Abstract
The herpes simplex virus type 1 (HSV-1) Us11 gene encodes a multifunctional double-stranded RNA (dsRNA)-binding protein that is expressed late in infection and packaged into the tegument layer of the virus particle. As a tegument component, Us11 associates with nascent capsids after its synthesis late in the infectious cycle and is delivered into newly infected cells at times prior to the expression of viral genes. Us11 is also an abundant late protein that regulates translation through its association with host components and contains overlapping nucleolar retention and nuclear export signals, allowing its accumulation in both nucleoli and the cytosol. Thus, at various times during the viral life cycle and in different intracellular compartments, Us11 has the potential to execute discrete tasks. The analysis of these functions, however, is complicated by the fact that Us11 is not essential for viral replication in cultured cells. To discover new host targets for the Us11 protein, we searched for cellular proteins that interact with Us11 and have identified PAT1 as a Us11-binding protein according to multiple, independent experimental criteria. PAT1 binds microtubules, participates in amyloid precursor protein trafficking, and has homology to the kinesin light chain (KLC) in its carboxyl terminus. The carboxyl-terminal dsRNA-binding domain of Us11, which also contains the nucleolar retention and nuclear export signals, binds PAT1, whereas 149 residues derived from the KLC homology region of PAT1 are important for binding to Us11. Both PAT1 and Us11 colocalize within a perinuclear area in transiently transfected and HSV-1-infected cells. The 149 amino acids derived from the KLC homology region are required for colocalization of the two polypeptides. Furthermore, although PAT1 normally accumulates in the nuclear compartment, Us11 expression results in the exclusion of PAT1 from the nucleus and its accumulation in the perinuclear space. Similarly, Us11 does not accumulate in the nucleoli of infected cells that overexpress PAT1. These results establish that Us11 and PAT1 can associate, resulting in an altered subcellular distribution of both polypeptides. The association between PAT1, a cellular trafficking protein with homology to KLC, and Us11, along with a recent report demonstrating an interaction between Us11 and the ubiquitous kinesin heavy chain (R. J. Diefenbach et al., J. Virol. 76:3282-3291, 2002), suggests that these associations may be important for the intracellular movement of viral components.
Juxtaposed between the nucleocapsid and the envelope lies the tegument, an electron-dense, structured region within herpesvirus particles (reviewed in references 26 and 36). Included among the RNA molecules (4, 42) and at least 15 polypeptides that populate this zone in HSV-1 virions is the product of the Us11 gene. As a tegument component, Us11 is delivered into the cytosol after fusion of the viral envelope with the host cell plasma membrane (39). Once inside the host cell, viral tegument components are free, in certain cases, to perform a diverse array of tasks prior to the onset of viral gene expression. Some tegument components associate with strategic host proteins, thereby modifying the cellular environment such that it supports more efficient viral replication. For example, to facilitate the loading of cellular ribosomes onto viral mRNAs, the HSV-1 Vhs nuclease selectively destabilizes mRNAs through an association with the translation initiation factor eIF4H (14, 15). Similarly, VP16, a potent transcriptional transactivator, assembles into a complex, along with the cellular transcription factors HCF-1 and Oct-1, to stimulate transcription from viral immediate-early promoters (48). Although introduced into the cytoplasm extremely early in the infectious cycle, Us11, like many tegument proteins, is synthesized late in the viral life cycle. In addition, since Us11 accumulates both in nucleoli and in the cytosol, it has the potential to perform discrete actions at different times in various cellular compartments (5, 40). Deciphering the functions executed by Us11 has proved difficult, since genetic analysis has revealed that the Us11 gene product is not required for productive viral growth in cultured cells (19, 25).
A role for Us11 in the regulation of viral protein synthesis emerged from the genetic analysis of HSV-1 γ34.5 mutants. In a variety of cultured cell lines, γ34.5 mutants grow poorly due to the premature cessation of translation prior to the completion of the viral life cycle (10). In the absence of the γ34.5 gene product, the activated cellular PKR kinase phosphorylates the translation initiation factor eIF2α, inhibiting the initiation of protein synthesis (9). Extragenic suppressor mutants selected for their restored ability to grow and synthesize proteins all overexpressed Us11 as an immediate-early protein (27, 29). Further analysis of Us11 mutants revealed that Us11 is also required for maximal rates of translation when expressed in its natural context late in infection (M. Mulvey and I. Mohr, unpublished data). Inhibition of PKR activation by the carboxyl-terminal 68 residues of Us11 is responsible for the observed effects on protein synthesis (34). This same segment of Us11 binds double-stranded RNA (dsRNA) through a novel RNA-binding motif (23), interacts with PKR and the PKR activator PACT (6, 32, 35), associates with 60S ribosomes and polysomes (40), contains an overlapping nucleolar retention and nuclear export signal that allows both nucleolar and cytoplasmic accumulation (5, 40), and can potentially bind to a variety of structured cellular and viral RNAs found in infected cells (1, 23, 37, 38). Although both the amino- and carboxyl-terminal portions of Us11 can be packaged into virions, the amino-terminal 40 amino acids have been reported to possess an activity similar to that of transactivator proteins encoded by complex retroviruses (11, 41). It is entirely possible that the catalogue enumerated above, although extensive, does not represent a comprehensive picture of Us11's activities. Our understanding of the role(s) played by Us11 molecules contained within the tegument, both extremely early and late in the infectious program, is also far from complete. Indeed, Us11 protein packaged in the tegument and subsequently discharged into the cytoplasm of a newly infected host cell is not sufficient to overcome the block to protein synthesis observed in cells infected with a γ34.5 mutant virus, even at very large multiplicities of infection (10). Thus, although ca. 600 to 1,000 molecules of Us11 per virion enter the cytosol, some of which associate with polyribosomes and enter the nucleolus (39), this population of Us11 molecules might engage in other actions that have yet to be documented.
To explore the possibility that Us11 might indeed contain other intrinsic activities, we searched for cellular proteins that associate with the Us11 polypeptide, since the interaction between viral and host proteins is important for many events in the viral life cycle. Here, we identify the cellular PAT1 polypeptide as a Us11 binding-protein by using multiple, independent assays. PAT1 binds microtubules, is involved in the intracellular trafficking of amyloid precursor protein (APP), and contains a region homologous to kinesin light chain (KLC) (49). This interaction requires the carboxyl-terminal dsRNA-binding domain of Us11 and a region of PAT1 that contains homology to the kinesin light chain. In addition, PAT1 and Us11 colocalize when ectopically expressed in transiently transfected cells and in HSV-1-infected cells. The association between a viral tegument protein and a cellular protein with the potential to interact with a molecular motor may be important for the intracellular movement of viral components.
MATERIALS AND METHODS
Cell culture.
293T and MDCK cells were propogated in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. Cos-1 cells were grown in identical media except 10% calf serum was used in place of FBS. The inducible MDCK585 and MDCK411 lines were grown in minimal essential medium supplemented with 1× minimal essential medium nonessential amino acids, 10% FBS, 2 mM l-glutamine, 3 μg of blasticidin/ml, 0.3 mg of zeocin/ml, 50 U of penicillin/ml, and 50 μg of streptomycin/ml as described previously (16). To induce the production of PAT1 and PAT1Δ412, 1 μg of tetracycline (Invitrogen)/ml and 2 mM sodium butyrate were added to the culture media for 18 h.
Construction of recombinant HSV-1 expressing a GFP-Us11 fusion protein.
The HSV-1 Patton strain was used throughout the present study. Nucleotide coordinates refer to the published strain 17 sequence (GenBank accession no. X14112). Derivatives of the plasmid pSXZY (27, 29) contain the Us10 −12 region, along with flanking sequences (nucleotides 143481 to 147040) and served to target the Us11 alleles to the homologous region in the viral genome. To replace Us11 with an open reading frame (ORF) that fuses enhanced green fluorescent protein (EGFP) coding sequences in frame to the first ATG of the Us11 gene, pSXZY was digested with XhoI, the 5′ overhangs were filled in with the Klenow fragment, and the plasmid was subsequently cleaved with BspEI. The plasmid pEGFP-Us11 contains the entire EGFP ORF fused to the first ATG of the Us11 gene and produces an EGFP-Us11 fusion protein under the control of the human cytomegalovirus promoter. After cleavage of pEGFP-Us11 with AgeI, the 5′ overhangs were filled in with the Klenow fragment, and the plasmid was digested with BspEI, releasing an 898-bp fragment that contains the EGFP-Us11 fusion gene. This fragment was ligated into XhoI (filled)- and BspEI-digested pSXZY to create pSXZY-GFP-Us11.
After cleavage with PvuII to release the HSV-1 sequences from the plasmid backbone, 10 μg of digested plasmid DNA was cotransfected, along with purified GFPΔUs11 viral DNA, into Vero cells. EGFP-positive plaques were subjected to two rounds of purification. The physical structure of the viral recombinants was confirmed by Southern analysis, and labeling of viral proteins was performed as described previously (27, 29).
Proteins.
GSTΔ1-87, GSTΔ88-155, and GST-RXP have been described (34, 35). In vitro-translated PAT1 was generated with the TNT quick-coupled reticulolysate system according to the manufacturer's instructions (Promega).
Plasmids.
The plasmids expressing PAT1 or PAT1Δ412 in pCDNA4TO have been described (16). To generate the amino-terminal fragments Δ281, Δ352, and Δ436, stop codons were introduced into the PAT1 coding sequence by PCR. PAT1 cDNA was excised from the pcDNA4TO vector with KpnI and XhoI and inserted into pBluescript SK(+) that had been digested with the same enzymes in order to produce a plasmid capable of synthesizing PAT1 RNA that could be translated in vitro.
Yeast two-hybrid screen.
The yeast two-hybrid assay was performed as described according to the MATCHMAKER two-hybrid user manual. The yeast strain EGY188 harboring the reporter genes LacZ and LEU2, under the control of the upstream LexA DNA-binding domain, was used in the assay. For pEG-202 (a bait vector containing the LexA DNA-binding domain) constructs, the entire ORF (155 residues) of HSV-Us11 were subcloned in frame into pEG-202. The bait-containing vector, pEG-202, and the human brain MATCHMAKER LexA cDNA library in the pJG4-5 plasmid were sequentially transformed into the yeast strain EGY188 and grown on minimal synthetic medium lacking tryptophan, histidine, leucine, and uracil to select potential interactors. The X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) indicator dye was included to screen transformants for lacZ expression. Approximately 3 × 106 clones were screened against the Us11 bait. Plasmid DNA isolated from potential positive clones was transformed into Escherichia coli KC8 cells. Bacterial colonies selected on M9 minimal medium lacking tryptophan contained the library plasmid, which was subsequently isolated, and its insert was sequenced. To map the interaction domains, deletion mutants of PAT1 were cloned into pJG4-5, and Us11Δ1-87 and Us11Δ88-161 were cloned into pEG-202. The pJG4-5 and pEG-202 constructs were cotransformed into yeast strain by using a small-scale LiAc/ssDNA/PEG transformation protocol. The transformants were plated onto appropriate minimum synthetic dropout media and were tested for β-galactosidase activity.
Preparation of protein extracts.
293T cells seeded in six-well dishes were transfected with either wild-type (WT) or mutant PAT1 expression plasmids by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. After 24 to 48 h, the cells were washed twice with 50 mM HEPES-KOH (pH 7.4)-150 mM NaCl-1 mM EDTA-5% glycerol and subsequently lysed in the same buffer supplemented with 0.5%% Triton X-100 and 200 μM phenylmethylsulfonyl fluoride. After 10 min on ice, the extract was clarified by centrifugation at 14,000 × g for 10 min at 4°C. The supernatant was used as the source of recombinant PAT1 protein.
In vitro binding assays.
A total 100 μl of a 10% glutathione-agarose slurry was equilibrated in binding buffer (50 mM HEPES-KOH [pH 7.4], 150 mM NaCl, 1 mM EDTA, 5% glycerol, 0.5% Triton X-100) and mixed with 1 μg of purified glutathione S-transferase (GST) fusion protein for 1 h on ice with periodic agitation. The beads were subsequently collected by centrifugation and suspended in 1 ml of binding buffer plus 3% BSA (fraction V). After the mixture was rocked for 1 h at room temperature, 3 μl of 35S-labeled PAT1 that was previously treated with micrococcal nuclease or various quantities of an extract prepared from 293T cells was added. Binding reactions were incubated for 30 min at 30°C and manually agitated every 5 min. The beads were subsequently collected by centrifugation and washed three times with binding buffer, and the associated proteins were fractionated by electrophoresis. Whereas labeled PAT1 was visualized after exposure of fluorophore-impregnated gels to film, samples containing FLAG-tagged PAT1 derivatives were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane and probed with an anti-FLAG monoclonal antibody (Sigma). Antigen-antibody complexes were visualized by using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (ECL; Amersham).
For far-Western analysis, 5-μg samples of GST, GST-US11 Δ1-87, and GST-Us11 Δ88-155 were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to a PVDF membrane. Renaturation of the immobilized polypeptides was accomplished by washing the membrane once for 10 min in PBB (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 100 μM phenylmethylsulfonyl fluoride), followed by an overnight incubation in PBB at 4°C. Nonspecific binding sites were next blocked in PBB plus 3% nonfat dry milk for 4 h at 4°C. The membrane was probed with in vitro-translated 35S-labeled PAT1 or luciferase for 24 h at 4°C. After five washes at room temperature with PBB plus 0.1% NP-40 (10 min for each wash), the membrane was air dried and exposed to either a phosphorimager screen or film.
Immunofluorescence.
Cells were seeded on coverslips and processed for immunofluorescence as described previously (49). Briefly, Cos-1 or MDCK cells were seeded onto glass coverslips for 24 h. After transfection of the appropriate expression plasmids with Lipofectamine 2000 (Invitrogen), the cells were fixed with 3.7% paraformaldehyde, quenched with 100 mM NH4Cl, and permeabilized with 0.1% Triton X-100. Samples were next incubated with anti-FLAG M2 primary antibody (Sigma) for 30 min at 37°C, followed by the addition of a Texas red-conjugated secondary antibody (Vector Laboratories). Nuclei were stained with Hoechst stain. Visualization, quantitative analysis, and photography were performed by using a Zeiss Axioplan 2 fluorescence microscope equipped with a filter for detecting rhodamine and fluoroscein. For confocal studies, Cos-1 cells were seeded onto 35-mm dishes (MatTek Corp.) and, when the cells achieved 70% confluence, they were transfected with plasmids expressing WT PAT1 or PAT1Δ412. At 48 h posttransfection, the cells were infected with GFP-Us11R at an MOI of 5. The samples were processed at various times postinfection as described above and observed by using a Zeiss Axiovert 100 M confocal microscope.
RESULTS
PAT1 is a Us11 interacting protein.
To identify Us11 interacting proteins, we performed a yeast two-hybrid screen. The entire Us11 ORF was fused to the LexA DNA-binding domain and cotransformed with a LacZ reporter plasmid into the EGY188 yeast strain. The yeast cells were subsequently transformed with a human brain cDNA library fused to a synthetic activation domain. Proteins that associate with Us11 will activate transcription of the lacZ gene via the fusion to the synthetic activation domain. Three million clones were screened, and four clones were isolated that interact with the full-length Us11 protein (M. Mulvey and I. Mohr, unpublished data). One of these clones fused amino acids 41 to 585 of PAT1 to the synthetic activation domain and was selected for further study.
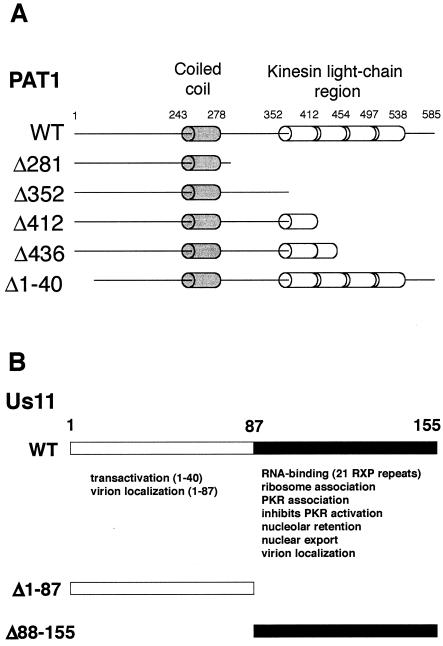
PAT1 (named for protein interacting with APP tail-1) was originally identified in a yeast two-hybrid screen as a protein that binds the basolateral sorting sequence contained in the APP (49). APP is a proteolytically processed transmembrane protein that is involved in the pathogenesis of Alzheimer's disease (33, 43). PAT1 binds microtubules and is involved in the intracellular trafficking of APP (49). Recent data indicate that PAT1 may also participate in intracellular signaling mediated by the cytoplasmic tail of APP (16). The central region of the 585-residue PAT1 protein contains a 35-amino-acid segment predicted to adopt the structure of a coiled coil, and the carboxyl-terminal portion includes a region with homology to kinesin light chain (Fig. 1A).
FIG. 1.
Summary of structural and functional motifs found in PAT1 and Us11. (A) Schematic representation of sequence motifs present in the WT PAT1 protein and the mutants used in the present study. The full-length PAT1 protein is composed of 585 amino acid residues, binds to microtubules, and is involved in the intracellular trafficking of APP. The protein contains a segment predicted to adopt the structure of a coiled coil (amino acids 243 to 278) and a region that shares homology with kinesin light chain (amino acids 352 to 538). Mutant PAT1 proteins used in the present study and the regions they contain are also illustrated. (B) Illustration of functional domains in the HSV-1 Us11 protein. The Us11 protein encoded by HSV-1 (Patton strain) is 155 amino acids long. The carboxyl-terminal 68-amino-acid domain contains a novel dsRNA-binding motif composed primarily of 21 repeats of the amino acid triplet RXP. The activities associated with the amino and carboxyl-terminal domains and a representation of the GST fusion proteins are shown.
PAT1 specifically associates with 68 residues within the carboxyl terminus of Us11.
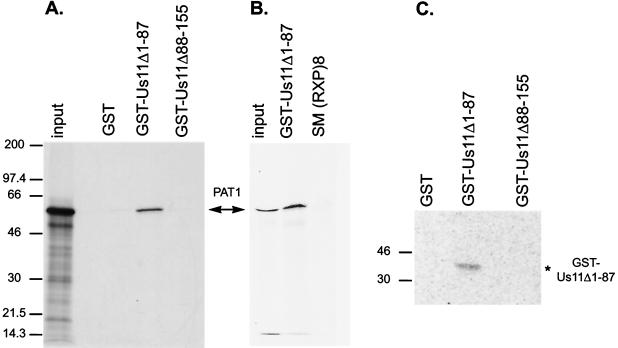
Many of the activities ascribed to the Us11 protein reside within the carboxyl-terminal 68 residues (Fig. 1B). A repetitive motif composed of the amino acids Arg-X-Pro comprises a novel dsRNA-binding domain (23) and is thought to fold into a type II polyprolyl alpha-helix (40, 41). This segment of Us11 also associates with ribosomes (40), interacts with PKR (6, 35) and PACT (32), inhibits PKR activation (34), and contains interwoven nucleolar retention and nuclear export signals, allowing nucleolar accumulation, along with subsequent translocation to the cytosol (5). To discern which segment of Us11 associates with PAT1, the ability of PAT1 to complex with a portion of the Us11 protein fused to GST was evaluated. Full-length PAT1 was translated in vitro and incubated with either purified GST, GST fused to the carboxyl-terminal 68 amino acids of Us11 (GST-Us11Δ1-87), or GST fused to the amino-terminal 87 amino acids of Us11 (GST-Us11Δ88-155). After the isolation of GST-containing proteins on glutathione-agarose beads, associated proteins were fractionated by SDS-PAGE and visualized by fluorography. Figure 2A demonstrates that the full-length PAT1 protein associates with a GST fusion protein that contains the carboxyl-terminal 68 amino acids of Us11 and not with GST-Us11Δ88-155 or GST. Labeled PAT1 also bound GST-Us11Δ1-87 in a far-Western assay. GST, GST-Us11Δ1-87, and GST-Us11Δ88-161 were subjected to SDS-PAGE, transferred to nitrocellulose, renatured, and probed with labeled PAT1. Only the immoblized GST-Us11Δ1-87 protein bound the PAT1 polypeptide (Fig. 2C). A labeled luciferase probe did not associate with any of the proteins immobilized on the filter (not shown). Together, these in vitro assays provide multiple, independent physical lines of evidence that confirm the PAT1-Us11 association discovered in the yeast two-hybrid screen and further delineate the PAT1 binding site on the Us11 protein. In addition, PAT1 does not interact with GST-RXP (Fig. 2B), a GST fusion protein containing 36 amino acids from the Epstein-Barr virus SM polypeptide. GST-RXP contains eight iterations of the RXP repeat sequence, binds RNA, associates with PKR, and inhibits PKR activation (35). Thus, although a different protein containing RXP repeats can execute a number of Us11-related functions, only the RXP segment derived from Us11 associates with PAT1.
FIG. 2.
PAT1 specifically interacts with the carboxyl-terminal 68 amino acids of Us11. (A) 35S-labeled PAT1 was incubated with purified GST fusion proteins containing the C-terminal domain of Us11 (Us11Δ1-87), the N-terminal domain of Us11 (Us11Δ88-155), or GST. After the complexes were isolated on glutathione-agarose beads, the proteins were separated by SDS-PAGE, and the fixed dried gel was exposed to Kodak film. The major translated product (input) corresponds to full-length PAT1. PAT1 specifically forms a complex with the C-terminal 87 amino acids of Us11. (B) GST fusion proteins were incubated with lysates prepared from 293T cells transfected with plasmids expressing FLAG-tagged, full-length PAT1. GST-containing complexes were collected on glutathione agarose beads, fractionated by electrophoresis in SDS-polyacrylamide gels, and transferred to a PVDF membrane. The membrane was probed with anti-FLAG antibody, followed by a horseradish peroxidase- conjugated secondary antibody, and PAT1 was visualized via enhanced chemiluminescence. (C) GST and the two GST-Us11 fusion proteins (GST-Us11Δ1-87 and GST-Δ88-155) were subjected to electrophoresis in SDS-polyacrylamide gels, blotted onto nitrocellulose membranes, renatured, and then probed with in vitro-translated [35S]methionine-labeled PAT1. The filter was washed and subsequently exposed in a phosphorimager cassette.
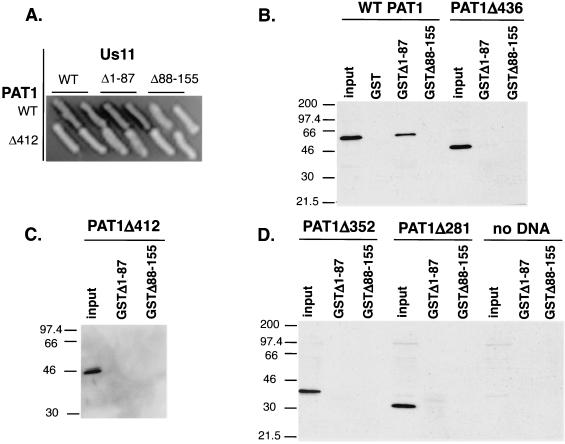
PAT1 residues 412 to 585 containing the kinesin light-chain homology region are required to interact with the C-terminal domain of Us11.
To determine the region(s) of the PAT1 protein important for binding to Us11, a series of PAT1 deletion mutants was constructed, and the resulting polypeptides were expressed both in yeast cells, as fusions to the synthetic activation domain, and in mammalian cells (Fig. 1A). PAT1 fusions expressed in yeast were examined for their ability to associate with LexA-Us11 fusion proteins and to activate transcription of a β-galactosidase reporter gene. Only the full-length, WT PAT1 protein and a 40-amino-acid amino-terminal deletion were able to associate with Us11 or Us11Δ1-87. Amino-terminal PAT1 fragments ranging in size from 281 to 436 amino acids, although expressed at similar levels (not shown), were unable to associate with Us11 and activate β-galactosidase transcription (Table 1 and Fig. 3A). Extracts prepared from mammalian cells expressing these FLAG-tagged PAT1 amino-terminal fragments were incubated with equal amounts of purified GST, GST-Us11Δ1-87, or GST-Us11Δ88-155. Glutathione-agarose-bound complexes were collected, fractionated in SDS-polyacrylamide gels, transferred to nitrocellulose, and incubated with an anti-FLAG antibody. Although similar amounts of FLAG-tagged PAT1 were present in the various extracts, only the full-length PAT1 protein was able to associate with GST-Us11Δ1-87 (Fig. 3B to D). Thus, sequences in the carboxyl-terminal 149 amino acids of PAT1 are important for binding to Us11.
TABLE 1.
Defining regions of PAT1 and Us11 required for interaction in the yeast two-hybrid system
| PAT1 protein | Interactiona of:
|
||
|---|---|---|---|
| WT Us11 | Us11Δ1-87 | Us11Δ88-155 | |
| WT | + | + | − |
| Δ281 | ND | − | − |
| Δ352 | ND | − | − |
| Δ412 | − | − | − |
| Δ436 | ND | − | − |
| Δ1-40 | + | + | − |
PAT1 proteins were expressed in yeast cells as activation domain fusions. Us11 derivatives were expressed in yeast fused to the LexA DNA-binding domain. Interaction between Us11 and PAT1 was detected by the ability to form blue colonies on indicator plates (+). −, white colonies; ND, not determined.
FIG. 3.
PAT1 residues 412 to 585 containing the kinesin light-chain homology region are required to interact with the C-terminal domain of Us11. (A) Full-length PAT1, PAT1 Δ281, Δ352, Δ412, Δ436, and Δ1-40 were expressed in yeast as activation domain fusions. WT Us11, Us11Δ1-87, and Us11Δ88-155 were expressed in yeast fused to the LexA DNA-binding domain. Interaction between Us11 and PAT1 was detected by the ability to form blue colonies on indicator plates. PAT1Δ412, which lacks amino acids 412 to 585 containing the kinesin light-chain repeats, does not interact with the C-terminal domain of Us11 and appears as a representative example. (B) Cell-free lysates were prepared from Cos cells transfected with plasmids expressing either FLAG-tagged WT PAT1 or PAT1 Δ281, Δ352, Δ412, or Δ436 (input). Equal amounts of purified GST, GST-Us11Δ1-87, or GST-US11Δ88-155 were incubated in the extracts. After the GST proteins were collected on glutathione-agarose beads, the complexes were then washed and fractionated by SDS-PAGE. The proteins were transferred to a solid support, reacted with antibodies, and visualized as described in the legend to Fig. 2.
Us11 expression alters the subcellular distribution of PAT1 in transfected cells.
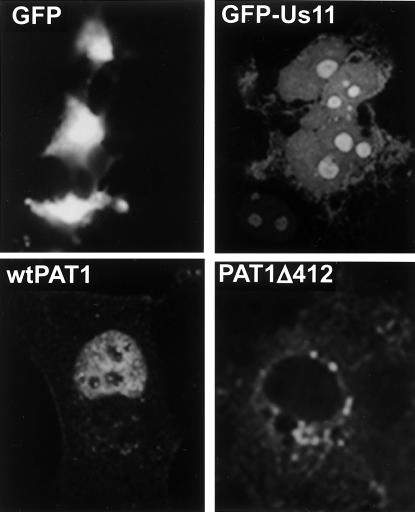
To discern whether the physical association of PAT1 and Us11 could affect the localization of the two polypeptides in intact mammalian cells, the subcellular distribution of the proteins, individually and in tandem, was evaluated by fluorescence microscopy. A fusion of Us11 protein coding sequences to EGFP was used to visualize Us11, whereas PAT1 tagged with a FLAG epitope was identified by indirect immunofluoresence. In contrast to the pattern displayed by EGFP after transfection into MDCK cells, EGFP-Us11 accumulated most intensely in the nucleolus and diffusely stained the nucleoplasm (Fig. 4). EGFP-Us11 was also present in the cytosol, albeit in much smaller amounts (Fig. 4). PAT1, however, was predominantly concentrated in the nucleus and exhibited nucleolar sparing. Numerous small, discrete PAT1 foci were observed in the cytosol as well (Fig. 4). A mutant derivative of PAT1 that lacks residues 412 to 585 (Δ412) accumulated diffusely in the cytosol and exhibited areas of more intense punctate staining in the perinuclear region (Fig. 4). These data, concerning the intracellular localization of each protein expressed individually, agree with previously published reports (5, 16, 40) and serve to validate our assay.
FIG. 4.
Subcellular localization of GFP Us11, PAT1, or PAT1Δ412 after expression of each protein individually in transfected cells. MDCK cells were fixed and permeabilized 24 to 48 h after transfection with plasmids expressing either GFP, GFP-Us11, FLAG-tagged PAT1, or FLAG-tagged PAT1Δ412. They were subsequently examined for intrinsic GFP fluoresence or processed for immunofluorescence to detect PAT1.
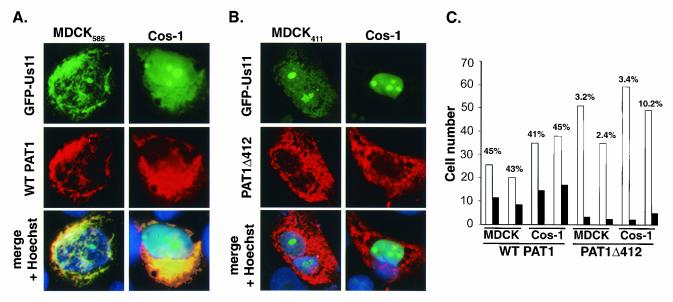
Experiments examining the localization of PAT1 and Us11 expressed together in the same cell were conducted in MDCK cell lines stably expressing either Flag-tagged WT PAT1 (MDCK585) or PAT1Δ412 (MDCK411) from an inducible promoter and in Cos-1 cells cotransfected with EGFP-Us11 and PAT1 derivatives. In MDCK585 cells, EGFP-Us11 accumulated predominantly in the perinuclear region of the cytosol, as did the WT PAT1 protein. Indeed, the two proteins colocalized in this area to a large extent in ca. 44% of cells that expressed both proteins (Fig. 5A and C). Interestingly, very little, if any, EGFP-Us11 is seen in the nucleolus, and the majority of the PAT1 protein appears to be excluded from the nucleus, its normal location (Fig. 4). In addition to colocalizing in cells expressing both polypeptides, coexpression of PAT1 and Us11 appears to mutually alter the subcellular distribution observed when each protein was expressed individually (Fig. 5A).
FIG. 5.
Redistribution of PAT1 and colocalization of PAT1 and Us11 require 173 C-terminal residues of PAT1 that contain homology to KLC. MDCK585 (A) or MDCK411 (B) cells were transfected with 7 μg of pEGFP-Us11. Cos-1 cells were cotransfected with 7 μg of pEGFP-Us11 and an additional plasmid (7 μg) expressing either FLAG-tagged WT PAT1 (A) or PAT1Δ412 (B) polypeptides. At 24 to 48 h after transfection, the cells were processed for immunofluoresence to detect PAT1. Nuclei were stained with Hoechst stain. The distribution of GFP-Us11 was monitored by its intrinsic fluorescence. (C) Cells expressing both GFP-Us11 and either PAT1 or PAT1Δ412 were counted (open bars), and the numbers of cells in which Us11 and PAT1 colocalized were recorded (solid bars). The percentage of cells that expressed both GFP-Us11 and PAT1 (WT or Δ412) in which colocalization was observed appears on top of the bar graph for each experiment. Whereas on average, 44% of cells expressing both GFP-Us11 and PAT1 demonstrated colocalization of both proteins, only 5% of cells that expressed GFP-Us11 and PAT1Δ412 exhibited colocalization of the polypeptides.
Importantly, this analysis was performed by directly observing a number of individual cells, unlike biochemical methodologies that yield results from entire populations. Therefore, the approach described here is subject to variations inherent within the overall population, which might reflect the asynchronous nature of the culture or genetic heterogeneity resulting from its continuous passage. It is well established that all of the cells within a population, especially those of established immortalized cell lines, do not respond identically to a variety of signals that regulate gene expression (24a). Likewise, in a population of cells that coexpress two polypeptides demonstrated to associate by biochemical techniques, it should come as no surprise that these proteins might not colocalize in 100% of the expressing cells. To demonstrate that the observed frequency of colocalization depends upon PAT1, the distribution of Us11 was evaluated in cells expressing a mutant PAT1 derivative that does not interact with Us11. Significantly, the PAT1 mutant derivative expressed in MDCK411 cells did not colocalize with EGFP-Us11 and displayed the punctate cytoplasmic distribution observed previously (Fig. 5B). PAT1Δ412 and Us11 colocalized in only 2.4 to 3.2% of cells expressing both polypeptides (Fig. 5C). The nucleolar accumulation of Us11 in MDCK411 cells was restored as well (Fig. 5B).
A similar picture emerged when the proteins were expressed transiently in Cos-1 cells. PAT1 again appeared to be excluded from the nuclear compartment and extensively colocalized with Us11 in the cytoplasmic and perinuclear area (Fig. 5A). Thus, Us11 expression can alter the subcellular distribution of PAT1 such that it is excluded from the nucleus; furthermore, this redistribution of WT PAT1 requires 149 amino acids in its carboxyl terminus that shares homology to kinesin light chain. Intriguingly, whereas Us11 is barely if at all detectable in the nucleoli of MDCK cells that express WT PAT1, the accumulation of Us11 in nucleoli of Cos-1 cells that express WT PAT1 is readily discernible (Fig. 5A). Although the exact reason(s) for this different behavior is not immediately obvious, it certainly could reflect species differences (canine versus monkey), cell state differences (nontransformed versus transformed), or the levels of the expressed proteins achieved in the different cells. In any event, whereas WT PAT1 appears to be capable of altering the subcellular distribution of Us11 in both MDCK and Cos-1 cells, it does not always prevent the nucleolar accumulation or retention of Us11 in transiently transfected Cos-1 cells.
Us11 and PAT1 colocalize in HSV-1 infected cells.
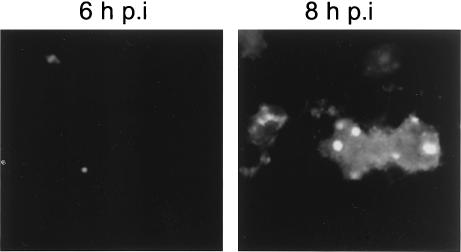
Although our studies indicate that Us11 can colocalize with PAT1 and alter its subcellular distribution in the absence of any other HSV-1 components, they have not addressed whether this can occur in infected cells. To evaluate the distribution of Us11 and PAT1 in infected cells, we first constructed a recombinant HSV-1 that fused EGFP coding sequences to the 5′ end of the Us11 gene (GFP-Us11R, Fig. 6D). This allowed us to localize Us11 expression in infected cells by monitoring EGFP fluorescence. Beginning with a Us11-null virus (ΔUs11 virus) in which the Us11 gene was replaced by the GFP ORF (Fig. 6C and 7C), we subsequently repaired the mutant Us11 allele with a GFP-Us11 fusion gene (Fig. 6D). Southern analysis of recombinant viral genomes demonstrated that Us11 coding sequences had been replaced with DNA encoding an EGFP-Us11 fusion protein (Fig. 7A). Aside from producing an EGFP-Us11 fusion protein displaying an apparent molecular mass of 50 kDa (Fig. 7C), the recombinant GFP-Us11R was otherwise indistinguishable from its WT counterpart in terms of its ability to direct viral protein synthesis in multiple cultured cell lines (Fig. 7B and M. Mulvey, unpublished results). EGFP-Us11 expression was first detected in nucleoli as early as 6 h postinfection and was clearly visible mainly in nucleoli and the surrounding nucleoplasm by 8 h postinfection, a finding consistent with its expression as a true late gene. Smaller quantities of Us11 were also detected in the cytosol (Fig. 8).
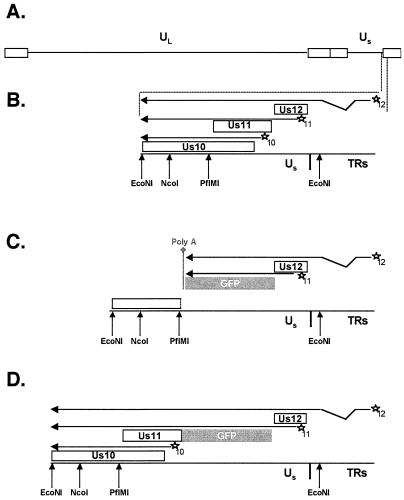
FIG. 6.
Structure of ΔUs11 and GFP-Us11 recombinant viruses. (A) The HSV-1 genome is depicted with the unique long (UL), unique short (Us), and repetitive segments (rectangles) delineated. (B) Expanded view of the WT Us-TRs junction region is shown along with pertinent restriction enzyme cleavage sites. The Us10, Us11, and Us12 ORFs appear as boxes. cis-Acting promoter elements are indicated by stars, and each promoter is normally associated with either the Us10, Us11, or Us12 ORF (denoted by the numbers 10, 11, or 12 at the lower right of each star). Arrows located above each ORF represent mRNA transcripts produced from each promoter. All three transcripts are polyadenylated at a common site downstream from the 3′ end of the Us10 ORF. The noncontiguous segment of the Us12 transcript denotes an RNA splicing event. (C) An expanded view of the Us-TRs junction in the ΔUs11 recombinant virus at which the Us11 ORF has been deleted and replaced with the GFP ORF, followed by a heterologous polyadenylation signal. As a consequence of this mutation, the Us10 ORF is disrupted (the breakpoint is denoted by a broken line at the right end of the box), the Us10 promoter embedded in the Us11 ORF is deleted, and an mRNA capable of encoding the Us10 polypeptide is not produced. (D) An expanded view of the Us-TRs junction in the GFP-Us11R recombinant virus that expresses a GFP-Us11 fusion protein. The GFP ORF has been fused to the 5′ end of the Us11 ORF such that a GFP-Us11 fusion protein is produced. Note that the Us10 ORF and promoter are intact and a Us10 mRNA is produced.
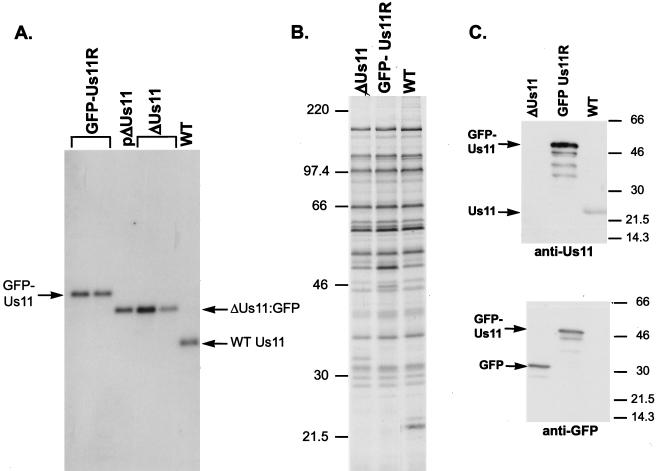
FIG. 7.
Construction and characterization of a recombinant HSV-1 expressing an EGFP Us11 fusion protein. (A) Southern analysis of the Us-TRs junction fragments. Viral (GFP-Us11R, ΔUs11, and WT) or plasmid (pΔUs11) DNA was digested to completion with EcoNI, fractionated by electrophoresis in a 1% agarose gel, and transferred onto a nylon membrane. After the blot was probed with a 32P-labeled NcoI-PflMI DNA fragment derived from the Us-TRs region shown in Fig. 6B, the washed membrane was exposed to X-ray film. The plasmid pΔUs11 was used to construct the recombinant virus ΔUs11. EcoNI-digested viral DNA from two different isolates of ΔUs11 appears as a standard. The genetic structure that surrounds the Us-TRs junction in both the plasmid (pΔUs11) and the ΔUs11 viruses is depicted in Fig. 6C. ΔUs11 was the parental virus from which the GFP-Us11R recombinants were constructed. Two plaque-purified isolates that contain a GFP-Us11 fusion gene as depicted in Fig. 6D are shown. (B) Pattern of late protein synthesis in cells infected with a recombinant virus that expresses a GFP-Us11 fusion protein. Vero cells were infected with WT HSV-1, the ΔUs11 mutant, or the GFP-Us11R virus. At 16.5 h postinfection, the cultures were labeled with 35S-labeled amino acids for 1 h, the samples were solubilized in SDS-PAGE loading buffer, and the proteins were fractionated in SDS-polyacrylamide gels. The fixed, dried gel was subsequently exposed to X-ray film. Molecular mass standards (in kilodaltons) appear to the left of the panel. (C) The GFP-Us11R recombinant virus expresses a GFP-Us11 fusion protein. Total protein was isolated from cells infected with either WT, ΔUs11, or GFP-Us11R HSV-1. After fractionation by SDS-PAGE, the polypeptides were electrophoretically transferred to a membrane and incubated with either anti-Us11 or anti-GFP antibodies. Proteins were detected by chemiluminescence after incubation with horseradish peroxidase-conjugated secondary antibodies. The relative migrations of the GFP-Us11 fusion protein, GFP, and Us11 appear to the left of the image. Molecular mass markers (in kilodaltons) are shown on the right.
FIG. 8.
Subcellular localization of Us11 in cells infected with a recombinant HSV-1 that expresses an EGFP Us11 fusion protein. Cos-1 cells infected with vEGFP Us11 were fixed at either 6 or 8 h postinfection, and the subcellular distribution of Us11 was monitored by the intrinsic fluorescence of GFP Us11.
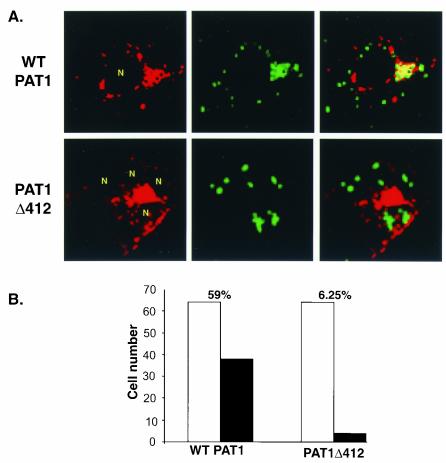
To assess the effects of viral Us11 expression on PAT1 localization, Cos-1 cells were transfected with vectors expressing either WT PAT1 or PAT1Δ412 and subsequently infected with GFP-Us11R 24 h later. At various times postinfection, cells were fixed and processed for indirect immunofluoresence to detect PAT1. Us11 localization was monitored by the intrinsic fluorescence of EGFP-Us11, and confocal images were obtained. In 59% of cells expressing WT PAT and Us11, PAT1 was excluded from the nucleus and colocalized (yellow signal) with the majority of the EGFP-Us11 polypeptide in a perinuclear zone and a cytosolic patch (Fig. 9). Only a small fraction of Us11 (green signal) did not colocalize with WT PAT1 in the merged panel (Fig. 9A). Although some PAT1 did not appear to colocalize with Us11 in the merged image (red signal), a large proportion of PAT1 clearly colocalized with Us11, as indicated by the yellow perinuclear area (Fig. 9A). Perhaps the association of both Us11 and PAT1 with other polypeptides accounts for the fraction of both proteins that does not colocalize. Indeed, both Us11 and PAT1 have been demonstrated to interact with other cellular proteins (6, 12, 32, 35, 49). Finally, Us11 did not accumulate in the nucleoli of PAT1-expressing cells (Fig. 9A).
FIG. 9.
Redistribution and colocalization of PAT1 and Us11 HSV-1-infected cells. (A) Cos-1 cells transfected with plasmids expressing either FLAG-tagged WT PAT1 or PAT1Δ412 were infected with a recombinant HSV-1, which encodes an EGFP Us11 fusion protein, at 24 h posttransfection. Infected cells were fixed at 10 h postinfection and processed for immunofluoresence to detect PAT1 proteins. Localization of Us11 was monitored by the intrinsic fluorescence of EGFP-Us11. Images were captured on a confocal microscope. N, infected cell nuclei. (B) Infected cells expressing both EGFP-Us11 and either FLAG-tagged WT PAT1 or PAT1Δ412 were counted (open bars) and the number of cells in which PAT1 and Us11 colocalized were recorded (solid bars). The percentages of infected cells that expressed both EGFP-Us11 and PAT1 (WT or Δ412) in which colocalization was observed appear on top of the bar graph for each experiment. PAT1 and Us11 colocalized in ca. 59% of infected cells that expressed both proteins, whereas PAT1Δ412 and Us11 only colocalized in 6.25% of infected cells that expressed both proteins.
In marked contrast to cells that express WT PAT1, Us11 amassed in the nucleoli of infected cells that expressed PAT1Δ412 and colocalized with the mutant PAT1 protein in only 6.25% of cells coexpressing the two polypeptides (Fig. 9). Thus, the subcellular distribution of PAT1 was altered in infected cells in a manner that required its carboxyl-terminal 149 residues. Moreover, Us11 can colocalize with PAT1 in HSV-1 infected cells and failed to accumulate in the nucleoli of infected cells expressing the WT PAT1 polypeptide.
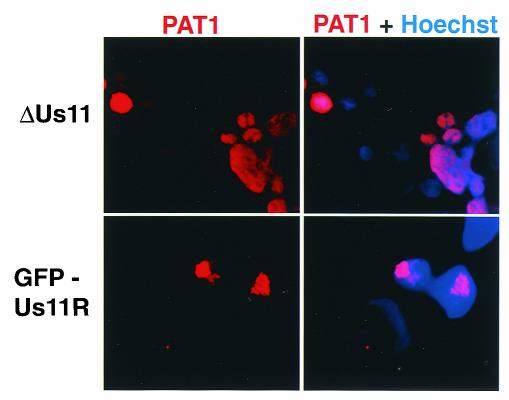
To determine whether Us11 was required for the redistribution of PAT1 in HSV-1 infected cells, Cos-1 cells transfected with a plasmid expressing WT PAT1 were infected with either GFP-Us11R or ΔUs11 recombinant viruses. At 10 h postinfection, the cells were fixed and processed for immunofluoresence to detect PAT1. All of the cells examined were infected as evidenced by their EGFP fluorescence (data not shown). Figure 10 demonstrates that PAT1 remains nuclear in cells infected with ΔUs11, as evidenced by visible nucleolar sparing and the coincidence of Texas red and DAPI (4′,6′-diamidino-2-phenylindole) stains. However, in cells infected with a virus that produces Us11, PAT1 is predominantly excluded from nuclei and concentrates in a perinuclear region. Thus, the redistribution of PAT1 from the nucleus to the cytosol in HSV-1-infected cells depends exclusively upon the Us11 gene product.
FIG. 10.
Redistribution of PAT1 in HSV-1-infected cells requires the Us11 gene product. Cos-1 cells transfected with a plasmid expressing FLAG-tagged WT PAT1 were infected with either GFPUs11R or ΔUs11 at 24 h posttransfection. Cells were fixed and processed for immunofluoresence to detect PAT1 at 10 h postinfection. Nuclei were stained with DAPI.
DISCUSSION
Us11 is a multifunctional dsRNA-binding protein produced late in the HSV-1 productive growth cycle that is required for maximal rates of late viral protein synthesis (23; Mulvey and Mohr, unpublished data). The carboxyl-terminal 68 amino acids binds a variety of structured RNAs (1, 11, 23, 37, 38); associates with 60S ribosomal subunits and polysomes (39); contains overlapping nucleolar retention and nuclear export signals, allowing nucleolar and cytoplasmic accumulation (5, 39, 40); binds the cellular PKR kinase (6, 35), as well as the PACT kinase activator (32); and can prevent activation of PKR (34). In addition, Us11 is a component of the viral tegument, packaged into the viral particle between the envelope and the nucleocapsid (39). Since genetic analysis of Us11 mutants is hampered by the fact that it is not essential for replication in any cultured cell lines examined to date (19, 25), we chose to search for host proteins that associate with Us11 as a means of identifying novel functions that Us11 might influence. We now report that the carboxyl-terminal segment of Us11 interacts with the cellular PAT1 polypeptide, a microtubule-binding protein that participates in the trafficking of APP (49). A region of PAT1 that contains homology to kinesin light chain is required for binding to Us11 and for the perinuclear colocalization of the two proteins in transfected and HSV-1-infected cells. The association between PAT1, a microtubule-binding protein involved in APP trafficking, and Us11, an RNA-binding tegument polypeptide, results in the redistribution of both polypeptides. Furthermore, this association could be important for the intracellular movement of a variety of viral components such as ribonucleoproteins or nucleocapsids with associated tegument proteins in infected cells.
While incoming HSV-1 capsids are propelled along microtubules from the cell periphery to the nucleus by dynein and dynactin (13), a recent study suggests that tegument proteins may be important for antereograde transport of unenveloped capsids in human neurons (12). In support of this hypothesis, Us11 was observed to redistribute from neuronal cell bodies into dendrites late in the infectious cycle and to physically associate with the ubiquitous kinesin heavy chain (μKHC). However, an interaction between kinesin light chain and Us11 was not detected in that study (12). Since discrete KLC isoforms appear to be important for associating with different cargo molecules (2, 21, 22), perhaps Us11 targets the KLC homology region of PAT1 for this purpose. This may reflect a natural function for PAT1 or represent another example of the virus redirecting host components for its own purposes. Additionally, different neurotrophic alphaherpesviruses may have devised alternative approaches to mobilize various virion components or subassemblies. Unlike the product of the Us9 gene which is conserved among alphaherpesviruses and important for the transit of glycoproteins into axons (44, 47), Us11 is only found in HSV-1, HSV-2, and simian B virus. Whether other members of the alphaherpesvirus family have targeted PAT1 or another KLC-like molecule remains to be explored. Finally, if the PAT1-Us11 complex is indeed capable of intracellular trafficking, it is possible that unenveloped capsids are not the only cargo transported. As an RNA-binding protein, Us11 can interact with a variety of structured RNA molecules and could distribute them to discrete sites in the cytosol. Movement of ribonucleoprotein components through an association with molecular motor molecules has been demonstrated in Drosophila melanogaster, and this in turn leads to localized translation (3, 8, 31). Since HSV is a neurotrophic virus, it is certainly worth noting that such localized translation of mRNAs has been described in neurons (reviewed in references 20 and 45), and dsRNA-binding proteins have been documented to play a role in the delivery of mRNAs to discrete neuronal subcellular destinations (46). Besides potentially transporting RNAs in infected cells, the localization of Us11 to discrete sites of translation may be important to prevent ribosome-bound PKR from becoming activated.
Our studies demonstrate that PAT1 and Us11 can colocalize in a perinuclear region in epithelial cells. In this regard, it is intriguing that overexpression of PAT1 seems to prevent the accumulation of Us11 in the nucleoli of infected cells. After their translation on ribosomes, the association of the two proteins might preclude each from reaching their respective compartments. Alternatively, their localization could be the result of a dynamic process in which overexpressed PAT1 binds to Us11 in the nucleoplasm, interfering with nucleolar retention while promoting nuclear export, resulting in the accumulation of both proteins in the perinuclear space. In support of this latter model, the impairment of host protein synthesis after infection with HSV-1 makes it likely that Us11 might need to first enter the nucleus in order to encounter the PAT1 protein in infected cells. Furthermore, the segment of Us11 that associates with PAT1 contains the overlapping nucleolar retention and nuclear export signals (5).
Perhaps PAT1 targets Us11 to a region where unenveloped capsids acquire tegument components in the cytosol. One proposed pathway for HSV-1 egress suggests enveloped capsids between the inner and outer leaflets of the nuclear membrane fuse with the outer nuclear membrane, releasing unenveloped capsids into the perinuclear region. These capsids transit through the cytosol and are thought to acquire tegument components by budding into a post-Golgi compartment prior to exiting the cell (17; reviewed in reference 26). Us11, in turn, might similarly foster the incorporation of the cellular PAT1 protein into the virion.
Despite its many activities, Us11 does not appear to contribute to viral neurovirulence in any of the existing animal models examined to date and remains nonessential for replication in cultured cells (19, 25, 30). It is certainly possible that some of the functions carried out by Us11 are important enough that more than one viral gene product might compensate for the loss of Us11. Different arrays of cellular functions expressed in different cell types could contribute to this as well. Indeed, the γ34.5 and Us11 polypeptides both regulate eIF2α phosphorylation in infected cells, albeit through different mechanisms (7, 18, 29, 32, 34). In a similar vein, it is conceivable that other viral proteins are able to compensate for the absence of Us11 in the tegument. For example, the VP5 capsid protein, along with the VP22 tegument protein, could potentially interact directly with μKHC as well (12). That other virion components would possess multiple functions would not be surprising. In addition to transactivating immediate-early promoters after its delivery into infected cells, at late times postinfection VP16 binds vhs, preventing indiscriminate mRNA destruction that could lead to the inhibition of protein synthesis (24). Furthermore, VP16 also plays an important essential role in viral egress distinct from either of these activities (28). Likewise, through its association with VP16, vhs inhibits the VP16-mediated activation of immediate-early genes late in infection (24).
It is likely that Us11, like other tegument proteins, participates in a spectrum of activities, some of which remain to be characterized. The inability of tegument-derived Us11 to surmount the translational block imposed by host defenses in cells infected with a virus deficient in γ34.5 functions (10), despite the fact that immediate-early expression of Us11 can effectively neutralize the equivalent cellular response (27, 29), is consistent with this hypothesis. Although the analysis of Us11 functions has proved complicated, perhaps insight into additional novel activities can be gleaned through continued investigation of the host components, such as PAT1 and μKHC, with which it associates.
Acknowledgments
We thank Adam Hittleman and Michael Garabedian for advice concerning the yeast two-hybrid system, Mark Phillips and David Sabatini for access to their confocal microscope, Diego Gravotta for generous assistance with the microscope, Bernard Roizman and Rich Roller for Us11 antisera, and David Khoo for technical assistance.
This work was supported by grants from the National Institutes of Health to I.M. and S.W.P. Additional funding to S.W.P. was provided by the Human Frontier Science Program and the Alzheimer's Foundation. M.M. was supported in part by an NIH predoctoral training grant.
REFERENCES
- 1.Attrill, H. L., S. A. Cumming, J. B. Clements, and S. V. Graham. 2002. The herpes simplex virus type 1 US11 protein binds the coterminal UL12, UL13, and UL14 RNAs and regulates UL13 expression in vivo. J. Virol. 76:8090-8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman, A. B., A. Kamal, B. W. Ritchings, A. V. Philp, M. McGrail, J. G. Gindhart, and L. S. Goldstein. 2000. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103:583-594. [DOI] [PubMed] [Google Scholar]
- 3.Brendza, R. P., L. R. Serbus, J. B. Duffy, and W. M. Saxton. 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289:2120-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 5.Catez, F., M. Erard, N. Schaerer-Uthurralt, K. Kinderbeiter, J. J. Madjar, and J. J. Diaz. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 22:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassady, K. A., and M. Gross. 2002. The herpes simplex virus type 1 Us11 protein interacts with protein kinase R in infected cells and requires a 30 amino acid sequence adjacent to a kinase substrate domain. J. Virol. 76:2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K., M. Gross, and B. Roizman. 1998. The herpes simplex virus Us11 proetin effectively compensates for the γ1(34.5) gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha, B. J., L. R. Serbus, B. S. Koppetsch, and W. E. Theurkauf. 2002. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4:592-598. [DOI] [PubMed] [Google Scholar]
- 9.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of an Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ1 34.5 mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, J., and B. Roizman. 1992. The γ34.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz, J. J., M. D. Dodon, N. Schaerer-Uthurralt, D. Simonin, K. Kindbeiter, L. Gazzolo, and J. J. Madjar. 1996. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 379:273-277. [DOI] [PubMed] [Google Scholar]
- 12.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohrer, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Valle, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, Y., and S. W. Pimplikar. 2001. The gamma-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc. Natl. Acad. Sci. USA 98:14979-14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2002. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of eukaryotic initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi, K., R. Fawl, R. J. Roller, and B. Roizman. 1993. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J. Virol. 67:2123-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Job, C., and J. Eberwine. 2001. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2:889-898. [DOI] [PubMed] [Google Scholar]
- 21.Kamal, A., and L. S. Goldstein. 2002. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14:63-68. [DOI] [PubMed] [Google Scholar]
- 22.Kamal, A., G. B. Stokin, Z. Yang, C. H. Xia, and L. S. Goldstein. 2000. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28:449-459. [DOI] [PubMed] [Google Scholar]
- 23.Khoo, D., C. Perez, and I. Mohr. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76:11971-11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 24a.Levsky, J. M., and R. H. Singer. 2003. Gene expression and the myth of the average cell. Trends Cell Biol. 13:4-6. [DOI] [PubMed] [Google Scholar]
- 25.Longnecker, R., and B. Roizman. 1986. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J. Virol. 58:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr, I. J., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 28.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulvey, M., J. Poppers, A. Ladd, and I. Mohr. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73:3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiyama, Y., R. Kurachi, T. Daikoku, and K. Umene. 1993. The Us 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419-423. [DOI] [PubMed] [Google Scholar]
- 31.Palacios, I. M., and D. S. Johnston. 2001. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17:569-614. [DOI] [PubMed] [Google Scholar]
- 32.Peters, G. A., D. Khoo, I. Mohr, and G. C. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pimplikar, S. W. 2002. AbetaPP, apoptosis and Alzheimer's disease. J. Alzheimers Dis. 4:39-40. [DOI] [PubMed] [Google Scholar]
- 34.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poppers, J., M. Mulvey, C. Perez, D. Khoo, and I. Mohr. 2003. Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation. J. Virol. 77:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins Co., Philadelphia, Pa.
- 37.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roller, R. J., and B. Roizman. 1991. Herpes simplex virus 1 RNA binding protein Us11 negatively regulates the accumulation of a truncated viral mRNA. J. Virol. 65:5873-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein Us11 is a virion component and associates with 60S ribosomal subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein Us11: mapping of the domain required for ribosomal and nuclolar association and RNA binding in vitro. J. Virol. 70:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaerer-Uthurralt, N., M. Erard, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 1998. Distinct domains in herpes simplex virus type 1 Us11 protein mediate posttranscriptional transactivation of human T-lymphotropic virus type 1 envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593-1602. [DOI] [PubMed] [Google Scholar]
- 42.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selkoe, D. J. 1998. The cell biology of amyloid precursor protein and presenilin in Alzheimers disease. Trends Cell Biol. 8:447-453. [DOI] [PubMed] [Google Scholar]
- 44.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steward, O., and E. M. Schuman. 2001. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 24:299-325. [DOI] [PubMed] [Google Scholar]
- 46.Tang, S. J., D. Meulemans, L. Vazquez, N. Colaco, and E. Schuman. 2001. A role for a rat homolog of Staufen in the transport of RNA to neuronal dendrites. Neuron 32:463-475. [DOI] [PubMed] [Google Scholar]
- 47.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 49.Zheng P, J. Eastman, S. Vande Pol, and S. W. Pimplikar. 1998. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc. Natl. Acad. Sci. USA 95:14745-14750. [DOI] [PMC free article] [PubMed] [Google Scholar]