Kpsn/TG* were calculated relative to those for PAN*. The values represent averages of relative binding affinities from three independent EMSAs, with the standard deviation shown. A diamond symbol (♦) indicates statistically significant difference in Rdbd binding between Kpsn* and Kpsn/TG* (P < 0.05, t test).
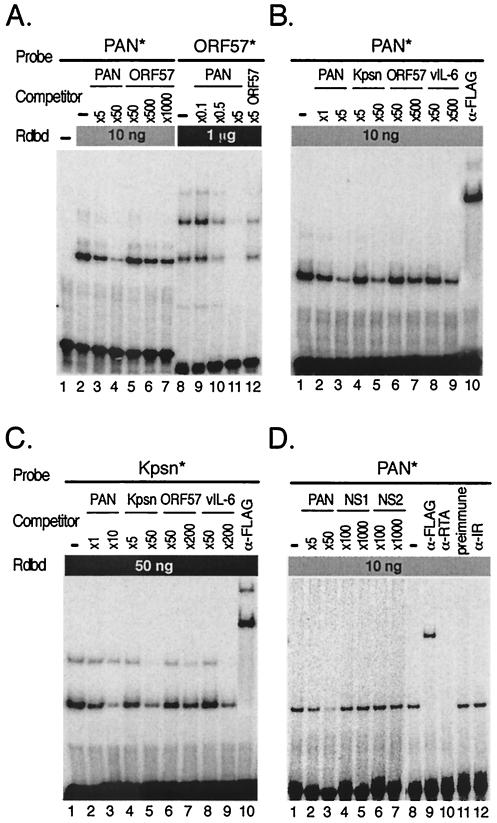
FIG. 7.
Cross-competition assays confirm relative affinities of Rdbd binding to the RREs. (A) Cross-competition assays between PAN and ORF57 RREs. Rdbd (10 ng) was incubated without (lane 2) or with excess unlabeled oligonucleotide, PAN (5- and 50-fold; lanes 3 and 4), or ORF57 (50-, 500-, and 1,000-fold; lanes 5 to 7), for competition assays prior to addition of PAN*. Indicated amounts of unlabeled oligonucleotide, PAN (0.1-, 0.5-, and 5-fold; lanes 9 to 11) or ORF57 (fivefold; lane 12), were mixed with 1 μg of Rdbd, followed by addition of ORF57*. Note that the amounts of the unlabeled oligonucleotide PAN used to compete for Rdbd binding were far less than those of the unlabeled oligonucleotide ORF57. (B) Cross-competition assays of PAN* (10 ng of Rdbd) were performed with a set of unlabeled oligonucleotides (PAN, Kpsn, ORF57, and vIL-6). As indicated, different amounts of cold competitors were used to produce similar levels of competition. Unlabeled oligonucleotides were preincubated with Rdbd, and an end-labeled probe, PAN* or Kpsn*, was added (lanes 2 to 9). A monoclonal antibody against the FLAG peptide was used to supershift protein-DNA complexes containing Rdbd (lanes 10). (C) Cross-competition assay of Kpsn* (50 ng of Rdbd) was performed as described for panel B. (D) Specificity of Rdbd binding to the RRE. The labeled PAN* probe were incubated in excess of unlabeled oligonucleotides NS1 and NS2, both of which contain nonspecific sequences, as negative controls. Coincubation of PAN* with unlabeled oligonucleotides containing its own sequence (PAN) resulted in efficient competition of the complex formation (lanes 2 and 3), while NS1 and NS2 did not show any significant competition (lanes 4 to 7). Supershift assays were performed with anti-FLAG antibody and polyclonal rabbit sera against RTA (lanes 9 and 10). Normal rabbit sera and polyclonal rabbit sera against irrelevant protein (lanes 11 and 12) were also included as negative controls.
RESULTS
Comparison of RTA binding affinities for PAN and Kaposin promoters. RTA has been shown to act as a strong transcription activator for several downstream genes of KSHV, including PAN RNA, ORF57, K-bZIP, Kaposin, vIL-6, K14, viral G-protein coupled-receptor, and thymidine kinase (8, 10, 27, 43, 59). We previously used a highly purified RTA binding domain protein (Rdbd) to demonstrate a very strong binding affinity of RTA for the RRE in the PAN promoter (pPAN RRE; 5′-GCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGGAAC-3′). Through sequence analysis of the KSHV genome, it was noted that the Kaposin promoter bears remarkable homology with the pPAN RRE sequence (Fig. 1A). The sequence of the Kaposin (Kpsn) promoter (5′-CCCGGGAAATGGGTGGCTAACCCCTACATAAGCAGTTTG-3′) contains a 25-bp homologous sequence of pPAN RRE (shown in italics) with a 16-bp consecutive sequence and 5-bp additional exact matches (shown in bold type).
We tested whether this sequence in the Kaposin promoter confers RTA binding affinity. End-labeled probes, PAN* and Kpsn* (asterisks indicate labeled oligonucleotides), were incubated with 20 or 50 ng of Rdbd and yielded a specific complex with Rdbd, suggesting that RTA binds to the Kaposin promoter (Fig. 1B, lane 5). Rdbd binding affinity for Kpsn* was fourfold lower than that for PAN* (Fig. 1C). Throughout the paper, we calculated percent binding of an RRE as a ratio of the bound probe to the total (bound plus unbound) probe using a software program (ImageQuant version 1.1) to quantitate signals from EMSA gels. Our previous detailed analysis of pPAN RRE showed that mutations at the 3′ end (nt −46 to −54) of the shared sequence with Kpsn significantly reduced Rdbd binding affinities in vitro compared to the wild type (wt). To test whether Kpsn* mimicked the pPAN RRE mutants, which have different nucleotides at the positions critical for Rdbd binding, we introduced a 2-bp mutation into Kpsn (CC→TG). Now, Kpsn/TG shares 27 bp matches, including the 19-bp consecutive sequence and 5 mismatches at nt −52, −49, and −46 to −44, of pPAN RRE (Fig. 1A). Once percent binding of each probe, PAN*, Kpsn*, or Kpsn/TG*, was calculated, we set PAN binding as 100% to show Kpsn* and Kpsn/TG* binding of RTA in a scale relative to that for PAN*. EMSA results with 32P-labeled Kpsn/TG* showed that an Rdbd binding affinity for Kpsn/TG* is higher than that for Kpsn* but still lower than that for PAN* (Fig. 1B, lanes 4 to 6, and 1C). This difference between Kpsn* and Kpsn/TG* is statistically significant (P < 0.05, t test) and consistent with the results from pPAN RRE mutagenesis studies of members of our group (44). MJmulti*, containing a 5-bp mutation in the PAN RRE (44), showed no significant Rdbd binding, indicating the importance of these nucleotides in RTA binding (Fig. 1B, lane 2). Although it contains four out of these five important nucleotides, the same as in pPAN RRE, Kpsn/TG showed approximately 40% relative binding of pPAN RRE, suggesting that other mismatches at nt −52, −49, and −46 might also contribute to reduced binding of pKpsn RRE. A single-base pair mutation at nt −52 (MJ52S: C→A) showed 60% relative binding of wt pPAN RRE. Two-base pair mutations at nt −49 and −48 (MJ49: AA→CC) or at nt −47 and −46 (MJ47: AT→CG) showed 40 to 50% of wt binding. Hence, Kpsn/TG behaves similarly to these pPAN RRE mutants in terms of reduced Rdbd binding, suggesting that the Kpsn sequence may be a derivative of a very strong RRE of the PAN promoter.
Transcript abundance and transcription initiation rates of RTA-responsive viral genes during lytic replication.
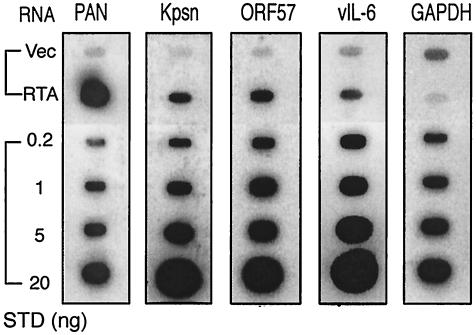
Others have reported that RTA expressed using a baculovirus expression system binds to the RREs in the ORF57 promoter (pORF57 RRE) (26). In addition, a highly homologous sequence of pORF57 RRE was noted in the KbZIP promoter and was shown to confer RTA binding activity (26). There is no apparent homology between pORF57 RRE and pPAN RRE (Fig. 6A). The RRE from the vIL-6 promoter (pvIL-6 RRE) was identified and shown to bind to bacterially expressed RTA protein (8). This sequence is unique among other known RREs, without any apparent homology (Fig. 6A). Thus, we undertook a comparative study of these RTA target genes to understand how RTA activates diverse sets of responsive elements. First, we sought to determine relative expression levels of these genes in virus-infected cells. We assessed the abundance of RTA-responsive gene transcripts in KS-1 cells during lytic replication, using slot blot analysis. Total RNAs were extracted from KS-1 and DG75 (a human B-cell line without viral infection) cells untreated or treated for 18 h with 3 mM sodium butyrate, a known strong activator of KSHV lytic replication. Total RNA (5 μg) immobilized on a membrane was hybridized with the probe specific to each sequence of the RTA-responsive genes tested. The membrane also included DNA of known amounts for each gene to serve as a standard.
Gene expression levels in KS-1 cells from the hybridized membranes were calculated and compared to the signals obtained from the standards (Fig. 2A). No cross-hybridization to RNA was detected from DG-75 cells (data not shown). There were relatively high levels of Kpsn and vIL-6 transcripts in uninduced cells. The amount of PAN RNA was estimated to be 3.9 ng out of 5 μg of total RNA from induced KS-1 cells. Assuming the portion of poly(A) RNA in total RNA in eukaryotic cells is 1 to 4% and the percentage of PEL cells undergoing lytic replication upon chemical induction is 10 to 20%, PAN RNA was calculated to constitute 10 to 80% of total polyadenylated RNA, consistent with our previous observation (48). Although expressions of other genes were somewhat comparable in induced KS-1 cells, besides PAN RNA being by far the most abundant, relative expression levels were in order of Kpsn, ORF57, and vIL-6.
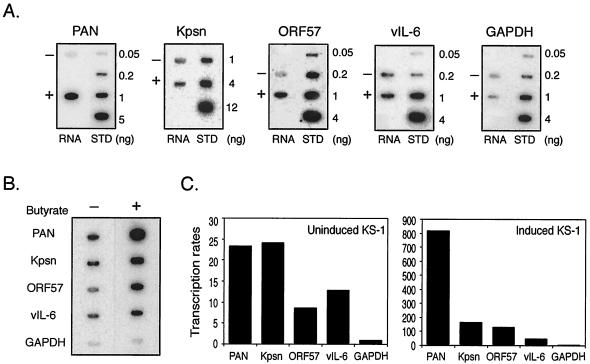
FIG. 2.
Transcript abundance and transcription rates of RTA-responsive genes in KS-1 cells during lytic replication. (A) Abundance of RTA-responsive genes during lytic replication. Total RNA was extracted from KS-1 cells untreated (−) or treated (+) with 3 mM sodium butyrate at 18 h postinduction and denatured with formaldehyde. In addition to 5 μg of RNA from KS-1 cells, different amounts of denatured DNA corresponding to the gene of interest were dotted onto nylon membranes and served as the standard (STD). The membranes were subjected to hybridization with gene-specific probes, including PAN RNA, Kpsn, ORF57, and vIL-6. GAPDH served as a control. (B) Nuclear run-on assays of KS-1 cells untreated (−) or treated (+) with 3 mM sodium butyrate. Nuclei were isolated from uninduced and induced KS-1 cells at 18 h postinduction and incubated with NTPs and [α-32P]UTP. Nylon membranes containing 5 μg of indicated plasmids, PAN, Kpsn, ORF57, vIL-6, and GAPDH (see Materials and Methods), were hybridized with nuclear run-on products. Equal amounts of run-on products (107 cpm) were added to each membrane. Representative images of the hybridized membranes are shown. (C) Quantitative analysis of nuclear run-on assays in uninduced (left panel) and induced (right panel) KS-1 cells. The intensity of a signal represents the amount of labeled nascent transcripts, which reflects the transcription initiation rate of the gene. The signal for GAPDH in uninduced KS-1 cells was set as 1, and the signals for other genes were analyzed relative to the value of GAPDH.
The expression level of a gene is determined by two factors, frequency of transcription initiation and RNA stability. Therefore, we further examined the transcription rates of the RTA-responsive viral genes, PAN RNA, Kpsn, ORF57, and vIL-6, using nuclear run-on assays.
Nuclear run-on assays provide a measure of the frequency of transcription initiation that contributes to regulated gene expression and is largely independent of RNA stability. The numbers of nascent transcripts on the gene are thought to be proportional to the numbers of paused RNA polymerase complexes on the DNA template, indicating the frequency of transcription initiation. Nuclei from KS-1 cells untreated or treated with 3 mM sodium butyrate for 18 h were isolated and incubated with ATP, CTP, GTP, and radiolabeled UTP. The majority of radiolabeled nascent transcripts from uninduced and induced KS-1 cells ran equivalent to the size of 200 to 300 nt, suggesting that the nuclear run-on reactions were efficient (data not shown). Radiolabeled newly synthesized RNAs from uninduced and induced KS-1 nuclei were hybridized with a membrane containing immobilized DNA templates of RTA-responsive viral genes, as well as GAPDH as a control. DNA templates included genomic sequences encompassing the ORF of each gene in a vector, as described in Materials and Methods, except PAN RNA and Kpsn. A template for PAN contains 1.1 kb of the PAN RNA genomic sequence, and that for Kpsn contains upstream sequences of GC direct repeats in addition to the Kpsn ORF (K12) (41). Results from the nuclear run-on assays are shown in Fig. 2B. For quantitative analysis of data, the signal for GAPDH in uninduced KS-1 cells was set as 1, and those for other genes were quantitated relative to the value of GAPDH. Compared with those from uninduced KS-1 cells, high levels of radiolabeled nascent transcripts of these genes were detected in induced KS-1 cells, which reflects the dramatic increase in transcription rates of these genes upon induction of lytic replication (Fig. 2C). Similar to results obtained from slot blot analysis, it was noted that there was limited but detectable radiolabeled viral lytic transcription from uninduced KS-1 cells (Fig. 2C, left panel). In induced KS-1 cells, the transcription initiation rates of tested viral genes were consistent with their RNA abundance, suggesting that expression of these RTA target genes is mainly regulated at the transcriptional level, rather than at the posttranscription levels (Fig. 2C, right panel).
For direct comparison of the effect of RTA on endogenous RTA-responsive gene expression, we also performed transfection of RTA in KSHV-infected cells and obtained similar results. Either pcDNA3 vector alone or pcDNA3/RTA was transfected into KS-1 cells. Total RNA was harvested at 40 h posttransfection and subjected to the slot blot analysis (Fig. 3). Upon RTA expression, viral transcripts in RTA-transfected cells were strongly induced and correlated with their transcription initiation rates. We also obtained similar results when we used a different KSHV-infected cell line, BCBL-1 (data not shown). Slot blot analysis of RNAs from KS-1 and BCBL-1 cells treated with 3 mM sodium butyrate for 40 h also showed comparable trends (data not shown). All together, these data suggest that RTA regulates its target gene expression primarily at the level of transcription of the promoters.
FIG. 3.
Expression of RTA-responsive genes in KS-1 cells transfected with RTA. Total RNA was extracted from KS-1 cells transfected with pcDNA3 vector alone (Vec) or with pcDNA3/RTA (RTA) at 40 h posttransfection and denatured with formaldehyde. In addition to 5 μg of RNA from transfected KS-1 cells, different amounts of denatured DNA corresponding to the gene of interest were dotted onto nylon membranes and served as the standard (STD). The membranes were subjected to hybridization with gene-specific probes, as described in the legend to FIG. 2.
The transcription activities of the RRE promoters mediated by RTA in reporter assays.
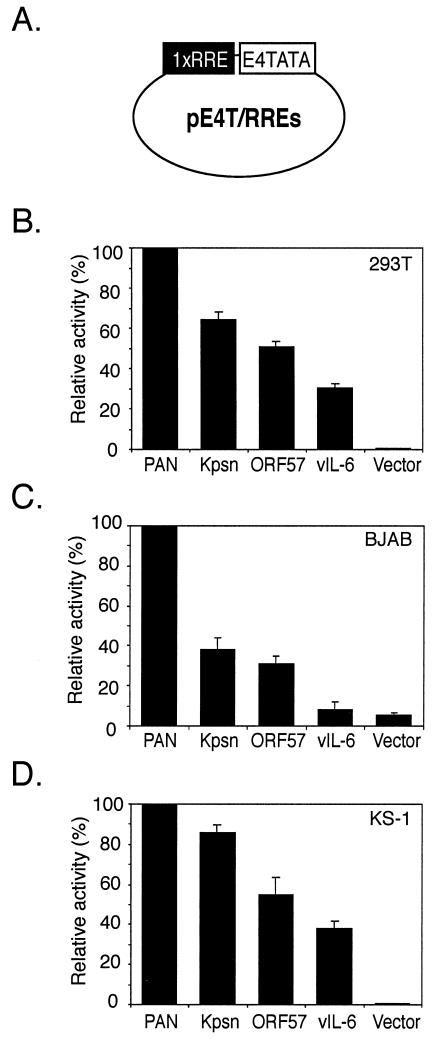
The RREs have been identified for all four promoters and shown to be critical for promoter activation by RTA (8, 26, 43, 44). We investigated whether promoter strengths of RTA-responsive genes would be a key factor for their gene expression. Since the distances between a TATA box and an RRE and putative binding sites for cellular proteins varied in their own promoter contexts, we tested the strength of the RREs in a uniform background. All the RRE sequences are listed in Fig. 6A, with numbers indicating genomic locations. These RREs were fused to a heterologous promoter, using a luciferase reporter construct, pE4T/Luc, containing the adenovirus E4 TATA box. A single copy of each RRE was cloned into the same locus of the reporter plasmid, pE4T/Luc, and termed pE4T/RREs (Fig. 4A). Each reporter construct (20 ng) was transfected into 293T cells with 100 ng of a vector, pcDNA3, or an RTA expression plasmid, pcDNA3/RTA. Plasmid pRLSV40 (10 ng), containing the coding sequence for Renilla luciferase under the control of a constitutively active SV40 enhancer/promoter, was included in each transfection and served as an internal control for transfection efficiency. At 24 h posttransfection, cell lysates were assayed for both firefly and Renilla luciferase activity. Firefly luciferase activity was normalized with the corresponding Renilla luciferase activity in each transfection. The normalized levels of luciferase activity of pcDNA3-transfected cells were subtracted from that of pcDNA3/RTA-transfected cells and then used as levels of the promoter activity. pE4T/PAN manifested the strongest promoter activity among the reporter constructs tested. Its fold activation by RTA was up to 3,300-fold, equivalent to results using our previous reporter construct containing a minimal PAN promoter, pLUC (−69) (43). The relative promoter activity was calculated as a ratio of the promoter activity of reporter constructs to that of pE4T/PAN (Fig. 4B). The transfections were performed at a suboptimal condition for RTA to avoid saturating the promoters. The promoter activities of the pE4T/RRE reporters in the presence of RTA had an order similar to that of transcription rates during reactivation of KSHV-infected cells.
FIG. 4.
Promoter strengths of reporters containing each RRE under a heterologous promoter, the E4 TATA box. (A) Schematic representation of a reporter construct. One copy of each RRE was cloned into a reporter construct containing the adenovirus E4 minimal TATA box. Reporter constructs were tested in 293T (B), BJAB (C), and KS-1 (D) cells, using a transient-transfection system. At 24 h posttransfection, transfected cells were harvested and subjected to dual luciferase assays. Promoter activities were calculated from levels of luciferase activity of reporters transfected with pcDNA3 or pcDNA3/RTA, in addition to a control vector, pRLSV40, levels of which constitutively expresses Renilla luciferase. Relative to that of pE4T/PAN, the promoter activities of the other pE4T/RREs were calculated. The values represent averages of at least three independent transfections, with the standard deviation shown.
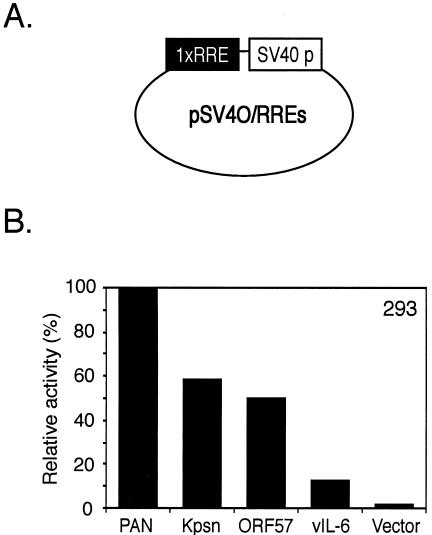
We then explored the possibility that B-cell-specific or viral factors are involved in activating these elements by testing the reporter constructs in BJAB, a human B-cell line with no viral infection, and in KS-1, a PEL cell line latently infected with KSHV. Reporter constructs (50 ng) were transfected into BJAB cells in the absence or presence of pcDNA3/RTA (250 ng). The ratio between the amount of reporter constructs and that of activator transfected into BJAB cells was the same as in 293T cells. pRLSV40 (10 ng) was cotransfected to serve as an internal control. KS-1 cells were also transfected in the same condition as BJAB cells. Promoter activity of pKpsn RRE in comparison of that of pPAN RRE was less strong in BJAB cells than in 293T or KS-1 cells (Fig. 3C). Differences among promoter activities of other RREs were less prominent in KS-1 cells than in 293T cells, suggesting that there might be other factors involved in activation of RREs from the Kpsn, ORF57, and vIL-6 promoters (Fig. 4D). However, the relative promoter strengths of pE4T/RREs in BJAB and KS-1 cells were similar to those in 293T cells, indicating that RTA is the main contributing factor to the promoter activity. We compared relative promoter strengths in a different promoter context by cloning a copy of each RRE into another heterologous promoter, such as SV40 promoter (Fig. 5A). Results from transient transfections in 293 cells were consistent with previous results showing RRE promoter strengths, although levels of the promoter activity were lower in 293 cells than in 293T cells, mainly due to high basal activity of the reporters (Fig. 5B). After all, these results suggest that the RREs may be a primary contributor in regulating gene expression, regardless of B-cell-specific or viral factors and types of heterologous promoters.
FIG. 5.
Promoter strengths of reporters containing each RRE under another heterologous promoter, the SV40 promoter. (A) Schematic representation of a reporter construct. One copy of each RRE was cloned into a reporter construct containing the SV40 promoter. (B) Reporter constructs were transiently transfected into 293 cells, and levels of luciferase activity were assayed as described in the legend to FIG. 4.
RTA binds viral RREs with various affinities.
Although each of four RREs has been shown to confer RTA binding using different protein expression systems by different groups, a recent study showing that RTA might be recruited by a cellular transcriptional factor, RBP-Jκ, to the ORF57 promoter, suggested a complicated mode of gene regulation by RTA. In addition, results from the mutagenesis study of pORF57 RRE indicated that there might be a sequence requirement for this promoter activation by RTA, other than a simple RBP-Jκ binding site (23, 26), thus serving as a composite site. Since both direct and indirect mechanisms were implicated in RTA transactivation of a target gene, the significance of DNA direct binding of RTA has yet to be revisited in terms of regulation of RTA-responsive gene expression. Therefore, we set out to test whether RTA can bind to these diverse sequences under the same conditions.
To compare relative binding affinities of RTA to these viral RREs, increasing amounts of Rdbd (0, 50, 150, and 500 ng) were incubated with the labeled oligonucleotides, including PAN*, Kpsn*, ORF57*, and vIL-6*. These labeled probes bound to the Rdbd protein in a dose-dependent manner, but the binding affinities for Rdbd varied, depending on the RRE sequence (Fig. 6B). For example, PAN* yielded nearly saturated binding to 150 ng of Rdbd (Fig. 6B, lane 3), while vIL-6* showed only limited binding to 500 ng of Rdbd (lane 16). The arrows indicated RTA-containing complexes, all of which were abolished by anti-RTA antibody and further supershifted by anti-FLAG antibody (data not shown). Distinct conformation of Rdbd proteins that migrated with slightly different mobility in the complexes (e.g., isomers) or different stoichiometry of the RTA protein in the complexes might account for the presence of these multiple complexes (8). Based on these results, the strength of Rdbd binding affinities is in the following order: PAN*, Kpsn*, ORF57*, and vIL-6*.
Relative Rdbd binding affinities were further confirmed by cross-competition assays (Fig. 7). To make direct comparisons of Rdbd binding between the two homologous groups of RREs, the labeled oligonucleotides, PAN* and ORF57*, were incubated with 10 ng or 1 μg of Rdbd, respectively, in the absence or presence of cold competitors. After several preliminary tests, various preselected amounts of the unlabeled oligonucleotides, PAN and ORF57, were used to compete with PAN* binding to Rdbd and competed efficiently (Fig. 7A). It was noted that the amounts of the unlabeled oligonucleotide, ORF57, that were required to cross-compete PAN* binding at similar levels were significantly higher than those of the unlabeled oligonucleotide, PAN, consistent with the previous observation that PAN has a higher binding affinity to Rdbd than ORF57. Binding of Rdbd to the labeled probe, ORF57*, was also competed more efficiently by the unlabeled PAN oligonucleotide than by the unlabeled oligonucleotides containing ORF57. These results confirmed that Rdbd has a higher binding affinity for pPAN RRE than for pORF57 RRE.
More comprehensive comparisons of relative Rdbd binding were made using PAN* with a panel of cold competitors corresponding to each RRE. A set of unlabeled oligonucleotides was incubated with PAN* in the presence of 10 ng of Rdbd (Fig. 7B). The amounts of cold competitors required to reduce complex formation at similar levels varied and correlated with the relative strengths of Rdbd binding affinities to these RREs. The weaker the binding affinity, the more a competitor was required. We also determined relative binding affinities, using the labeled oligonucleotides with weaker binding affinities than PAN*, to test whether the strength of the RRE used as a probe would influence relative cross-competitions with other RREs. Competition assays for the labeled oligonucleotide, Kpsn*, in the presence of 50 ng of Rdbd were performed as described for Fig. 7B. The competition data for Kpsn* correlated with the results from competition assays using PAN* to determine relative binding affinities (Fig. 7C). The specificities of the complexes were also shown by supershift analysis with anti-FLAG antibody (Fig. 7B and C, lane 10). Likewise, the Rdbd-DNA complexes with other RREs, including ORF57* and vIL-6*, were supershifted by anti-FLAG antibody, indicating the specificity of these complexes (data not shown). An unlabeled oligonucleotide, vIL-6, when used in large amounts, appeared to compete more efficiently for Rdbd binding to PAN* and Kpsn* than ORF57 (Fig. 7B and C, lanes 7 and 9). However, our own previous study showed that Rdbd binding affinity of pORF57 RRE was comparable to that of pvIL-6 RRE overall as determined by cross-competition assays against each other (8). Binding of Rdbd to these elements was sequence specific, since excess of nonspecific competitors (NS1 and NS2) with unrelated sequences failed to compete this binding (Fig. 7D, lanes 4 to 7). In addition, polyclonal rabbit sera against RTA inhibited formation of the protein-DNA complex while anti-FLAG antibody supershifted the complex, indicating the specificity of the complex (Fig. 7D, lanes 9 and 10) (37). As negative controls for supershift assays, normal rabbit sera and polyclonal rabbit sera against an irrelevant protein were used and neither altered nor supershifted the formation of the complex (Fig. 7D, lanes 11 and 12). Taken together, these results indicate that RTA binds to the RREs in a sequence-specific manner with various binding affinities, which might contribute to transactivation of RTA target genes.
DISCUSSION
In our efforts to understand how RTA, a single viral transactivator, concomitantly activates many target genes to various expression levels during lytic replication, we compared four RTA-responsive viral early genes containing RREs in their promoter regions. We undertook several independent approaches to measure steady-state RNA levels, transcription rates, and promoter strengths to examine how RTA regulates target gene expressions. We found that the strength of RREs in the promoters may be one of the major contributors in expression of the four target genes tested during lytic replication. Our data also suggest that DNA direct binding of RTA to the RREs with different affinities at least partially account for their various levels of gene expression. This leads us to hypothesize that varying binding affinities serves as one of the mechanisms for the virus to express multiple genes at various levels, using a single transcription activator. This is intriguing, given the difference in the molecular switch system of reactivation between EBV (gamma1) and KSHV (gamma2); while KSHV seems to make a main use of RTA, another human gamma herpesvirus, EBV utilizes two viral transactivators, ZEBRA and RTA, in regulation of viral gene expression. Delineating a mechanism underlying RTA regulation of KSHV viral genes may highlight similarities and differences in viral gene regulation of two similar herpesviruses.
The roles of cellular factors involved in regulating RTA-target gene expression are not fully understood at present. Our results indicating that relative strengths among the RTA-responsive promoters varied, depending on the assays used, suggest that in conjunction with RTA, there are likely to be cellular or other viral factors mediating the activation of these viral genes in vivo. Since each assay is designed to examine a different aspect of gene regulation, it would not be surprising to find variations among these assays. For instance, we detected dramatic differences among the relative affinities of Rdbd binding to the pPAN RRE and other known RREs. To achieve a similar level of competition, 10- to 100-fold more of the unlabeled oligonucleotide ORF57 than the unlabeled oligonucleotide PAN was required (Fig. 7). Given that the Kd of Rdbd for the pPAN RRE was determined to be approximately 8 × 10−9 M (44), the Kd of Rdbd for other RREs are in a 10−8 to 10−6 M range, which is considered to be a reasonable binding affinity for a transcription activator. However, we considered these results to determine relative affinities of Rdbd binding in a qualitative fashion rather than in a quantitative one, because cold competitors may not compete in a linear manner.
In contrast, in terms of promoter strengths, differences in reporter analysis results between pPAN RRE and pORF57 RRE were less than twofold, which is much less than that by EMSAs (Fig. 4B and D), suggesting that not only RTA binding but also other cellular or viral factors may play a role in these gene expressions regulated by RTA. For example, a recent study showed that an RTA-interacting cellular protein, RBP-Jκ, recognizes a part of pORF57 RRE and plays a critical role in RTA transactivation of pORF57, while RBP-Jκ does not seem to work on pPAN RRE in the same manner (23). As a viral protein, K-bZIP, the positional homologue of EBV/ZEBRA, is shown to repress RTA transactivation of the ORF57 promoter through RTA-KbZIP protein-protein interactions but not of the PAN promoter (17). In addition to promoter-specific factors, a cellular or viral factor common to target genes may play an important role in RTA transactivation. The defined RREs tested in this study were 30 to 50 bp in length, which could accommodate a binding site of other factors in conjunction with RTA, although interaction of RTA with other factors that do not directly bind to DNA is also a possible mechanism. Alternatively, a cellular protein might facilitate recruitment of RTA and other coactivators to its responsive regions without direct interaction with RTA (33). Recently we have found that a non-histone DNA architectural protein, HMG-1, can further increase KSHV RTA binding to all of these target sites (M. J. Song, S. Hwang, W. Wang, J. Round, B. Wang, R. C. Johnson, M. Carey, and R. Sun, unpublished results). Importantly, enhancement of RTA binding by HMG-1 was greater to low-affinity binding sites than to high-affinity ones, which, at least in part, would account for smaller differences seen in reporter assays in vivo than in binding assays in vitro.
While binding assays and reporter analyses utilized artificial systems, nuclear run-on assays were performed in vivo in a natural genomic context. Transcription rates of PAN and ORF57 showed more than a sixfold difference in nuclear run-on assays (Fig. 2). The distances between each RRE and the TATA box in its own promoter, the efficiency of promoter demethylation, and the local concentrations of activators during lytic replication could account for larger differences in this assay than in reporter analyses. The Kpsn locus was transcriptionally more active than the ORF57 locus during lytic replication, but fold activation in the transcription rate of ORF57 was higher than that in Kpsn compared with their expressions during latency (Fig. 2). This might be due to a complicated transcriptional regulation pattern of the Kpsn locus, depending on the viral life cycle, as discussed later. Nevertheless, despite the quantitative variations, the results showing the relative strengths of the promoters, as determined from all assays, are consistent and strongly support our hypothesis that DNA binding of Rdbd makes significant contributions to expression levels of PAN RNA, Kspn, ORF57, and vIL-6.
We used slot blot analysis to quantitate steady-state RNA levels. Compared to nuclear run-on assays, this assay enabled us to determine whether any posttranscriptional regulation may contribute to RNA abundance. Since the ORF57 product was shown to enhance PAN RNA expression as a posttranscriptional activator, the expression level of PAN RNA could be further increased (21). Members of our group and others previously performed Northern analysis, demonstrating highly specific hybridization patterns for all the genes tested (21, 22, 41, 43, 49, 50). No splicing event was found to be involved in expression of PAN RNA and vIL-6 (8, 48). Northern analysis with an ORF57-specific probe and a Kpsn-specific probe gave rise to a single size of the transcript, although ORF57 and Kspn appear to undergo splicing in induced PEL cells (21, 22); therefore, it would not affect our interpretation of the results. Thus far, relative expression levels of these genes have not been well addressed. Although global analyses of transcription in KSHV-infected cell lines using an array technique have provided excellent insights on expression kinetics of each gene over time (18, 36), comparison of relative abundance among different genes would be difficult, possibly due to various hybridization efficiencies of different genes. For example, depending on detection methods employed to determine gene expression levels, i.e., array versus Northern analysis, relative abundance of one gene seems to peak at different time points, suggesting technical differences in different methods (36). Therefore, our study of four different genes set out to compare relative abundance of RTA-responsive genes using various amounts of DNA standard for each gene.
Results from both slot blot analysis and nuclear run-on assays showed high levels of Kpsn and vIL-6 expression in uninduced KS-1 cells (Fig. 2). This may reflect a low level of lytic replication in a small subset of KS-1 cells, resulting from spontaneous reactivation. However, relative to PAN and ORF57 in induced KS-1 cells, we observed high transcription rates of Kpsn and vIL-6 in uninduced KS-1 cells, consistent with our slot blot analysis data, suggesting that there might be ongoing transcription of Kpsn and vIL-6 during latency (Fig. 2B). Kpsn was first identified as the most abundant latent transcript in KS tumor samples, but transcription at this locus was further activated during lytic replication (60). Kpsn was also expressed in some of the PEL cell lines in a similar pattern (41). However, it is not well understood whether Kpsn utilizes the same transcription unit during both latency and lytic replication. Our study is mainly focused on the relevant Kpsn transcription unit proximal to Kpsn ORF that is responsive to RTA (28). Interestingly, a recent study reported an unusual spliced Kpsn transcript in PEL tumor and identified a novel Kpsn promoter (nt 124,242 to 123,842), located farther upstream of the known Kpsn transcription unit (22). A reporter analysis of the novel Kpsn promoter (−400 + 317Luc) showed a constitutive promoter activity ∼150-fold over that of the vector alone in a B-cell line. The result suggests that this region might be active during latency, since its activation does not require any other stimulation, such as by 12-O-tetradecanoylphorbol 13-acetate or RTA. Overall, the kpsn gene locus shows very complicated patterns of transcription as well as translation, depending on tumor types and cell lines tested (22, 41), and remains to be further elucidated. Constitutive expression of vIL-6 in PEL cells was reported in both primary tumor samples and cell lines, and its expression increased during lytic replication (8, 46, 50). Thus, these observations are consistent with our interpretation that Kpsn and vIL-6 might be latently transcribed.
In conjunction with the fact that PAN RNA is the most abundant lytic transcript of KSHV, the transcription initiation rate of PAN RNA was the highest of four viral genes tested during lytic replication, suggesting that very strong promoter activity may be the main contributor to the abundance of PAN RNA. This is also consistent with interpretation of our previous Northern analysis result showing that the luciferase transcript, driven by the minimal PAN promoter, was expressed at a level comparable to that of PAN RNA driven by the same size of PAN promoter in a transient-transfection system (43). In addition, the relative abundance of Kpsn, ORF57, and vIL-6 gene transcripts ranks similarly to their relative transcription rates. Therefore, the transcription initiation rates, rather than RNA stability, are likely to be a key factor in determining the transcript abundance of these RTA-responsive genes.
Although the RTA-binding sequences tested here are quite diverse, pPAN RRE is strikingly homologous to pKpsn RRE. With variances in the 5′ and 3′ flanking sequences of these two RREs, the PAN and Kpsn genes showed the highest RTA-binding affinities, promoter strengths, abundance, and transcription rates. Given that PAN RNA and Kpsn genes are unique in KSHV among gammaherpesviruses, it is interesting that the virus utilizes similar regulatory elements to efficiently control expression of these two unique genes during lytic replication. An independent study also reported importance of the conserved regulatory sequence between pRAN RRE and pKpsn RRE (3). Combined deletion of 5′ and 3′ flanking sequences in pKpsn RRE failed to show RTA binding and reduced the promoter activity, suggesting that these flanking sequences also contribute to RTA binding as well as transactivation of pKpsn RRE.
In conclusion, our comparative study of RTA-responsive gene expression revealed that direct binding of RTA to the RREs makes a significant contribution to activation of the promoters and gene expression during lytic replication. These results led us to hypothesize that when the virus undergoes lytic replication, RTA, a single viral transactivator, activates the different levels of viral gene expression by modulating its binding affinities to the diverse responsive elements. Although viral gene expression is regulated in much more complicated ways than just RTA direct binding alone, our results affirmatively suggest that modulating DNA binding affinity of RTA to various target sites is at least one of the mechanisms by which KSHV RTA regulates viral gene expression during lytic replication.
Acknowledgments
We thank W. Aft for editing the manuscript.
This work was supported by NIH grants CA83525, CA91791, and DE14153, the Stop Cancer Foundation (R.S.), and the California Cancer Research Committee. H.D. is a Lymphoma Research Foundation Fellow, and M.J.S. was supported by a UCLA Chancellor's Dissertation Year Fellowship.
REFERENCES
- 1.Cannon, J. S., J. Nicholas, J. M. Orenstein, R. B. Mann, P. G. Murray, P. J. Browning, J. A. DiGiuseppe, E. Cesarman, G. S. Hayward, and R. F. Ambinder. 1999. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J. Infect. Dis. 180:824-828. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darr, C. D., A. Mauser, and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 10.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 11.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensoli, B., G. Barillari, and R. C. Gallo. 1992. Cytokines and growth factors in the pathogenesis of AIDS-associated Kaposi's sarcoma. Immunol. Rev. 127:147-155. [DOI] [PubMed] [Google Scholar]
- 13.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, Y. Q., J. J. Li, M. H. Kaplan, B. Poiesz, E. Katabira, W. C. Zhang, D. Feiner, and A. E. Friedman-Kien. 1995. Human herpesvirus-like nucleic acid in various forms of Kaposi's sarcoma. Lancet 345:759-761. [DOI] [PubMed] [Google Scholar]
- 17.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedda, M. A., L. Margolius, M. C. Kew, C. Swanepoel, and D. Pearson. 1996. Kaposi's sarcoma-associated herpesvirus in Kaposi's sarcoma occurring in immunosuppressed renal transplant recipients. Clin. Transplant. 10:429-431. [PubMed] [Google Scholar]
- 21.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H., T. Komatsu, B. J. Dezube, and K. M. Kaye. 2002. The Kaposi's sarcoma-associated herpesvirus K12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J. Virol. 76:11880-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linial, M., N. Gunderson, and M. Groudine. 1985. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science 230:1126-1132. [DOI] [PubMed] [Google Scholar]
- 26.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 29.Manet, E., H. Gruffat, B. M. C. Trescol, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, D. F., J. P. Dunn, J. L. Davis, J. S. Duker, R. E. Engstrom, Jr., D. N. Friedberg, G. J. Jaffe, B. D. Kuppermann, M. A. Polis, R. J. Whitley, R. A. Wolitz, and C. A. Benson. 1999. Use of the ganciclovir implant for the treatment of cytomegalovirus retinitis in the era of potent antiretroviral therapy: recommendations of the International AIDS Society-USA panel. Am. J. Ophthalmol. 127:329-339. [DOI] [PubMed] [Google Scholar]
- 31.Memar, O. M., P. L. Rady, and S. K. Tyring. 1995. Human herpesvirus-8: detection of novel herpesvirus-like DNA sequences in Kaposi's sarcoma and other lesions. J. Mol. Med. 73:603-609. [DOI] [PubMed] [Google Scholar]
- 32.Miles, S. A., O. Martinez-Maza, A. Rezai, L. Magpantay, T. Kishimoto, S. Nakamura, S. F. Radka, and P. S. Linsley. 1992. Oncostatin M as a potent mitogen for AIDS-Kaposi's sarcoma-derived cells. Science 255:1432-1434. [DOI] [PubMed] [Google Scholar]
- 33.Mitsouras, K., B. Wong, C. Arayata, R. C. Johnson, and M. Carey. 2002. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol. 22:4390-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 35.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 36.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragoczy, T., and G. Miller. 1999. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, J. A., R. G. Nador, D. Spaulding, Y. Tani, E. Cesarman, and D. M. Knowles. 1998. Demonstration of Kaposi's sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposis sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood 91:3825-3832. [PubMed] [Google Scholar]
- 41.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by RTA in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 46.Staskus, K. A., R. Sun, G. Miller, P. Racz, A. Jaslowski, C. Metroka, H. Brett-Smith, and A. T. Haase. 1999. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J. Virol. 73:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swenson, J. J., E. Holley-Guthrie, and S. C. Kenney. 2001. Epstein-Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J. Virol. 75:6228-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Santen, V. L. 1993. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J. Virol. 67:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 55.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zacny, V. L., J. Wilson, and J. S. Pagano. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J. Virol. 72:8043-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 60.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]