Abstract
The herpes simplex virus UL15 and UL28 genes are believed to encode two subunits of the terminase involved in cleavage and packaging of viral genomes. Analysis of the UL15 protein sequence and its herpesvirus homologues revealed the presence of 20 conserved regions. Twelve of the twenty regions conserved among herpesviruses are also conserved in terminases from DNA bacteriophage. Point mutations in UL15 were designed in four conserved regions: L120N (CR1), Q205E (CR2), Q251E (CR3), G263A (CR3), and Y285S (CR4). Transfection experiments indicated that each mutant gene could produce stable UL15 protein at wild-type levels; however, only one mutant (Q251E) was able to complement the UL15-null virus. Each mutation was introduced into the viral genome by marker transfer, and all mutants except Q251E were unable to form plaques on Vero cells. Furthermore, failure to form plaques on Vero cells correlated with a defect in cleavage and packaging. Immunofluorescence experiments indicated that in cells infected with all mutant viruses the UL15 protein could be detected and was found to localize to replication compartments. Although wild-type and mutant Q251E were able to produce A, B, and C capsids, the rest of the mutants were only able to produce B capsids, a finding consistent with their defects in cleavage and packaging. In addition, all mutant UL15 proteins retained their ability to interact with B capsids. Therefore, amino acid residues 120, 205, 263, and 285 are essential for the cleavage and packaging process rather than for association with capsids or localization to replication compartments.
Herpes simplex virus type 1 (HSV-1) DNA cleavage and packaging involves at least seven essential gene products: UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (reviewed in references 6 and 10). This complex process has been compared to the better-studied bacteriophage, T4 and lambda, in which monomeric genomes are released from the concatemeric DNA by cleavage and subsequently packaged into preassembled capsids (reviewed in references 8 and 13). In the lambda phage system, gpA and gpNu comprise a two-subunit terminase that binds to and cleaves concatemeric DNA at specific sequences. The terminase, bound to the DNA, docks onto the prohead at a unique portal vertex. Translocation of the DNA into the capsid requires ATP hydrolysis. The DNA is cleaved at specific sites, and the terminase, with the remaining DNA still attached, detaches from the prohead (13). Other phage proteins such as gpD and gpW act to stabilize DNA in the capsids (8). Recent experiments with HSV indicate that the herpesviruses may encode similar cleavage and packaging machinery.
Newcomb et al. recently reported that the HSV-1 UL6 protein localizes to a unique vertex on procapsids and forms a 12-member ring in vitro (24); these results suggest that UL6 forms a unique portal through which the viral DNA enters the capsid. After packaging, UL25 may play a role in stabilizing DNA inside the capsid (22, 31). UL17 and UL32 may act to transport capsids to replication compartments (RCs) that are believed to be the sites of cleavage and packaging (20, 32). UL15 and UL28 are believed to comprise two subunits of the terminase (2, 5, 36). The function of UL33 is unknown (3), but it has recently been reported to associate with UL28 and UL15 (7).
By analogy with phage, the terminase complex would be expected to bind viral DNA, transiently associate with the capsids at a unique portal vertex, carry out site specific cleavage of viral DNA, and translocate the DNA inside the capsids in an ATP-dependent fashion. Several lines of evidence support the notion that UL15 and UL28 make up two subunits of the terminase. Protein sequence analysis reveals homology between UL15 and gp17 (a subunit of the T4 terminase), including the Walker A and B boxes characteristic of ATP-binding domains (14, 23). A point mutation in the Walker A box in the HSV-1 UL15 gene prevents cleavage and packaging, suggesting that ATP hydrolysis is required for the process (36). In vitro experiments indicate that UL28 and its homolog in human cytomegalovirus (HCMV) bind to a packaging (pac) site, which is believed to comprise part of the cleavage recognition sequence (2, 9). Furthermore, UL15 and UL28 transiently associate with capsid intermediates (27, 29, 37) and associate with each other as determined by coimmunoprecipitation and copurification (1, 18). Under certain experimental conditions, UL28 requires the presence of UL15 to be transported to the nucleus (1, 18). Mutations in UL15 and UL28 homologues from HCMV have been shown to promote resistance to the same antiviral compounds (11, 15, 19), indicating a possible interaction between the two proteins. Taken together, these lines of evidence suggest UL15 and UL28 may comprise two subunits of the putative terminase.
We used sequence analysis and site-directed mutagenesis here to provide additional evidence that UL15 is part of the terminase complex and to elucidate regions of UL15 that are essential in the cleavage and packaging process. The homology alignments reveal regions that are highly conserved among UL15 homologues in the herpesvirus family and bacteriophage terminases. It seems likely that such evolutionarily conserved regions will be important for biological function. Mutations in exon I of the UL15 gene were engineered in highly conserved regions and then analyzed for biological activities exhibited by the wild-type UL15 protein, including (i) the ability to localize to RCs, (ii) participation in cleavage and packaging, and (iii) transient association with capsid intermediates.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero; American Type Culture Collection, Rockville, Md.) were propagated and maintained as described previously (34). MV cells are a Vero cell line expressing a full-length UL9 containing mutation G354A in motif V. MV cells potentiate HSV-1 infection and have been used in the present study to enhance viral infection (B. Martincheva et al., unpublished data). C2 cells, which express the UL15 gene products and are permissive for UL15 mutants, were propagated as described previously (35). The KOS strain of HSV-1 was used as the wild-type virus, and UL15-null viruses, hr81-1 and hr81-2, which contain the ICP6::lacZ mutagen cassette inserted in UL15 exon I and exon II, respectively, were described previously (35). Four point mutations in UL15 were introduced into the viral genome as described below. The G263A mutant with a mutation in the Walker A Box of UL15 was previously described (36). An Sf21 cell extract containing the UL15 protein expressed from a recombinant baculovirus was a kind gift from Daniel Tenney (Bristol-Myers Squibb).
Alignments.
Using the HSV-1 UL15 protein sequence, a PSI-BLAST search of the NCBI database was performed to identify possible homologues (4). The eight human herpes UL15 homologues were aligned using CLUSTALW (Blosum algorithm) within MacVector (33). Using the lambda family bacteriophage (HK97) gp2 (terminase subunit) protein sequence, a PSI-BLAST search of the NCBI database was performed to identify homologues from other bacteriophage and the herpesvirus family (4). Homologues gp2 (HK97), UL15 (HSV-1), gp17 (kvp40), and gp17 (T4) and the consensus sequence from the above alignment were aligned using CLUSTALW (identity algorithm) within MacVector (33).
Sequential PCR mutagenesis.
The G263A mutation in plasmid pcDNAUL15C (containing a CMV promoter and the genomic UL15 sequences, including exon I, UL16, UL17, and exon II) has been previously reported (36). In the present study, four new point mutations were introduced into the UL15 gene. For mutagenesis, pUC119-UL15GE1 plasmid, containing exon I of UL15, was used as the template for two-step PCR with the oligonucleotides listed in Table 1. In each case, the primers were designed to introduce a silent mutation resulting in the introduction of a new restriction enzyme site. The PCR products were subcloned into pUC119-UL15GE1, replacing the existing wild-type UL15 sequences. These mutant clones were confirmed by sequence analysis and detection of the new restriction site within the plasmid. Subsequently, exon I from the pUC119-UL15GE1 mutant plasmids was subcloned into pcDNA-UL15C, replacing the wild-type exon I. The pcDNA-UL15C mutant plasmids were used for transient-transfection and transient-complementation assays, and the pUC119-UL15GE1 plasmids were used for marker transfer.
TABLE 1.
Primers used in two-step PCRa
| Mutant | Upper primer sequence | Lower primer sequence | Restriction site introduced |
|---|---|---|---|
| L120N | AAACCGtctTCGCGCCACCGC | CGAagaCGGTTTAAGGAGTGC | BbsI |
| Q205E | CATTTTttcGAACAGCTCCAG | CTGTTCgaaAAAATGATCCTTATG | BstBI |
| Q251E | GCGCtcgcgaAAGTGGCGCACGGC | CACTTtcgcgaGCGCGCCACCGTG | NruI |
| Y285S | CGCaGTGTAGgcGATCTTGAT | ATCgcCTACACtGCGCACATCCGC | FspI |
Primers were designed to introduce a point mutation within the UL15 protein and a new restriction site within the gene. Lowercase letters indicate sequences that were changed to create the mutation and introduce the restriction site. All oligonucleotides are written in a 5′-to-3′ direction.
Transient transfection and complementation.
Vero cells were transfected with 3 μg of pcDNA-UL15C plasmids containing wild-type or mutant versions of the UL15 gene by use of Lipofectamine Plus according to the manufacturer's protocol (Gibco-BRL). Cells were harvested at 20 h posttransfection and subjected to Western blot analysis. Transient-complementation assays were performed as previously described (38). Briefly, Vero cells were transfected with 3 μg of pcDNA-UL15C plasmids containing wild-type or mutant UL15 genes driven by a CMV promoter and superinfected at a multiplicity of infection (MOI) of 3 PFU/cell with UL15-null virus (hr81-1 or hr81-2). Progeny virus were harvested and assayed on the complementing cell line (C2). The transient-complementation index was determined by dividing the viral titer resulting from the mutant plasmid by the viral titer resulting from the empty plasmid.
Construction of UL15 mutants by marker transfer.
Marker transfer experiments were carried out as described previously (16). Vero cells were transfected with pUC119-UL15GE1 plasmids and infectious hr81-1 DNA (containing a lacZ insertion in exon I of the UL15 gene). Plaques were stained with neutral red in the presence of the chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside); white plaques were purified three times on C2 cells before viral stocks were prepared. Recovery of the desired mutant was verified by Southern blot analysis to detect the introduction of new restriction sites within the viral genome.
Preparation of cell extracts.
Vero cells, infected with KOS or UL15 mutant viruses at an MOI of 10 PFU/cell, were harvested and prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis as previously described (35), except that samples were sonicated prior to boiling.
Capsid preparation.
Capsid preparation was essentially as previously described (29) except that six large (225-cm2) flasks of MV cells were infected at an MOI of 5 PFU/cell for each virus. Sucrose gradients (20 to 50%) were prepared with a Gradient Master (Biocomp). Sucrose gradient fractions were collected with a piston gradient fractionator (Biocomp). After fraction collection, the samples were precipitated with trichloroacetic acid, and the pellets were washed with ethanol and then resuspended in loading buffer (200 mM Tris [pH 8.8], 1% [vol/vol] 2-mercaptoethanol, 2% [wt/vol] SDS, 10% [vol/vol] glycerol, 0.001% [wt/vol] bromphenol blue). The samples were boiled for 5 min and stored at −20°C.
Western blot analysis.
Samples (10 to 30 μl) were subjected to SDS-PAGE on 10 or 12% polyacrylamide gels and transferred to enhanced chemiluminescence (ECL) membranes. To detect UL15 gene products, αAS9 rabbit polyclonal antibody (generously provided by Daniel Tenney at Bristol-Myers Squibb), was used at a 1:2,000 dilution in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween) overnight at 4°C (35). To detect VP5 gene products, α-HSV MCP ICP5 monoclonal antibody (ABI, Columbia, Md.) was used at a dilution of 1:2,000 in TBST overnight at 4°C. Membranes were developed either by ECL (Amersham) or by using alkaline phosphatase (AP; Promega or Bio-Rad) as instructed by the manufacturer.
DNA isolation and Southern blot hybridization.
Vero cells (1.5 × 106) were infected at an MOI of 10 PFU/cell for 18 h and harvested, and the total DNA was isolated, as described previously (21, 28). DNA was then digested with BamHI and subjected to electrophoresis and Southern blot analysis (35). Membranes were probed with a 32P-labeled BamHI SQ junction fragment (21).
Immunofluorescence and imaging.
Immunofluorescence was carried out as described by Burkham et al. (12), except as noted. Cells were fixed in 1.5% formaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized in cold acetone at −20°C for 2 min. The primary antibodies were applied for 3 h; 39S, a monoclonal antibody (ICP8) (30), was used in 3% normal goat serum (NGS) in PBS (1:200) to detect RCs, and αAS9, a polyclonal antibody in 3% NGS in PBS (1:150), was used to detect UL15. Secondary antibodies, i.e., fluorescein isothiocyanate-conjugated goat anti-mouse and Texas red-conjugated goat anti-rabbit in 3% NGS in PBS (1:200), were applied for 1 h. Images were arranged and labeled by using Adobe Photoshop 5.0 and Illustrator 7.0.
RESULTS
UL15 is highly conserved among herpesviruses.
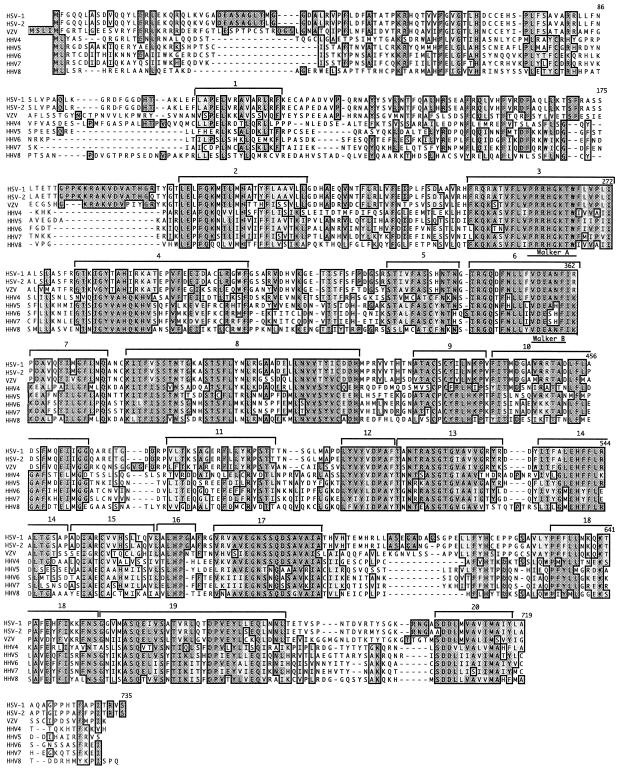
As previously reported, the HSV-1 UL15 protein sequence is highly conserved within the Herpesviridae family (14). To initiate a structure-function analysis of UL15, we identified potentially important residues by extending the sequence comparison between HSV-1 UL15 and other Herpesviridae family members. More than 50 UL15 homologues from human and animal herpesviruses were identified. Figure 1 shows an alignment of UL15 homologues from all eight human herpes viruses. Twenty highly conserved regions (CRs) were designated within the HSV-1 UL15 protein coding sequence and its homologues. These regions were chosen on the basis of the following criteria: a region must either exhibit 70% homology (similarity and identity) among all eight human herpesviruses or it must exhibit 70% homology to a CR of a bacteriophage terminase. Nineteen CRs were identified that met the first criterion (CR2 to CR20). CR1 exhibited 64% homology among the herpesviruses, but a subregion of CR1 revealed 70% similarity compared to bacteriophage terminases (see below). Although it would have been possible to combine some of the CRs into larger regions, this was not done because we wanted to distinguish between CRs that demonstrated homology to bacteriophage terminases and those that did not. The high degree of sequence conservation probably indicates important regions for biological function. Of the 20 CRs, the most highly conserved are CR3 and CR6 containing the Walker A and B boxes, respectively, which are found in ATP-binding proteins.
FIG. 1.
UL15 homology alignments: UL15 is highly conserved among the herpesvirus family. A PSI-BLAST search was performed on the HSV-1 UL15 protein sequence to obtain all its homologues. The eight human herpesvirus homologues were aligned using CLUSTALW (Blosum algorithm) within MacVector. We assigned 20 CRs between UL15 and its homologues based on the fact that many but not all of these regions are also conserved in bacteriophage terminase proteins (see Fig. 2). The Walker A and B boxes of the putative ATP-binding domain are indicated.
CRs of UL15 are also conserved in terminases of bacteriophage.
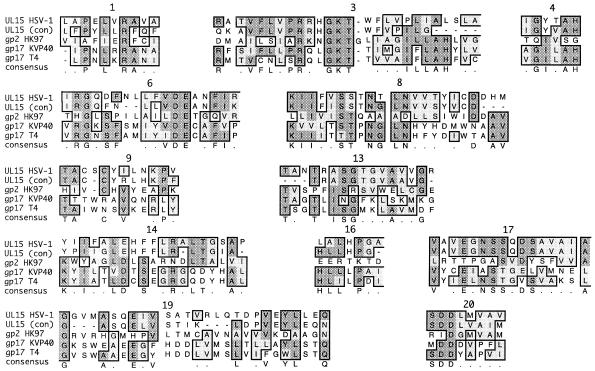
Previous protein sequence analysis suggested a relationship between the UL15 protein of channel catfish herpes virus and the gp17 protein encoded by bacteriophage T4 (14). More recent work has suggested homology between HSV-1 UL15 and terminases in the T4 family (23). This homology was confined primarily to the Walker A and B boxes that make up the ATP-binding domain. Although the previous analysis of amino acid sequence conservation between bacteriophage terminases has not revealed extensive homology outside the ATP-binding domain, we have now extended this analysis by using the identity algorithm within the CLUSTALW program. The large subunit of the terminase from several members of the lambda and T phage families, including gp2 (HK97), gp17 (T4), and gp17 (KVP40), were aligned with HSV-1 UL15 (Fig. 2). In confirmation of previous results, the proteins were homologous in the Walker A and B box regions but, in addition, 10 other regions of homology were identified. These include CRs 1, 3, 4, 6, 8, 9, 13, 14, 16, 17, 19, and 20 (Fig. 1). This high degree of homology suggests an evolutionary relationship between phage and herpesvirus terminases and may help to identify key residues within the HSV-1 UL15 that are essential for terminase function.
FIG. 2.
UL15 and phage alignments. A PSI-BLAST search was performed on gp2 from HK97. UL15 (HSV-1), the UL15 consensus sequence from Fig. 1 [UL15(con)], gp2 (HK97), gp17v (KVP40), and gp17 (T4) were aligned by using CLUSTALW (identity algorithm) within MacVector. Of the 20 CRs shown in Fig. 1, 12 are also conserved between HSV and the phage terminases. The numbers above each CR correspond to the CRs shown in Fig. 1.
Point mutations in exon I.
In order to determine whether the CRs in UL15 play a role in terminase function, point mutations were designed in the first four CRs, three of which are also conserved in bacteriophage. In the present study we report five mutations: one in CR1 (conserved in bacteriophage and herpesviruses), one in CR2 (conserved in herpesviruses only), two in CR3 (one conserved in herpesviruses and one conserved in bacteriophage), and one in CR4 (conserved in bacteriophges and herpesviruses) (Table 2). Our intention was to identify residues that are either intrinsically important for terminase activity in general or are specifically important for herpesviruses. Mutations were engineered into the pcDNA plasmid containing the genomic version of the UL15 gene driven from the CMV promoter.
TABLE 2.
Designed point mutations in the UL15 proteina
| CR | Sequenceb | Mutationc | Position |
|---|---|---|---|
| CR1 | GX6LX6L | N | 120 |
| CR2 | LELFQKMILMHATYFA | E | 205 |
| CR3 (mutation 1) | FRQRATVFLVPRRHGKT | E | 251 |
| CR3 (mutation 2) | TVFLPRRHGKTWFLVP | A | 263 |
| CR4 | IGYTAHIRKATEP | S | 285 |
By using PCR, point mutations were introduced into the UL15 protein. These mutations were designed in CRs 1 to 4 of the UL15 protein.
The residues that underwent mutagenesis are underlined.
The underlined residues from the “sequence” column were replaced with the residues shown in this column.
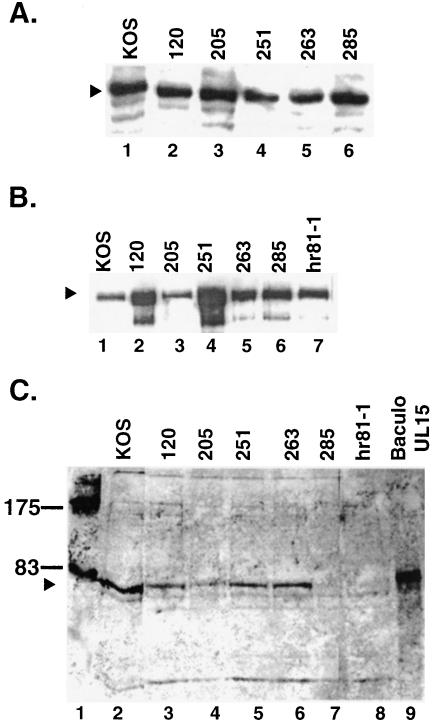
To determine whether the altered genes could produce stable full-length UL15 protein, Vero cells were transfected with mutant and wild-type versions of the UL15 gene in the pcDNA-UL15 plasmid. At 20 h posttransfection, cells were harvested and subjected to Western blot analysis. Figure 3A shows that wild-type and mutant proteins were expressed at similar levels. This indicates that all mutant UL15 proteins were relatively stable when expressed in the absence of other viral proteins.
FIG. 3.
UL15 detected by Western blot analysis. (A) Western blot analysis of transiently transfected cells. Vero cells (2.1 × 106) transfected with a pcDNA plasmid containing a wild-type or mutant version of the UL15 gene were harvested 20 h posttransfection, subjected to SDS-PAGE on a 12% gel, and transferred to an ECL membrane. Western blot analysis was performed by using αUL15 (1:2,000) and AP Immunestar development (Bio-Rad). The arrowhead indicates the band corresponding to the 81-kDa UL15 protein (lane 1). (B and C) Western blot analysis of infected cell lysates. Vero cells (2.1 × 106) were infected with wild-type or mutant virus at an MOI of 10 PFU/cell and harvested 20 h postinfection. The samples were subjected to SDS-PAGE on a 12% gel and then transferred to an ECL membrane. Western blot analysis was performed by using αVP5 (1:2,000) and ECL development (Amersham Pharmacia) (B) and αUL15 (1:2,000) and AP development (Promega) (C). In panel B, the arrowhead indicates the position of VP5. In panel C, the arrowhead indicates the position of the 81-kDa form of UL15. An extract from insect cells infected with a UL15 expressing baculovirus was used as a control for the presence of the 81-kDa form of UL15 (lane 9). The positions of molecular mass markers (in kilodaltons) are shown in lane 1.
Four of the five exon I mutants exhibited severe growth defects.
To determine whether the mutant UL15 genes could complement the UL15-null viruses (hr81-1 and hr81-2), transient-complementation assays were performed as described in Materials and Methods. Vero cells were transfected with the pcDNA-UL15C plasmids containing wild-type or mutant versions of the UL15 gene and superinfected with either hr81-1 or hr81-2; the titers of the progeny were determined for the complementing cell line (Table 3). Only the wild-type and mutant Q251E proteins were able to efficiently complement both exon I and exon II UL15-null viruses. As previously reported, the G263A mutation was unable to complement UL15 null mutants (36; data not shown). In addition, the L120N, Q205E, and Y285S mutations failed to complement UL15 null mutants in this assay (Table 3). Three mutants—Q205E, G263A, and Y285S—exhibited complementation indices significantly less than that of the empty plasmid (Table 3) (36). This may indicate that these mutant proteins exert a transdominant effect. Further experiments will be necessary to explore this possibility.
TABLE 3.
Transient-complementation assaya
| Mutant virus and plasmid | Assay resultb in:
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| Titer | CI | Titer | CI | |
| hr81-1 | ||||
| pcDNA-amp | 3.1 × 105 | 1 | 8.3 × 105 | 1 |
| pcDNA-UL15G | 2.2 × 107 | 73 | 2.9 × 107 | 35 |
| pcDNA-L120N | 1.4 × 104 | 0.05 | 1.2 × 103 | 0.14 |
| pcDNA-Q205E | 2.4 × 104 | 0.08 | 4.3 × 104 | 0.05 |
| pcDNA-Q251E | 2.5 × 107 | 83 | 2.6 × 107 | 31 |
| pcDNA-Y285S | 3.8 × 104 | 0.13 | 5.5 × 104 | 0.07 |
| hr81-2 | ||||
| pcDNA-amp | 4.8 × 104 | 1 | 2.0 × 105 | 1 |
| pcDNA-UL15G | 1.9 × 107 | 396 | 3.2 × 107 | 160 |
| pcDNA-L120N | 1.1 × 105 | 2.3 | 3.0 × 105 | 1.5 |
| pcDNA-Q205E | 1.0 × 104 | 0.21 | 4.5 × 104 | 0.23 |
| pcDNA-Q251E | 2.0 × 107 | 417 | 3.2 × 107 | 160 |
| pcDNA-Y285S | 3.2 × 104 | 0.7 | 4.0 × 104 | 0.2 |
The transient-complementation assay was performed as described in Materials and Methods.
The titer was determined on permissive C-2 cells as described in Materials and Methods. The transient-complementation index (CI) was also determined as described in Materials and Methods. Two different UL15-null mutant viruses were used: hr81-1 (insertion in exon I) and hr81-2 (insertion in exon II).
To determine the effect of UL15 mutations in the context of viral infections, each UL15 mutant gene was transferred into the viral genome by marker transfer as described in Materials and Methods. Progeny viruses were plaque purified three times and verified by gain of a restriction site within the UL15 gene, and viral stocks were made. Only mutant Q251E virus was able to form plaques on noncomplementing Vero cells (Table 4). All of the mutants (L120N, Q205E, Q251E, G263A, and Y285S) were able to form plaques on the complementing cell line, C2, indicating that the growth defects were due to a UL15 mutation and not to a second mutation in another gene.
TABLE 4.
Virus growth assaya
| Virus | Virus growth (PFU/ml) in:
|
|
|---|---|---|
| Vero cells | C2 cells | |
| KOS | 3 × 109 | 5 × 109 |
| L120N | <1 × 102 | 2 × 108 |
| Q205E | <1 × 102 | 4 × 108 |
| Q251E | 7 × 108 | 1 × 109 |
| G263A | <1 × 102 | 5 × 108 |
| Y285S | <1 × 102 | 1 × 109 |
| hr81-1 | <1 × 102 | 1 × 109 |
Wild-type (KOS) and mutant viruses were tested for their ability to form plaques on noncomplementing (Vero) and complementing (C2) cell lines.
Mutant UL15 proteins were detected in infected cell lysates.
To determine whether stable protein could be produced in the context of viral infection, Western blot analysis of cells infected with each mutant virus was performed as described in Materials and Methods. Antibody to VP5 was used as an internal control to demonstrate that efficient infection had occurred. Figure 3B shows that all mutant viruses (lanes 2 to 7) produced wild-type (lane 1) levels of capsid protein VP5 (marked with an arrowhead). Figure 3C shows that all mutants except Y285S (lane 7) produced wild-type levels of UL15. Interestingly, Y285S could produce stable protein after overexpression by transfection (see Fig. 3A, lane 6). The Y285S mutant protein could, however, be detected either in infected cells by immunofluorescence or in capsids by sucrose gradient centrifugation and Western blot analysis (described below). Thus, the Y285S protein is made in transfected and infected cells; however, in the context of infection may be somewhat unstable. On the other hand, the synthesis of wild-type levels of mutant proteins by viruses L120N, Q205E, Q251E, and G263A indicates that no gross alterations in protein formation leading to global instability have occurred (Fig. 3A, lanes 2, 3, 4, and 5, respectively).
Cleavage but not DNA synthesis was inhibited by UL15 mutants.
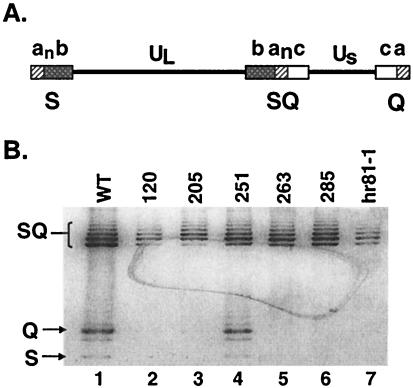
Previously, it has been shown that UL15-null viruses are able to produce viral DNA but not able to carry out cleavage and packaging (25, 35). To determine whether viruses bearing point mutations were also impaired in DNA cleavage, Vero cells (1.5 × 106) were infected with wild-type and mutant viruses at an MOI of 10 PFU/cell. Total DNA was isolated and subjected to Southern blot analysis, as described in Materials and Methods. The cleavage of the viral DNA was detected by the presence of S and Q fragments (Fig. 4A). All viruses were able to produce viral DNA as assessed by the presence of the joint SQ fragments. On the other hand, however, S and Q fragments were only observed in DNA harvested from cells infected with wild-type and mutant Q251E viruses (Fig. 4B, lanes 1 and 4). This indicates that genomic cleavage does not occur in cells infected with the remaining mutant viruses (L120N, Q205E, G263A, Y285S, and hr81-1) (Fig. 4B, lanes 2, 3, 5, 6, and 7, respectively).
FIG. 4.
Southern blot analysis of viral DNA from cells infected with wild-type and mutant viruses. (A) The HSV-1 genome consists of two unique regions, UL and US, which are flanked by repeated sequences a, b, and c. The subscript n in “an” indicates multiple copies of the a sequence. The BamHI fragments corresponding to S, Q, and SQ junction fragments are indicated. (B) Vero cells (107) were infected with the indicated virus at an MOI of 10 PFU/cell for 18 h. Total DNA was digested with BamHI, subjected to electrophoresis, and transferred to a GeneScreen Plus membrane. The membrane was probed with a 32P-labeled BamHI SQ junction fragment. Arrows indicate the junction fragments SQ, Q, and S (from top to bottom). DNA from cells infected with wild-type and null mutant virus (hr81-1) are shown in lanes 1 and 7, respectively. Lanes 2 to 6 contained DNA from cells infected with mutants L120N, Q205E, Q251E, G263A, and Y285S, respectively. Although mutants L120N, Q205E, and hr81-1 appear to contain a reduced amount of SQ DNA compared to wild type and the other mutants, this difference was not reproducible, and we do not feel that it reflects a true reduction in levels of viral DNA synthesis.
All UL15 mutant proteins were detected in RCs.
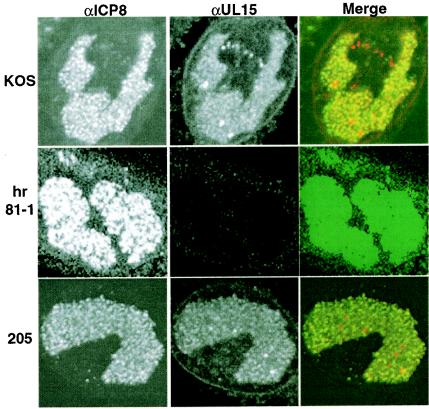
We and others have shown that viral gene expression, DNA replication, and encapsidation occur in large globular domains in the nucleus called RCs (17, 20, 26). RCs are detected with antibody to the major single-stranded DNA-binding protein (ICP8). It was previously reported that wild-type UL15 localized to RCs within the nucleus of infected cells (37). Immunofluorescence was performed to determine whether mutant forms of UL15 behaved like the wild type with respect to nuclear localization. In cells infected with wild-type virus, UL15 was detected in RCs, confirming previous results (Fig. 5, row 1). In cells infected with UL15-null virus (hr81-1), RCs were present, as detected by anti-ICP8 antibody, but UL15 was not detected (Fig. 5, row 2). In cells infected with all mutant viruses, UL15 was detected in RCs. Cells infected with mutant Q205E, are shown in Fig. 5, row 3, as a representative example. Thus, although UL15 protein from cells infected with mutant Y285S was undetectable by Western blot, UL15 was clearly detectable by immunofluorescence, although it exhibits a somewhat fainter staining pattern than that observed in cells infected with wild-type virus (data not shown). This finding suggests that all mutant proteins are synthesized, folded properly, and correctly transported to the RCs, but in the case of mutant Y285S, the UL15 proteins maybe somewhat unstable.
FIG. 5.
Immunofluorescence analysis. Vero cells were infected with wild-type virus (KOS) (top row), UL15-null virus (hr81-1) (middle row), or the UL15 mutant virus Q205E (bottom row) at an MOI of 10 PFU/cell and fixed for immunofluorescence at 8 h postinfection. Infected cells were stained with anti-ICP8 monoclonal antibody (39S) (left panels, green) and anti-UL15 polyclonal antibody (αAS9) (middle panels, red). The right-hand column labeled Merge shows the merged images. The color was adjusted on the hr81-1 panels to demonstrate the lack of UL15 in these cells.
Transient capsid association of UL15 was maintained in all of the UL15 mutant viruses.
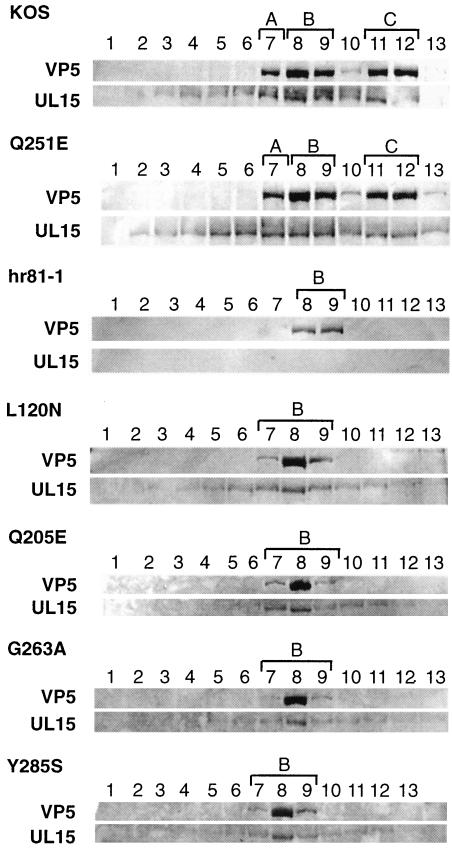
Multiple bands that react with the UL15 antibody, αAS9, were previously reported to associate with capsids. The 81-kDa UL15 protein was found to be associated more strongly with B capsids than with C capsids (27, 29, 37). We wanted to examine whether the UL15 point mutations would alter the ability of the 81-kDa UL15 protein to bind to capsids. Western blot analysis with αAS9 (UL15) was performed on capsids isolated from MV cells infected with mutant or wild-type viruses. Capsid bands were purified by sucrose gradient centrifugation and collected with a piston gradient fractionator (Biocomp) as described in Materials and Methods. Figure 6 shows Western blots of sucrose gradient fractions for wild-type (KOS) and mutant viruses. In the top panel for each virus, VP5 was detected, indicating which fractions contained capsids. This was confirmed visually as fraction samples were collected (data not shown). Mutant Q251E was able to produce A, B, and C capsids similar to wild type (Fig. 6). All other mutants produced only B capsids similar to the UL15-null virus (hr81-1). In the bottom panel for each virus, the 81-kDa UL15 protein was detected by use of antibody αAS9 (Fig. 6). The wild-type and Q251E 81-kDa UL15 proteins were found to associate more strongly with B capsids than C capsids. The UL15-null virus, hr81-1, did not produce UL15 protein. UL15 protein produced in cells infected with mutant viruses L120N, Q205E, G263A, and Y285S was also found to associate with B capsids (Fig. 6). This suggests that CRs 1 to 4 are not essential for the association of UL15 protein with B capsids. In summary, it appears that the failure of mutants L120N, Q205E, G263A, and Y285S to cleave and package viral DNA is not due to the inability of UL15 to associate with B capsids.
FIG. 6.
Western blot analysis of wild-type and mutant capsids. MV cells were infected with the indicated virus at an MOI of 5 PFU/cell for 20 h. Sucrose gradient fractions were collected as described in Materials and Methods. Gradient fractions for each virus were subjected to SDS-PAGE on a 10% gel, and Western blot analysis was performed by using αAS9 (αUL15) and AP development (Promega) as described in Materials and Methods. The αAS9 antibody cross-reacts with VP5 proteins, and this cross-reaction aids in the detection of fractions that contain capsids (top panel for each virus). We previously reported an 87-kDa protein which associated with C capsids that was detected by αAS9 (29, 37). In the experimental conditions used in the present study, the 87-kDa protein was not observed. The bottom panel for each virus shows the position of the 81-kDa form of UL15. Fractions containing A, B, and C capsids are marked with brackets.
DISCUSSION
HSV-1 DNA cleavage and packaging is a complex process that bears many similarities to the cleavage and packaging process in the better-studied bacteriophage. By analogy with bacteriophage, HSV-1 appears to encode a terminase that is essential for encapsidation and the production of progeny virus. HSV-1 UL15 has been identified as a putative component of the terminase. By using sequence analysis, we were able to identify regions in the HSV-1 UL15 protein sequence conserved among homologues in the herpesvirus family. Even more striking was the observation that 12 of the 20 CRs were also found in HK97, T4, and Kvp40 bacteriophage terminases. This analysis extended the original work of Davison et al. and Mitchell et al. showing that UL15 was similar to T4 gp17 primarily in the region of the Walker A and B boxes of the ATP-binding domain (14, 23). We propose that these evolutionarily conserved regions may be important for the biological functions of the terminase. In the present study, we introduced point mutations into the HSV-1 UL15 coding gene to determine which regions are important for function.
By analogy with bacteriophage systems, if UL15 is a component of the terminase, we might expect that it takes part in several functions during encapsidation, including binding and transporting concatemeric DNA to the capsid, docking at the unique portal vertex of the capsid, cleavage and packaging of the viral DNA, and disassociation from the capsid. Direct biochemical analysis of the putative terminase subunits, UL15 and UL28, has been hampered by the insolubility of these proteins expressed in heterologous expression systems. Expression systems tested to date include insect cells infected with recombinant baculoviruses and bacteria transformed with expression vectors (D. Tenney, unpublished data). In addition to the alignment data described above, the strongest evidences to date that UL15 and UL28 are components of the terminase complex are their ability to transiently associate with B capsids during encapsidation (27, 37); the requirement of UL6, the portal protein (24, 27, 37), for the transient association; and the ability of UL28 to bind to pac sequences (2, 9). It was anticipated that the introduction of subtle mutations in CRs of UL15 may result in mutants with a partial phenotype that would be able to carry out some but not all of the functions of the putative terminase. For instance, it might be possible to isolate a mutant that could cleave at the UL terminus but not the US terminus. Such data would support the assignment of UL15 as a component of the terminase.
Several key residues and regions of UL15 were identified that are essential for cleavage and packaging. Mutations in amino acids 120 (CR1), 205 (CR2), 263 (CR3), and 285 (CR4) resulted in the inability to cleave viral DNA, indicating that these residues (and possibly the CRs they are a part of) may be important for this aspect of terminase activity. On the other hand, all mutant UL15 proteins were able to localize to RCs and to associate with B capsids. Thus, the localization to RCs and association with capsids are functions separable from the ability to participate in cleavage and packaging. These point mutations may affect downstream activity such as initiation of packaging, cleavage of the first termini, translocation of the DNA into the capsid, and the ATP hydrolysis necessary for the translocation of the DNA. The mutant G263A (CR3) results in a change in the invariant glycine in the Walker A box of the ATP-binding domain of the UL15 protein. This mutant protein may be defective in the binding and/or hydrolysis of ATP. Another mutation in CR3, Q251E, behaves like wild-type virus, indicating that a conservative change in this CR is tolerated. It is possible that a less conservative change at residue 251 would not be tolerated. Although highly conserved among herpesvirus homologues, amino acid 251 was not conserved among bacteriophage. This may indicate that amino acid 251 is not essential for terminase function per se. Mutant Y285S (CR4) is defective in cleavage and packaging. Interestingly, mutant Y285S protein was detected by immunofluorescence and was found to be associated with capsids but was not readily visible on Western blots of cells infected with mutant virus. The mutation at amino acid 285 may cause the mutant protein to be somewhat unstable in infected cell lysate; however, it may be stabilized by its association with capsids. Alternatively, mutant UL15 may be enriched in the capsid preparation, perhaps by binding better than wild-type UL15 to capsids or by binding irreversibly. It will be of interest to determine whether this mutant exerts a transdominant effect. Because of the apparent instability of the mutant protein in cell extracts, we cannot say whether the defect in cleavage and packaging is a result of the specific mutation in CR4 or due to the relative instability of the protein. The presence of the mutant protein in capsids, however, may indicate the former.
Mutants L120N, Q205E, G263A, and Y285S fail to cleave and package viral DNA despite the fact that they can localize and associate with capsids in a fashion similar to wild type. These results suggest that residues in CRs 1 to 4 are involved in the DNA cleavage reaction. In summary, the mutational analysis described here has resulted in the identification of three separable functions of UL15: localization to RCs, association with capsids, and cleavage of viral DNA. The similarity of the cleavage phenotype in mutants in CRs 1 to 4 indicates that this reaction may be carried out in a synchronized fashion involving multiple regions of the protein.
Acknowledgments
We are grateful to all of the members of our laboratory for critical discussions during this study and in preparation of the manuscript. We thank Amy K. Sheaffer and Daniel J. Tenney for providing the UL15 antisera, the insect cell extract containing UL15 expressing baculovirus, and helpful comments. We especially thank Alan Davidson for the suggestion of using HK97 gp2 for the UL15 and phage alignment.
This work was supported by National Institute of Health grant AI37549.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.al-Kobaisi, M. F., F. J. Rizon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. J. Virol. 180:380-388. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., A. P. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., and S. K. Weller. Cleavage and packaging of herpes simplex virus 1 DNA. In C. E. Catalano (ed.), Viral genome packaging, in press. Landes Bioscience, Boulder, Colo.
- 7.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black, L. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 9.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, J. C., M. A. McVoy, and F. L. Homa. 2001. Packaging DNA into herpesvirus capsids, p. 111-153. In A. Holzenburg and E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum, London, England.
- 11.Buerger, I., J. Reefschilaeger, W. Bender, P. Eckenberg, A. Popp, O. Weber, S. Graeper, H.-D. Klenk, H. Ruebsamien-Waigmann, and S. Hallenberger. 2001. A novel nonnucloside inhibitor specifically targets cyomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type I prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J. Gen. Virol. 73:2709-2713. [DOI] [PubMed] [Google Scholar]
- 15.Evers, D. L., G. Komazin, D. Shin, D. D. Hwang, L. B. Townsend, and J. C. Drack. 2002. Interactions among antiviral drugs acting late in the replication cycle of human cytomegalovirus. Antiviral Res. 56:61-72. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, D., and S. K. Weller. 1988. Herpes simplex virus type-1 induced ribonuclease reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knipe, D., D. Senechek, S. Rice, and J. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W. H. Feng, R. K. Jain, R. G. Trak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drack. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, M. S., S. Matsuzaki, S. Imai, and V. B. Rao. 2002. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30:4009-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon, A. P., and B. Roizman. 1993. Characterization of a temperature sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan, M., L. Chen, and D. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 27.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 29.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115-125. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weller, S. K., K. J. Lee, D. J. Sabourin, and P. A. Schaffer. 1983. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J. Virol. 45:354-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, D., A. K. Sheaffer, D. J. Tenney, and S. K. Weller. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 71:2656-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]
- 37.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, L. A., and S. K. Weller. 1992. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J. Virol. 66:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]