Abstract
Heterologous lentiviral vectors (LVs) represent a way to address safety concerns in the field of gene therapy by decreasing the possibility of genetic recombination between vector and packaging constructs and the generation of replication-competent viruses. Using described LVs based on human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus MAC251 (SIVMAC251), we asked whether heterologous virion particles in which trans-acting factors belonged to HIV-1 and cis elements belonged to SIVMAC251 (HIV-siv) would behave as parental homologous vectors in all cell types. To our surprise, we found that although the heterologous HIV-siv vector was as infectious as its homologous counterpart in most human cells, it was defective in the transduction of dendritic cells (DCs) and, to a lesser extent, macrophages. In DCs, the main postentry defect was observed in the formation of two-long-terminal-repeat circles, despite the fact that full-length proviral DNA was being synthesized and was associated with the nucleus. Taken together, our data suggest that heterologous HIV-siv vectors display a cell-dependent infectivity defect, most probably at a post-nuclear entry migration step. As homologous HIV and SIV vectors do transduce DCs, we believe that these results underscore the importance of a conserved interaction between cis elements and trans-acting viral factors that is lost or suboptimal in heterologous vectors and essential only in the transduction of certain cell types. For gene therapy purposes, these findings indicate that the cellular tropism of LVs can be modulated not only through the use of distinct envelope proteins or tissue-specific promoters but also through the specific combinatorial use of packaging and transfer vector constructs.
Lentiviral vectors (LVs) are useful tools for the delivery of genes with potential therapeutic applications. The major advantage of LVs over other gene delivery systems lies in their ability to pass through an intact nuclear membrane barrier and to integrate into the host genome. For gene therapy purposes, this translates into the possibility of integrating the transgene of interest, ensuring its durable expression, and the ability to target highly differentiated cells (46).
LVs have now been developed from primate lentiviruses, such as the human and simian immunodeficiency viruses (HIV-1, HIV-2, SIVMAC251, and SIV1A11) (33, 36, 38, 41, 52), as well as from nonprimate lentiviruses (6, 25, 34, 40). Common to all such vectors is the physical separation of functions normally encoded by the retroviral genome into separate constructs: one or more packaging constructs that encode the trans-acting factors required for particle formation and infectivity and a gene transfer vector. The latter represents essentially a miniviral genome, devoid of viral open reading frames but bearing the cis elements required for viral infectivity. As a consequence, the life cycle of LVs mirrors that of lentiviruses in a single-round infection but LVs are themselves nonreplicative.
A major concern in the use of primate LVs is the pathogenicity of their parental viruses in humans. Although LVs are nonreplicative, the possibility exists that replication-competent viruses will be generated following recombination between vector and packaging constructs during the phases of vector production (7, 37, 55). It is clear that the possibility of such an event, although small, may temper enthusiasm over the use of LVs in gene therapy. Theoretically, a possible strategy to avoid formation of replication-competent viruses would be to lower the homology of regions shared between the packaging construct and the transfer vector, thus decreasing the likelihood of recombination. This may be achieved, for example, by engineering heterologous LVs in which packaging and vector constructs are derived from different but related viruses.
It is well documented that particles of certain viruses can cross package and mobilize the genomic RNA of a related virus, giving rise to heterologous virion particles, or RNA pseudotypes, as in the case of the spleen necrosis virus (20) and more recently HIV-1, SIV, and the feline immunodeficiency virus (6, 41, 42, 50, 52). Cross packaging is the result of a promiscuous interaction between elements present in the different viral species and involved in viral RNA packaging in cis (the packaging sequence, PSI) and in trans (the nucleocapsid domain of the Gag polyprotein, NC). This interaction is in most cases nonreciprocal. Indeed, spleen necrosis virus virion particles incorporate and mobilize murine leukemia virus viral genomic RNA, while the opposite does not occur. Similarly, HIV-1 has been shown to cross package HIV-2 and SIV RNAs (13, 28, 41, 42), although the reciprocal situation does not occur (in the case of HIV-2) (28) or occurs inefficiently (in the cases of SIV and feline immunodeficiency virus) (6, 52).
Since HIV-1's cross packaging of SIV RNA occurs efficiently (42), we used previously derived LVs (33, 36, 38) to develop heterologous LVs in which trans-acting factors belonged to HIV-1 and cis elements in the transfer vector belonged to SIVMAC251 (HIV-siv). Heterologous HIV-siv LVs have been previously engineered and shown to be able to transduce aphidicolin-arrested cells (50). However, a close inspection of such vectors in comparison with homologous LVs and in a larger spectrum of cells had not been carried out (50). This analysis is required if such vectors are to be used in gene therapy applications.
In this study, we analyzed the behavior of heterologous HIV-siv LVs in close comparison with that of homologous LVs in different human cell types. We show that despite the fact that the infectivity of heterologous HIV-siv LVs was similar to that of homologous LVs in most cells, they were, unlike the latter, incapable of transducing differentiated dendritic cells (DCs) and, to a lesser extent, macrophages. We characterized the infectivity defect of HIV-siv vectors in DCs and were able to show that the major impairment occurs in the formation of two-long-terminal-repeat (2LTR) circles, despite the fact that full-length (FL) proviral DNA was synthesized and associated with the nucleus.
MATERIALS AND METHODS
Cells.
The human monocyte-like cell line U937 (45) and the human myeloid leukemia cell line HL60 (12) were obtained through the centralized facility for AIDS reagents supported by European Union program EVA/MRC and the United Kingdom medical research council. Primary lymphocyte and monocyte fractions were obtained from peripheral blood mononuclear cells of healthy donors at the Etablissement Français du Sang de Lyon as described previously (19). Briefly, after isolation of peripheral blood mononuclear cells by centrifugation in lymphocyte separation medium (Eurobio, Les Ulis, France), cells were layered onto a four-step discontinuous density gradient (75 to 50.5 to 40 to 30% Percoll at a density of 1.130 g/ml; Pharmacia, Uppsala, Sweden) to separate monocytes from peripheral blood lymphocytes (PBLs). After centrifugation at 1,000 × g for 25 min, monocytes and lymphocytes were recovered as low- and high-density fractions, respectively. Prior to freezing, monocytes were further purified by negative selection by using a cocktail of hapten CD3, CD7, CD19, CD45RA, and CD56 anti-immunoglobulin E antibodies coupled to MACS microbeads by following the manufacturer's instructions (Miltenyi Biotec, Paris, France). U937 and HL60 cells, cells from the human T-cell line Jurkat (51), and PBLs were cultured in complete RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). Human 293T and HeLa P4P5 fibroblasts (HeLa cells expressing the CD4 and CCR5 receptors and bearing an HIV-LTR-β-galactosidase reporter cassette, obtained from Pierre Charneau, Pasteur Institute, Paris, France) were maintained in complete Dulbecco modified Eagle medium supplemented with 10% FCS. For stimulation, PBLs were treated for 24 h with 1 μg of phytohemagglutinin (PHA; catalog no. L8902; Sigma)/ml supplemented with 150 U of human recombinant interleukin-2 (IL-2; catalog no. 136; obtained through the National Institutes of Health [NIH] AIDS reagent and reference program)/ml or with 25 ng of IL-7 (R&D Systems)/ml. DCs were differentiated from monocytes upon culture for 4 to 6 days in granulocyte-macrophage colony-stimulating factor (GM-CSF; 100 ng/ml; Schering-Plough) and IL-4 (100 ng/ml; R&D Systems) in complete RPMI 1640 medium plus 5% FCS or human AB serum, as described previously (43). At the end of the differentiation period, cells were CD14− (CD1a+ in FCS), had increased surface expression of HLA-DR, CD80, and CD86, and upon maturation with lipopolysaccharide, CD40L, or tumor necrosis factor alpha, had upregulated CD83. DCs differentiated in this manner were fully competent in phagocytosis of latex beads and in the mixed lymphocyte reaction. Macrophages were obtained from purified monocytes. Briefly, monocytes were washed and plated at 106/ml/well of a 6-well plate in complete RPMI 1640 medium in the absence of serum for 3 h at 37°C. By this time the majority of monocytes had adhered to the plate, and FCS and GM-CSF were added at final concentrations of 10% and 100 ng/ml, respectively. Macrophages were cultured for 6 or 17 days under these conditions.
LV DNA constructions.
The HIV-1- and SIVMAC251-based LV systems have been described elsewhere (in references 21 and 36 and in reference 32, respectively). The packaging constructs, SIV3+ and 8.2, are hereafter referred to as SIV and HIV, respectively. Both constructs express the complete sets of structural and accessory viral proteins with the exception of Env and Vpu (in HIV-1) and are devoid of the packaging sequence, PSI, to impair incorporation of FL viral genomic RNA into virion particles. The lentivirus-derived vector genomes gae-pgk and pRRLsin.PPT.hPGK.GFPpre are hereafter referred to as siv and hiv, respectively. The vesicular stomatitis virus G (VSVg) envelope protein that pseudotypes lentiviral particles efficiently and allows for broad cell tropism was expressed from the plasmid MD.G (36).
Virion particle production and purification.
LVs were produced by calcium phosphate DNA transfection of 293T cells and were pseudotyped with the VSVg envelope. Forty-eight hours after transfection, supernatant of transfected cells was harvested and purified essentially, as described before (11). Briefly, supernatant was filtered (pore size, 0.45 μm) and purified through a double-step sucrose cushion (45 to 25% [wt/vol]). After ultracentrifugation for 2 h at 25,000 rpm in a Beckman centrifuge, the interface between the 45 and 25% sucrose was harvested, diluted three times in phosphate-buffered saline, and recentrifuged under the same conditions onto a single 25% sucrose cushion. At the end of the ultracentrifugation, virions were resuspended in RPMI 1640 medium devoid of serum and supplemented with 10 mM MgCl2 and 200 μM deoxynucleoside triphosphates, aliquoted, and frozen. Normalization of virions contained in the different viral preparations was carried out either by normalization for protein content by using the exogenous reverse transcriptase assay as described previously (11) or by determining the infectious titers after transduction of HeLa or 293T cells, which are used as standard cells in our laboratory. Generally, quantifications determined by protein content and infectious titers agreed.
Cell transduction with LVs.
A total of 105 cells were used for transduction with LVs. Adherent cells were plated onto 12-well plates (24-well plates for macrophages) the day before transduction. Nonadherent cells were instead plated onto 96-well plates the same day as transduction. Transductions were carried out in the presence of 6 μg of polybrene (Sigma)/ml for 2 h at 37°C. Cells were then spun directly in the plate; the medium was removed and replaced with fresh medium. Cell transduction was examined 3 to 4 days posttransduction by flow cytometry by determining the percentage of cells expressing the enhanced green fluorescent protein (eGFP) reporter gene carried by the LVs. When growth arrested, cells were γ-irradiated at 3,000 rad. To distinguish and discard highly contaminated viral preparations, control infections were routinely performed in the presence of nucleoside analogs zidovudine (at 10 μg/ml; catalog no. 3485) and 2′,3′-dideoxycytidine (at 20 μg/ml; catalog no. 220), both obtained through the AIDS reagent and reference program, NIH. These infections allow for the distinction between eGFP transgene expression following lentiviral transduction and eGFP protein carryover and internalization (transduction as opposed to pseudotransduction).
Western blot analysis and antibodies.
Cell and virion lysates were prepared and analyzed by using standard procedures. A monoclonal anti-CA antibody that recognized both SIV and HIV CA (catalog no. 3537; obtained from the AIDS reagent and reference program, NIH) was used; the monoclonal anti-VSVg antibody was clone P5D4 (Sigma).
Slot blot analysis of viral genomic RNA.
Slot blot analysis of viral genomic RNA was carried out essentially as described before (11). Briefly, virions produced by calcium phosphate transfection of 293T cells were purified by ultracentrifugation through sucrose as described above, normalized by using exogenous reverse transcriptase activity or infectious titers, and transferred onto a nylon membrane by using a slot blot apparatus. The membrane was incubated overnight at 42°C in a solution containing 10% polyethylene glycol, 1.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 7% sodium dodecyl sulfate (SDS), and 100 μg of salmon sperm DNA/ml with a 32P-end-labeled DNA probe that hybridizes to the eGFP gene sequence 5′-GGCCGTTTACGTCGCCGTCCAGC-3′ present on both hiv- and siv-derived vector genomes. The membrane was washed in 0.1% SDS-0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and exposed for phosphorimager analysis.
Analysis of reverse transcriptase intermediates in transduced cells.
For PCR analysis, DCs and HeLa cells were analyzed in parallel. Transduction was carried out at multiplicities of infection (MOIs) ranging from 0.5 to 2, as described above. Cell aliquots were harvested at 2 and 24 h posttransduction and lysed in a solution containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.5% NP-40, 0.5% Tween-20, and 120 μg of proteinase K/ml for 1 h at 60°C. Proteinase K was inactivated for 30 min at 95°C, and lysates were used directly for PCR analysis (in fivefold dilutions). PCRs were carried out with 2.5 U of Taq DNA polymerase (Promega) in the presence of a solution containing 1.5 mM MgCl2, 5 mM Tris-HCl (pH 8.0), 10 mM NaCl, 0.01 mM EDTA, 0.1 mM dithiothreitol (DTT), 5% glycerol, 0.1% Triton X-100, 200 μM deoxynucleoside triphosphates, and 10 pmol of each primer in a final volume of 50 μl. Amplifications were carried out by using 30 to 40 cycles as follows: 92°C for 30 s, 55°C for 30 s, and 72°C for 45 s. Primer sequences were as follows (from 5′ to 3′; nucleotide numbers within brackets refer to the complete SIVMAC251 sequence [accession number D01065]): minus-strand strong stop (MSSS) primers PE103, AGTCGCTCTGCGGAGAGGCTG (nucleotides [nt] 507 to 527), and PE83, TGCTAGGGATTTTCCTGC (nt 789 to 807); FL product primers 39, CCGTCGTGGTTGGTTCCTGCCG (nt 878 to 899), and 40, GCTAGATACCCAGAAGAGTTGGAAG (nt 294 to 309); 2LTR primers PE107, AGCTGCCATTTTAGAAGTAAGCC (nt 664 to 686), and PE151, TCTGACAGGCCTGACTTGC (nt 318 to 336); mitochondrial DNA (as in reference 5) primers 98, GAATGTCTGCACAGCCACTTTCCAC, and 99, GATCGTGGTGATTTAGAGGGTGAAC; and actinup, CGAGAAGATGACCCAGATC, and actindown, TGCCGCCAGACAGCACTGTG. Probe sequences were, respectively, primer PE107; primer PE107; primer 40; probe 100, TGGGGTTTGGCAGAGATGT; and actinprobe, GGAGAAGAGCTACGAGCTGC. The products obtained after PCR amplification were transferred onto a nylon membrane and hybridized with the specific 32P-end-labeled probe. The intensity of the signals obtained after hybridization was determined by phosphorimager analysis.
Subcellular fractionation.
Cells were transduced and analyzed 24 h posttransduction by following a procedure described previously, with minor modifications (53). Briefly, cells were washed in cold phosphate-buffered saline and then resuspended in a solution containing 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, 10 mM KCl, and 2 mM DTT. Cells were vortexed and incubated for 1 min on ice. Cells were then spun at 5,500 rpm for 5 min, and the supernatant was transferred to a new tube (fraction 1, cytoplasmic fraction). The fraction was clarified by centrifugation at 14,000 rpm in a Hedaeus centrifuge for 10 min. The nuclear fraction was washed twice in the buffer mentioned above, and the supernatant was pooled and clarified as described above (fraction 2, control for integrity of the nuclear fraction). The pellet containing the nuclei was lysed and vortexed in a solution containing 10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 160 mM KCl, 1 mM DTT, 0.6% SDS, and glass beads (fraction 3, nuclear fraction). All fractions were adjusted for identical salt concentrations and incubated at 60°C for 1 h after addition of 120 μg of proteinase K/ml. Lysates were treated for PCR analysis, as described above.
RESULTS
Cell-specific infectivity defect of an HIV-1 LV packaging an siv-derived genome.
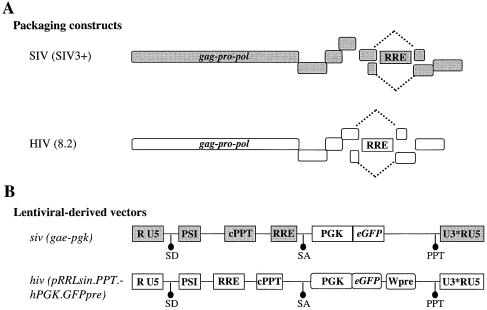
To develop heterologous LVs with HIV-1 packaging an siv-derived genome, we used comparable, although not identical, versions of HIV-1 and SIVMAC251 LVs (21, 33, 36, 38) (Fig. 1A and B). The HIV-1 and the SIVMAC251 packaging constructs, referred to as HIV and SIV, respectively, are devoid of the packaging sequences PSI and env, code for Gag-Pro-Pol, and express the complete set of accessory viral genes. Both lentivirus-derived vector genomes, referred to as hiv and siv, bear an eGFP expression cassette. Besides this, they carry the cis sequences required for transcription, splicing (the major viral splice donor and acceptor sites), nuclear export (the Rev-responsive element [RRE]), packaging (PSI), and reverse transcription (primer binding site [PBS] and 3′ polypurine tract [PPT]) of the vector genome. They also bear a central polypurine tract-central termination sequence (cPPT-CTS). The cPPT increases the efficacy of cell transduction by LVs (21, 54), although the function of this sequence is actually debated (18, 30). The woodchuck hepatitis virus posttranscriptional regulatory element (Wpre) that increases transgene expression by stabilizing mRNAs in cis (16) is present on hiv but not on siv. The presence of this element increases transgene expression in HIV-1 vectors (56), but for unknown reasons it is detrimental during the transduction of certain cell types by SIV LVs (32). As such, Wpre was not present in our siv-derived vector genome.
FIG. 1.
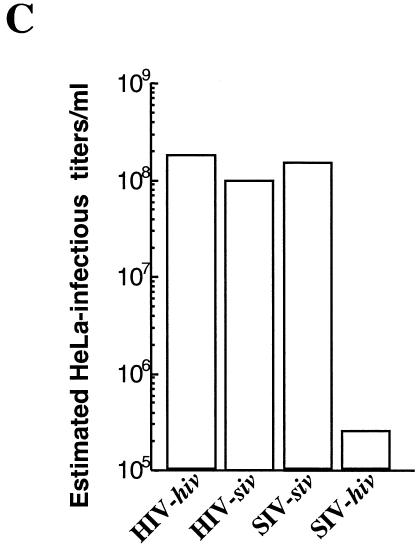
Schematic representation of the SIVMAC251- and HIV-1-based LVs used. Structures of the SIVMAC251 and HIV-1 packaging constructs (A) and of the respective vector genomes (B) carrying an eGFP reporter gene under the control of the phosphoglycerate kinase 1 promoter (PGK). Original names of the constructs are reported within parentheses (capital letters for the packaging constructs, italic small letters for the vector genomes). SD and SA, major splice donor and acceptor sites, respectively. The U3s of both siv and hiv are self-inactivating and result in an impaired U3 viral promoter (U3*). cPPT actually denotes a composite cPPT-CTS element (10, 21, 54). Accessory genes carried by the packaging constructs are not detailed here. (C) Viral preparations were normalized by exogenase reverse transcriptase activity and used to transduce HeLa cells. The estimated infectious titers per milliliter are reported.
Homologous HIV-hiv and SIV-siv LVs were produced along with the heterologous HIV-siv vector by calcium phosphate transfection of 293T cells with the DNAs shown in Fig. 1. All LVs were pseudotyped with the VSVg envelope to allow for broad cell tropism of viral particles. As a first step towards the characterization of heterologous LVs, virions were normalized by exogenous reverse transcriptase activity and their infectious titers were determined upon transduction of HeLa cells. While all vectors had similar levels of infectivity, SIV-hiv LV titers were drastically reduced (Fig. 1C). Given the data described in the literature, this defect is not surprising and it is most probably caused by the inability of SIV virions to cross package an hiv genome (28). For this reason, SIV-hiv LVs were not used any further in subsequent experiments.
To normalize the different viral preparations, we routinely determined their infectious titers in 293T or HeLa cells, highly permissive to both SIV and HIV transduction (14). This method very closely, albeit not identically, mirrored normalization by protein content, as determined by exogenous reverse transcriptase activity and Western blotting (data not shown). To exclude the possibility of pseudotransduction, i.e., the presence of intracellular eGFP signal in target cells due to protein carryover or phagocytosis rather than viral transduction, control test infections were routinely performed in the presence of reverse transcriptase inhibitors.
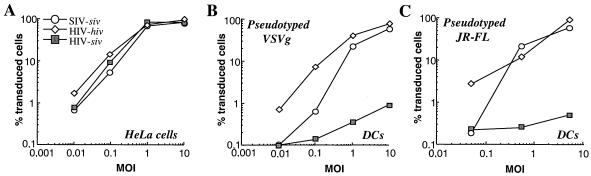
Normalized amounts of virions were used for parallel transduction of HeLa cells, as a control, and DCs, the latter being an important cell target in gene therapy and in HIV pathogenesis. HeLa cells and DCs were transduced with the same MOIs in logarithmic dilutions (Fig. 2). As expected, HeLa cells were equally well transduced by all vectors, irrespective of their cycling status (data not shown). DCs were more refractory to transduction with LVs than were HeLa cells, as indicated by the lower percentages of transduced cells for the same MOIs. However, both homologous SIV and HIV vectors transduced DCs well and routinely; transduction efficiencies of around 60 to 90% were attained with an MOI of 10. MOIs higher than 10 were avoided, since we noticed a negative impact on DC viability and morphology under these conditions (data not shown). The homologous SIV vector was between five- and 10-fold less infectious than its HIV-1 counterpart in human DCs, a defect present at low but not at high MOIs. This observation is reminiscent of the Fv1-like cell restriction recently described for primate lentiviruses and may explain why HIV-1 vectors transduce human cells better than their SIV counterparts (see below).
FIG. 2.
Cell-specific infectivity defect of HIV-siv vectors in DCs. VSVg (A and B)- and JR-FL (C)-pseudotyped LVs were produced by calcium phosphate DNA transfection into 293Tcells and purified by ultracentrifugation through sucrose, and their infectious titers were determined upon transduction of target HeLa or 293T cells. HeLa cells (105) and DCs were then transduced in parallel with logarithmic dilutions of homologous SIV-siv and HIV-hiv vectors along with the heterologous HIV-siv vector. Successful transduction events were analyzed by flow cytometry by determining the percentage of eGFP-positive cells 72 to 96 h posttransduction. Results obtained from a representative experiment are shown here, with the percentages of transduced eGFP-positive cells as the ordinates and the MOIs as the abscissas.
In contrast to homologous SIV and HIV LVs, transduction of DCs with the HIV-siv vector was severely impaired (at least 100-fold). This impairment cannot be explained by loss of infectivity of heterologous HIV-siv LVs, as these do transduce HeLa cells efficiently. As certain postentry blocks may be determined by the entry pathway used by the virus (29), LVs were pseudotyped with an HIV-1 R5 envelope protein (JR-FL) and used for DC transduction (Fig. 2C). Similar to what was observed with VSVg-pseudotyped LVs, heterologous HIV-siv LVs were unable to transduce DCs. These results show a drastic infectivity defect of HIV-siv LVs in DCs that is independent from the viral entry pathway used.
The infectivity defect of the HIV-siv vector is specific for DCs and, to a lesser extent, macrophages.
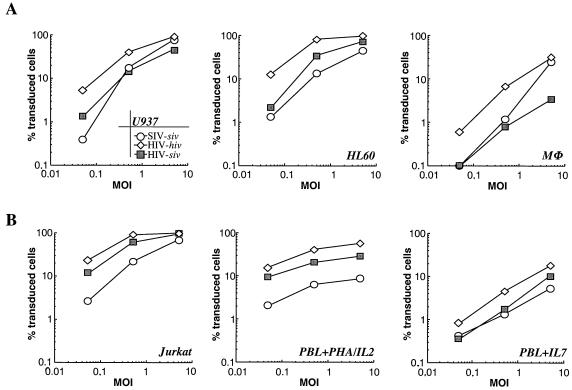
To determine whether the infectivity defect of HIV-siv virions was specific for DCs, different human cell lines and primary blood cells were analyzed for their susceptibility to transduction with LVs. Virions were purified as described above and used in the transduction of the human monocytic cell lines U937 and HL60 and primary macrophages at MOIs of 5, 0.5, and 0.05 (Fig. 3A). Similarly, identical amounts of virions were used to transduce the lymphocytoid Jurkat cell line and primary PBLs that had been activated with PHA-IL-2 or IL-7 for 24 h (Fig. 3B). As transduction of unstimulated PBLs did not yield appreciable transduction rates, it was not examined further (data not shown).
FIG. 3.
Transduction of primary cells and different cell lines with homologous HIV, SIV, and heterologous HIV-siv LVs. Virions, produced as described in the legend to Fig. 2, were used to transduce monocyte/macrophage-like cell lines and primary macrophages (A) and lymphoid Jurkat cells along with primary PBLs stimulated with PHA-IL-2 or IL-7 (B). Results from representative experiments are depicted. The percentages of transduced eGFP-positive cells (ordinates) are shown as a function of MOIs (abscissas).
All the cell lines tested in this study were transduced more efficiently than primary cells, regardless of the cycling statuses of the cells (data not shown). Generally, the homologous HIV-hiv vector had an advantage compared with SIV in the transduction of all the human cells tested in this study. The magnitude of this advantage varied among the cell lines (from five- to eightfold) and was more marked at low rather than high MOIs, at which SIV and HIV transduction rates were close to equal. With primary lymphocytes instead, HIV maintained its advantage over SIV at all MOIs.
Contrary to what was observed with DCs, heterologous HIV-siv virion particles were able to efficiently transduce all the other cells tested in this study (with the exception of primary macrophages; see below). The infectivity of HIV-siv virions neared that of homologous HIV-hiv particles and was consistently higher than that of SIV. The only difference among homologous and heterologous HIV vectors was a slightly lower infectivity of the latter (one- to threefold), probably due to intrinsic differences between the hiv and siv genomic constructs. With primary macrophages however, the difference between homologous and heterologous HIV vectors reached 10-fold. This defect was not as marked as it was in DCs but was nevertheless readily distinguishable. Overall, these results show that the infectivity defect of HIV-siv virion particles is restricted to DCs and, to a lesser extent, macrophages, suggesting a cell-specific infectivity defect.
Characterization of HIV-siv virion particles.
The fact that HIV-siv virion particles efficiently transduced different cell types argues against an intrinsic defect of such particles. However, to exclude completely this possibility, homologous HIV and SIV virions were compared with HIV-siv virion particles with respect to their protein profiles and genomic RNA contents. Normalized amounts of virion particles were analyzed by Western blotting by using an anti-VSVg and an anti-CA antibody that recognizes both SIV and HIV-1 CAs (Fig. 4A). As expected, virions contained similar amounts of Gag and VSVg proteins regardless of the vector genome incorporated (Fig. 4A, lanes 2 and 3 and 5 and 6). Gag processing by the viral protease was also unmodified in HIV particles, regardless of the vector genome packaged.
FIG. 4.
Characterization of HIV-siv virion particles. (A) Western blot analysis of cell lysates (cell-assoc.; lanes 1 to 3) and virion particles (virion-assoc.; lanes 4 to 6) produced after calcium phosphate transfection of DNA into 293T cells. Virion particles were purified onto a double-step sucrose gradient prior to analysis. Western blots were probed by using an anti-CA antibody that recognizes a common epitope present on both HIV and SIV CA and with an antibody that recognizes the VSVg envelope, as indicated. The positions of migration of molecular mass markers (in kilodaltons) are indicated on the right. The products generated upon viral protease processing of SIV (p57-p45-p27) and HIV-1 (p55-p41-p24) Gag polyproteins and recognized by the anti-CA antibody are shown on the left. (B) Genomic-vector RNA incorporation as determined by slot blot analysis on normalized amounts of virion particles by using a probe that hybridizes to the common eGFP gene sequence present on both siv and hiv vector genomes. RNase A treatment of viral RNA preparations was included as a control for DNA contaminations. Twofold dilutions of viral RNA were used as a control for the linearity of the assay. Std. dilutions, standard dilutions.
Next, the amount of viral genomic RNA incorporated into the particles was determined by slot blot analysis by using procedures described previously (11). RNA derived from MOI-normalized amounts of virion particles was transferred onto a nylon membrane by using a slot blot apparatus and hybridized with a 32P-end-labeled antisense probe specific for the common eGFP gene sequence (Fig. 4B). As controls for plasmid DNA carryover, viral RNA preparations were treated with RNase A. Densitometry analysis revealed no major differences among the levels of hiv- and siv-derived vector genomes incorporated into HIV virion particles. Taken together, our results indicate that there is no major structural difference among homologous and heterologous HIV-1 virion particles.
Characterization of the early steps of transduction with HIV-siv LVs by PCR.
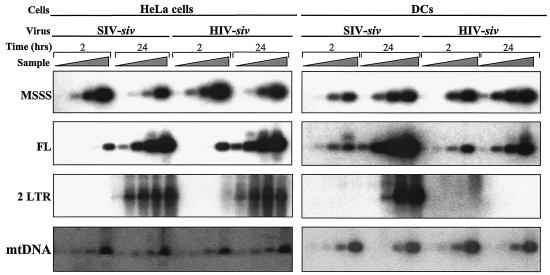
As the transduction defect of HIV-siv LVs was more dramatic in DCs, we decided to focus our efforts on these cells. To precisely characterize this defect, accumulation of reverse transcriptase intermediates was determined by semiquantitative PCR after cell transduction. To this end, the homologous SIV-siv vector and the HIV-siv vector were compared side by side. As the two vectors contain the same genome, PCR analysis is performed by using the same primers and the results obtained for the two vectors can be directly compared. HeLa cells and DCs were transduced in parallel at MOIs between 0.5 and 2 in different experiments, i.e., at MOIs at which differences between HIV and SIV vectors were minor in DCs. Cell aliquots were harvested at 2 and 24 h posttransduction and lysed, and intermediates representing the MSSS, the FL, and 2LTR circles were analyzed by PCR by using primers designed to distinguish each of these forms. Cell lysates were amplified in fivefold dilutions, and PCR was performed in linear conditions for semiquantitative analysis. The products obtained after PCR were transferred onto a nylon membrane and hybridized to a 32P-labeled internal probe specific for each amplified product (Fig. 5).
FIG. 5.
Semiquantitative PCR analysis of reverse transcription intermediates upon cell transduction. Normalized amounts of SIV-siv and HIV-siv vectors were used to transduce 105 HeLa cells and DCs, as indicated. Cell aliquots were harvested at 2 and 24 h posttransduction, lysed, and analyzed by PCR. PCR analysis was conducted with fivefold serial dilutions of starting material by using primers that recognized specifically the MSSS, the FL, and the 2LTR forms produced during the reverse transcription process, as indicated. Amplification of mitochondrial DNA (mtDNA) was used for DNA input and quality control. PCR products were transferred onto a nylon membrane and hybridized with P32-labeled specific internal probes prior to phosphorimager analysis.
Consistent with the similar infectivities of SIV-siv and HIV-siv virion particles, accumulation of MSSS, FL, and 2LTR circle products was equivalent following transduction of HeLa cells. In DCs, the amounts of MSSS were also equivalent following transduction with the SIV-siv and HIV-siv vectors. FL products accumulated normally at 2 h, but levels of these products were six- to eightfold lower at 24 h posttransduction with HIV-siv than they were with SIV-siv. FL products were, however, readily obtained after transduction of DCs with HIV-siv. In contrast, accumulation of 2LTR circles was drastically reduced in DCs transduced with the heterologous HIV-siv vector (at least 100-fold compared to that in DCs transduced with SIV). Our results indicate that HIV-siv virion particles display a slight defect in FL proviral DNA accumulation and a major defect in the formation of 2LTR circles specifically in DCs. A defect in 2LTR circle formation may be explained by several possibilities: instability of proviral DNA, impairment of nuclear entry, or block at a post-nuclear migration step.
Proviral DNA of HIV-siv LVs is associated with the nucleus following subcellular fractionation of transduced DCs.
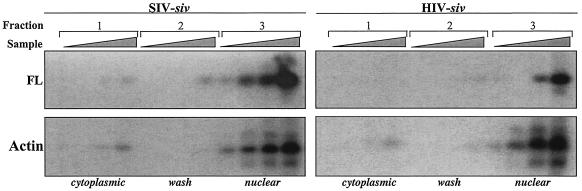
To begin to distinguish among the hypotheses mentioned above, PCR analysis was performed with cytoplasmic and nuclear cell fractions obtained 24 h posttransduction of DCs (Fig. 6). Nuclear and cytoplasmic extracts were obtained according to procedures previously established (53), and the presence of FL proviral DNA was determined by limiting-dilution PCR, as described above. Amplification of actin DNA was used as a control for the fractionation procedure and nuclei integrity.
FIG. 6.
Subcellular fractionation and localization of proviral DNA in DCs. DCs were transduced with SIV-siv and HIV-siv vectors and fractioned 24 h later into cytoplasmic (fraction 1), wash (fraction 2), and nuclear (fraction 3) fractions, as indicated. PCR was then conducted with serial dilutions of samples by using a primer specific for FL proviral DNA and one specific for actin as a control for nuclear integrity. The amplification products obtained after PCR analysis were transferred onto a nylon membrane and hybridized with P32-labeled specific internal probes prior to phosphorimager analysis.
As expected, actin DNA was amplified essentially from the sole nuclear fraction, and only trace amounts could be found in the cytoplasmic and nuclear wash fractions, probably as a result of mechanical breakage of a small proportion of nuclei. When FL proviral DNA was examined, the amount was six- to eightfold lower following transduction with HIV-siv than that following transduction with the SIV-siv vector (as also shown in Fig. 5), but in both cases, nearly all of the FL proviral DNA was associated with the nucleus. These results suggest that the preintegration complex (PIC) of the heterologous HIV-siv vector is able to access the nucleus, although it remains formally possible that proviral DNA might be tightly tethered outside the nuclear membrane rather than inside it.
DISCUSSION
In addition to its purpose with regard to gene therapy, one objective of this study was to determine whether functions specified by cis elements and trans-acting factors of closely related lentiviruses were conserved during the transduction of different human cell types. The data collected here confirm earlier findings, obtained by using heterologous LVs on chemically arrested cells and primary human macrophages, that this is the case (13, 41, 50) but add the notion that the extent of function conservation may be influenced on a cell-specific basis.
Comparison of HIV-1 and SIVMAC251 LVs in the transduction of human cells.
Although the data presented here are not meant to constitute an exhaustive comparative study of HIV-1 and SIVMAC251 vectors, they provide some points of interest towards this end. Generally, HIV-1 LVs (both heterologous and homologous) have an advantage over SIV LVs in the transduction of all the human cells tested in this study, and reciprocally, SIV LVs seem more infectious in simian cells (Vero cells) (data not shown). However, with the exception of lymphocytes, the difference in transduction efficiencies between HIV and SIV lowers with increasing MOIs. This observation may reflect general adaptation of these lentiviruses for replication in their natural host and their ability to overcome restrictions such as the one specified by the resistance factor 1 (Ref1) and the lentiviral resistance gene (Lv-1) in primate cells (3, 4, 14, 23, 26, 35) that target incoming retroviral capsid proteins at different points postentry. Although this may partly explain the relative transduction efficiencies of SIV and HIV LVs in human cells, we ignore at present the question of whether the phenomenon observed in this study is specified through similar mechanisms. At any rate, an Lv-1-like restriction cannot explain the difference between homologous and heterologous HIV-1 LVs in DCs, as they share identical capsids and should thus be subjected to the same restrictions.
Cell-dependent transduction defect of HIV-siv LVs.
We have determined that the transduction defect of HIV-siv LVs observed in DCs is independent from several parameters: the viral entry pathway, as similar results were obtained whether LVs were pseudotyped with VSVg or HIV-1 R5 (JR-FL) envelope proteins; the presence or absence of viral accessory proteins of HIV-1; the specific transgene expression cassette; the Wpre element; the degree of differentiation of DCs; and the blood donor (data not shown). In addition, the HIV-siv transduction defect was not rescued by treatment of DCs with As2O3, a drug that enhances retroviral transduction efficiency by stimulating reverse transcription (2, 47; data not shown). One possible explanation for the transduction defect of HIV-siv LVs may be their inability to transduce nondividing cells. We believe this explanation unlikely for several reasons. HIV-siv vectors are fully capable of transducing growth-arrested γ-irradiated or aphidicolin-treated cells (data not shown) or PBLs treated with IL-7, a cytokine that promotes G0-to-G1b transition but not cell proliferation (15, 17, 48); their proviral DNA is associated with the nuclei of transduced DCs, suggesting that PICs cross the nuclear membrane; and the degrees of transduction inhibition are different between DCs and macrophages, although both are nondividing cells, suggesting involvement of other factors besides cell cycle arrest, as recently suggested for lymphocytes (15, 17, 48). Interestingly, both macrophages and DCs are derived from monocytes, a cell population completely refractory to lentiviral transduction (39; data not shown). In these cells, LV transduction results in normal levels of FL proviral DNA in the absence of 2LTR circles (39). Thus, it is possible to speculate that monocytes bear a restrictive cell environment that eases with differentiation into DCs or macrophages and becomes accessible to a certain extent to transduction with LVs. However, these cells may still express a repressive activity (or lack a facilitating activity) that homologous but not heterologous HIV-siv LVs are able to surmount, probably because one or more viral functions required for transduction are suboptimal in the latter.
Viral determinants of the cell-specific transduction defect of HIV-siv LVs.
We believe that in addition to a cellular determinant, the defect of heterologous HIV-siv LVs in DCs is due to the swapping of cis and trans viral functions. Indeed, when these functions belong to the same virus, as in homologous LVs, transduction of DCs proceeds normally. Thus, we hypothesize the existence of an interaction between a cis element(s) and a trans-acting viral factor(s) required during transduction with LVs and normally influenced by cellular factors, hence its dependence on the cell environment. In its heterologous configuration, the interaction is suboptimal, resulting in a transduction defect only in restrictive cells. The identity of such interacting elements is currently unknown. As viral accessory proteins of HIV-1 are not necessary for transduction of DCs, we believe that a product of the structural Gag-Pro-Pol polyprotein may be involved in the interaction with a cis element (39, 49; data not shown). The siv vector contains six major viral elements: PBS, PPT, cPPT-CTS, PSI, RRE, and LTRs. PBS and PPT are nearly identical between HIV-1 and SIVMAC251, while PSI and RRE (which interact with NC and Rev, respectively) are required for assembly of infectious virion particles apparently normal for heterologous LVs. On the other hand, both the cPPT and the LTRs play a role during the early phases of transduction. The cPPT induces a structure on proviral DNA termed DNA flap and augments transduction efficiency of LVs (21, 31, 44, 49, 54), although the underlying mechanism remains controversial (10, 18, 30, 54). Whether the cPPT acts by recruiting viral trans-acting factors and/or cellular proteins is currently unknown. The LTRs interact with the viral integrase (IN) through short attachment sequences (att) of about 10 nt positioned at their ends (1, 24). The IN-att interaction displays sequence specificity, although elements of closely related viruses functionally recognize one another, as indicated by the infectivity of heterologous retroviral particles (6, 13, 41, 42, 52; this study). However, it is possible that the affinity of the IN-att interaction and thus the extent of its functional conservation is influenced by competition with factors differentially expressed in human cells, only a few of which are actually known (9, 27, 47).
Postentry block after transduction of DCs with HIV-siv LVs.
Our data suggest that the defect of HIV-siv LVs in DCs occurs at a post-nuclear migration step prior to integration. Indeed, postintegration restrictions in several cells have been described before for wild-type lentiviruses, but these do not apply to LVs whose expression, once integrated, proceeds independently of all viral elements. The first defect observed upon transduction of DCs with HIV-siv LVs is a lower accumulation of FL proviral DNA that may result from lower efficiency of reverse transcription or from increased degradation of proviral DNA ends, as described for certain murine leukemia virus and HIV-1 NC mutants (8, 22). We believe the latter unlikely. Indeed, similar kinetics of FL proviral DNA accumulation were obtained by using PCR primers at different positions in the 5′ LTR (data not shown), indicating that if end degradation occurs, it is similar for both homologous and heterologous LVs. Once formed, FL proviral DNA is associated with the nucleus, where formation of 2LTR circles is severely reduced.
Impairment of 2LTR circles has been generally taken as an indirect measure of a nuclear import defect. However, if the presence of 2LTRs is a measure of proviral DNA entry into the nucleus, as 2LTRs form only after ligation by nuclear ligases, their absence may in principle be due to impairment at a post-nuclear migration step. The fractionation data presented here argue in favor of this hypothesis and are supported by findings of similar defects in p12 mutants of the Moloney murine leukemia virus (53) and by results of infecting human macrophages with certain strains of SIV (29) (with the due caveat that the current fractionation techniques do not allow us to unequivocally distinguish between DNA inside the nucleus and DNA tethered outside it). Our understanding of the events that, after nuclear migration of PICs, lead to integration is still incomplete, as is our understanding of the identity of cellular proteins and the role that these proteins may play (47). Thus, it is possible that once in the nucleus the path that results in integration is a complex one and blocks at these stages may result in sequestration of proviral DNA in the nucleus in a form that does not allow its integration.
In conclusion, the finding described here raises the possibility that an interaction between cis elements and trans-acting viral factors exists that is required for early steps of viral infection and that is modulated to different extents by cellular factors. For a more applicative approach, the results presented here show that restriction of transgene expression by LVs may be attained not only through the use of Env proteins or cell-specific promoters but also with the choice of specific combinations of packaging and transfer vectors. As HIV-siv LVs are impaired in the transduction of professional antigen-presenting cells but not other cell types, these vectors could be used to deliver genes of therapeutic interest, possibly lowering the risks of an immune response against the transgene itself.
Acknowledgments
We thank members of the service of immunology of the Etablissement Français du Sang for technical help with primary cells, Christophe Caux from Schering-Plough for providing GM-CSF, and Bernard Dubois for initial counseling on DCs. We thank members of the Darlix laboratory and François-Loïc Cosset for sharing of material and helpful discussion. Specific thanks to Chantal Guiet and Christelle Daudé for technical help.
A.C. was supported by an INSERM poste vert fellowship and is now supported by Marie Curie grant MCFI-2001-00784. We acknowledge the NIH AIDS Research and Reference Reagent Program and the centralized facility for AIDS reagents supported by European Union program EVA/MRC and the United Kingdom Medical Research Council for providing cells and antibodies used in the present study.
REFERENCES
- 1.Basu, S., and H. E. Varmus. 1990. Moloney murine leukemia virus integration protein produced in yeast binds specifically to viral att sites. J. Virol. 64:5617-5625. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Berthoux, L., G. J. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As(2)O(3) enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 5.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning, M. T., R. D. Schmidt, K. A. Lew, and T. A. Rizvi. 2001. Primate and feline lentivirus vector RNA packaging and propagation by heterologous lentivirus virions. J. Virol. 75:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchschacher, G. L., Jr., and F. Wong-Staal. 2000. Development of lentiviral vectors for gene therapy for human diseases. Blood 95:2499-2504. [PubMed] [Google Scholar]
- 8.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman, F. D. 1999. Host proteins in retroviral cDNA integration. Adv. Virus Res. 52:301-317. [DOI] [PubMed] [Google Scholar]
- 10.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, S. J., R. C. Gallo, and R. E. Gallagher. 1977. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature 270:347-349. [DOI] [PubMed] [Google Scholar]
- 13.Corbeau, P., G. Kraus, and F. Wong-Staal. 1998. Transduction of human macrophages using a stable HIV-1/HIV-2-derived gene delivery system. Gene Ther. 5:99-104. [DOI] [PubMed] [Google Scholar]
- 14.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dardalhon, V., S. Jaleco, S. Kinet, B. Herpers, M. Steinberg, C. Ferrand, D. Froger, C. Leveau, P. Tiberghien, P. Charneau, N. Noraz, and N. Taylor. 2001. IL-7 differentially regulates cell cycle progression and HIV-1-based vector infection in neonatal and adult CD4+ T cells. Proc. Natl. Acad. Sci. USA 98:9277-9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donello, J. E., J. E. Loeb, and T. J. Hope. 1998. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 72:5085-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducrey-Rundquist, O., M. Guyader, and D. Trono. 2002. Modalities of interleukin-7-induced human immunodeficiency virus permissiveness in quiescent T lymphocytes. J. Virol. 76:9103-9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eljaafari, A., K. Duperrier, S. Mazet, C. Bardin, J. Bernaud, B. Durand, L. Gebuhrer, H. Betuel, and D. Rigal. 1998. Generation of stable monocyte-derived dendritic cells in the presence of high concentrations of homologous or autologous serum: influence of extra-cellular pH. Hum. Immunol. 59:625-634. [DOI] [PubMed] [Google Scholar]
- 20.Embretson, J. E., and H. M. Temin. 1987. Lack of competition results in efficient packaging of heterologous murine retroviral RNAs and reticuloendotheliosis virus encapsidation-minus RNAs by the reticuloendotheliosis virus helper cell line. J. Virol. 61:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 22.Gorelick, R. J., W. Fu, T. D. Gagliardi, W. J. Bosche, A. Rein, L. E. Henderson, and L. O. Arthur. 1999. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J. Virol. 73:8185-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishimoto, L. K., M. Halperin, and J. J. Champoux. 1991. Moloney murine leukemia virus IN protein from disrupted virions binds and specifically cleaves its target sequence in vitro. Virology 180:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Johnston, J. C., M. Gasmi, L. E. Lim, J. H. Elder, J. K. Yee, D. J. Jolly, K. P. Campbell, B. L. Davidson, and S. L. Sauter. 1999. Minimum requirements for efficient transduction of dividing and nondividing cells by feline immunodeficiency virus vectors. J. Virol. 73:4991-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolicoeur, P., and D. Baltimore. 1976. Effect of Fv-1 gene product on synthesis of N-tropic and B-tropic murine leukemia viral RNA. Cell 7:33-39. [DOI] [PubMed] [Google Scholar]
- 27.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 28.Kaye, J. F., and A. M. Lever. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S. S., X. J. You, M. E. Harmon, J. Overbaugh, and H. Fan. 2001. Use of helper-free replication-defective simian immunodeficiency virus-based vectors to study macrophage and T tropism: evidence for distinct levels of restriction in primary macrophages and a T-cell line. J. Virol. 75:2288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manganini, M., M. Serafini, F. Bambacioni, C. Casati, E. Erba, A. Follenzi, L. Naldini, S. Bernasconi, G. Gaipa, A. Rambaldi, A. Biondi, J. Golay, and M. Introna. 2002. A human immunodeficiency virus type 1 pol gene-derived sequence (cPPT/CTS) increases the efficiency of transduction of human nondividing monocytes and T lymphocytes by lentiviral vectors. Hum. Gene Ther. 13:1793-1807. [DOI] [PubMed] [Google Scholar]
- 32.Mangeot, P. E., K. Duperrier, D. Negre, B. Boson, D. Rigal, F. L. Cosset, and J. L. Darlix. 2002. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther. 5:283-290. [DOI] [PubMed] [Google Scholar]
- 33.Mangeot, P. E., D. Negre, B. Dubois, A. J. Winter, P. Leissner, M. Mehtali, D. Kaiserlian, F. L. Cosset, and J. L. Darlix. 2000. Development of minimal lentivirus vectors derived from simian immunodeficiency virus (SIVmac251) and their use for gene transfer into human dendritic cells. J. Virol. 74:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitrophanous, K., S. Yoon, J. Rohll, D. Patil, F. Wilkes, V. Kim, S. Kingsman, A. Kingsman, and N. Mazarakis. 1999. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 6:1808-1818. [DOI] [PubMed] [Google Scholar]
- 35.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 37.Naldini, L., and I. M. Verma. 2000. Lentiviral vectors. Adv. Virus Res. 55:599-609. [DOI] [PubMed] [Google Scholar]
- 38.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 39.Neil, S., F. Martin, Y. Ikeda, and M. Collins. 2001. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol. 75:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onuma, M., E. Koomoto, H. Furuyama, Y. Yasutomi, H. Taniyama, H. Iwai, and Y. Kawakami. 1992. Infection and dysfunction of monocytes induced by experimental inoculation of calves with bovine immunodeficiency-like virus. J. Acquir. Immune Defic. Syndr. 5:1009-1015. [PubMed] [Google Scholar]
- 41.Poeschla, E., P. Corbeau, and F. Wong-Staal. 1996. Development of HIV vectors for anti-HIV gene therapy. Proc. Natl. Acad. Sci. USA 93:11395-11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi, T. A., and A. T. Panganiban. 1993. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J. Virol. 67:2681-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romani, N., D. Reider, M. Heuer, S. Ebner, E. Kampgen, B. Eibl, D. Niederwieser, and G. Schuler. 1996. Generation of mature dendritic cells from human blood. An improved method with special regard to clinical applicability. J. Immunol. Methods 196:137-151. [DOI] [PubMed] [Google Scholar]
- 44.Sirven, A., F. Pflumio, V. Zennou, M. Titeux, W. Vainchenker, L. Coulombel, A. Dubart-Kupperschmitt, and P. Charneau. 2000. The human immunodeficiency virus type-1 central DNA flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood 96:4103-4110. [PubMed] [Google Scholar]
- 45.Sundstrom, C., and K. Nilsson. 1976. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17:565-577. [DOI] [PubMed] [Google Scholar]
- 46.Trono, D. 2000. Lentiviral vectors: turning a deadly foe into a therapeutic agent. Gene Ther. 7:20-23. [DOI] [PubMed] [Google Scholar]
- 47.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 48.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VandenDriessche, T., L. Thorrez, L. Naldini, A. Follenzi, L. Moons, Z. Berneman, D. Collen, and M. K. Chuah. 2002. Lentiviral vectors containing the human immunodeficiency virus type-1 central polypurine tract can efficiently transduce nondividing hepatocytes and antigen-presenting cells in vivo. Blood 100:813-822. [DOI] [PubMed] [Google Scholar]
- 50.Wagner, R., M. Graf, K. Bieler, H. Wolf, T. Grunwald, P. Foley, and K. Uberla. 2000. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum. Gene Ther. 11:2403-2413. [DOI] [PubMed] [Google Scholar]
- 51.Weiss, A., and J. D. Stobo. 1984. Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 160:1284-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, S. M., M. Renda, N. Y. Nam, E. Klimatcheva, Y. Zhu, J. Fisk, M. Halterman, B. J. Rimel, H. Federoff, S. Pandya, J. D. Rosenblatt, and V. Planelles. 1999. Lentivirus vectors using human and simian immunodeficiency virus elements. J. Virol. 73:2832-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan, B., A. Fassati, A. Yueh, and S. P. Goff. 2002. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J. Virol. 76:10801-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 55.Zufferey, R. 2002. Production of lentiviral vectors. Curr. Top. Microbiol. Immunol. 261:107-121. [DOI] [PubMed] [Google Scholar]
- 56.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]