Abstract
The persistence of herpes simplex virus (HSV) and the diseases that it causes in the human population can be attributed to the maintenance of a latent infection within neurons in sensory ganglia. Little is known about the effects of latent infection on the host neuron. We have addressed the question of whether latent HSV infection affects neuronal gene expression by using microarray transcript profiling of host gene expression in ganglia from latently infected versus mock-infected mouse trigeminal ganglia. 33P-labeled cDNA probes from pooled ganglia harvested at 30 days postinfection or post-mock infection were hybridized to nylon arrays printed with 2,556 mouse genes. Signal intensities were acquired by phosphorimager. Mean intensities (n = 4 replicates in each of three independent experiments) of signals from mock-infected versus latently infected ganglia were compared by using a variant of Student's t test. We identified significant changes in the expression of mouse neuronal genes, including several with roles in gene expression, such as the Clk2 gene, and neurotransmission, such as genes encoding potassium voltage-gated channels and a muscarinic acetylcholine receptor. We confirmed the neuronal localization of some of these transcripts by using in situ hybridization. To validate the microarray results, we performed real-time reverse transcriptase PCR analyses for a selection of the genes. These studies demonstrate that latent HSV infection can alter neuronal gene expression and might provide a new mechanism for how persistent viral infection can cause chronic disease.
A fascinating attribute of herpes simplex virus (HSV) is its ability to enter a quiescent state and establish a lifelong latent infection in sensory neurons that innervate the site of primary, productive infection. Following productive infection by HSV at the site of inoculation, the virus spreads to and enters sensory neurons, where it establishes a latent infection. Latent infection forms a reservoir of virus for recurrent infection, disease, and transmission to other individuals (91). HSV type 1 (HSV-1) is usually associated with primary infections of the orofacial area and latent infection of the trigeminal ganglion, while HSV-2 is usually associated with genital infections and latent infection in sacral ganglia. Although both primary and recurrent infections are usually self-limited, HSV can cause serious diseases such as neonatal disseminated herpes, viral encephalitis, and blinding keratitis (91). Also, genital herpes infection has been associated with an increased risk for human immunodeficiency virus infection (83, 90). Additionally, long-term neurological symptoms have occasionally been associated with HSV infection (14, 25, 45, 46, 77).
Latent HSV infection entails repression of the productive cycle of gene expression (68, 73). Although the viral locus encoding the latency-associated transcripts (LATs) contributes to this repression (10, 23), host functions are also likely to play a role in the repression of viral gene expression. In mouse models of latent HSV infection, the presence of infiltrating immune cells and cytokines in latently infected ganglia (7, 9, 28, 53, 78) suggests that a local immune response may contribute to the maintenance of latent infection. Low-level expression of productive-cycle genes during latency (22, 41) may provide an antigenic stimulus for immune effectors that could repress HSV gene expression (9, 52). Some evidence suggests that neuronal functions also help maintain latency (33, 44, 92). Thus, latent HSV infection appears to involve functions encoded by the pathogen and host functions including nonspecific and specific immune responses and neuronal functions.
Control of transcription is likely to be a central process in the molecular interactions between HSV and its mammalian host. Transcript profiling with DNA microarrays provides an opportunity to identify host genes whose expression is affected by the presence of a latent HSV infection. Subsequent analyses of these genes could potentially deepen our understanding of specific mechanisms of virus-host interactions. Microarrays have been used to study productive HSV infection in cell culture, both to monitor viral gene expression (85) and to detect HSV-induced changes in cellular gene expression (60). Microarray approaches have also been used to identify changes in cellular gene expression under conditions known to reactivate HSV from latent infection in experimental animal models (33, 35, 89). However, changes in host gene expression during the maintenance of latency were not reported in these studies.
Analysis of gene expression in infected tissues by using microarrays can be complicated greatly by cellular heterogeneity (15). Latent HSV genomes reside in the nuclei of neuron cell bodies in trigeminal ganglia. However, trigeminal ganglia comprise not only clusters of sensory neurons and their nerve fibers but also nerve fibers derived from cells located outside the trigeminal ganglion that pass through or terminate within the ganglion and small glial cells called satellite cells that completely envelop the neuronal cell bodies (47). Also contained in trigeminal ganglia are Schwann cells, endothelial (capsular) cells of the microvasculature, blood cells, and motor neurons. During the first several days following corneal inoculation of mice with HSV, the virus replicates in corneal epithelial cells and is then cleared by the host immune response. During this period, HSV enters nerve terminals and is transported to neuronal cell bodies, where it undergoes acute replication. Nonspecific and specific immune effector cells infiltrate along the trigeminal nerve and form foci around individual infected neurons (79). Although viral replication is not detectable and most infiltrating cells are cleared by 30 days after corneal inoculation, low levels of immune effector cells remain in ganglia harboring latent HSV (53, 78).
We set out to use microarrays to test the hypothesis that HSV perturbs host gene expression in latently infected ganglia. To address the challenge of identifying gene expression changes in neurons that harbor latent HSV within the context of a complex tissue, we have performed microarray analyses in replicate experiments and applied statistical tests to identify those host genes whose expression changes significantly with latent HSV infection. We found significant changes in the expression of the immune response and, of particular interest, in the expression of neuronal genes, including several with roles in the regulation of gene expression and neural transmission.
MATERIALS AND METHODS
Cells and virus.
The KOS strain of HSV was propagated and assayed on Vero cell monolayers as previously described (48).
Infection protocol.
Seven-week-old male HSD/ICR mice (Harlan Sprague-Dawley) were anesthetized and either infected with 2 × 106 PFU of HSV per eye or mock infected with a medium by inoculation onto scarified corneas as previously described (48). At the indicated times postinoculation, the animals were sacrificed and trigeminal ganglia were removed, rapidly frozen in dry ice, and stored at −80°C. Mice were housed in accordance with institutional and National Institutes of Health (NIH) guidelines on the care and use of animals in research, and all procedures were approved by the Institutional Animal Use Committee of Harvard Medical School.
Microarray hybridization.
RNA was isolated from trigeminal ganglia by using RNA STAT-60 (Tel-Test, Friendswood, Tex.) and was pooled for each treatment group. Synthesis of [33P]-labeled cDNA, hybridization conditions, and data collection were exactly as described previously (12), except for the use of mouse total RNA (15 μg) and mouse C0t 10 DNA (10 μg) in hybridizations to mouse cDNA arrays. Array hybridizations performed in duplicate were highly reproducible, as reflected by the fact that the coefficient of variation (calculated as the standard deviation divided by the mean) averaged less than 0.2 for genes whose intensities were above a detection threshold, as reported by Chiang et al. (12). The dynamic range of detection spanned 3 orders of magnitude. Spot intensities were normalized to the median array spot intensity for each individual filter (global normalization). Data preprocessing was performed, and scatterplots were generated with Expression Explorer (Millennium proprietary software).
Statistical analysis.
To obtain estimates of measurement error, we assumed an error model with a fixed error component (e.g., background variation) and a proportional error component (e.g., variation in the amount of spotted DNA). Measurement error (E) was modeled separately for each group of arrays corresponding to an individual tissue sample. We modeled the fixed error (A) as being the median standard deviation of the expression level for genes in the 5th percentile of expression (an average over replicates) and the coefficient for proportional error (C) as the coefficient of variation (standard deviation/mean) for genes in the 95th percentile of mean expression. The measurement error estimate for each gene within each group of arrays from a given sample was then calculated as  , where A and C are defined above and x is the replicate-averaged expression level of the gene. The observed standard deviation was then regularized with the estimated measurement error by using the method of Baldi and Long (3, 54) with their default value of 10 pseudocounts.
, where A and C are defined above and x is the replicate-averaged expression level of the gene. The observed standard deviation was then regularized with the estimated measurement error by using the method of Baldi and Long (3, 54) with their default value of 10 pseudocounts.
For each gene in each of the three independent experiments, we obtained a P value from the two-sided Student t test comparing latently infected and mock-infected samples. The P values from independent experiments were then combined by using Fisher's method (81). A Bonferroni correction based on the number of genes tested (81) was then applied so that the single-hypothesis-test P value corresponding to a significant difference (multiple-hypothesis-corrected α = 0.05) was ≤ 9.78 × 10−6 and that for a highly significant difference (multiple-hypothesis-corrected α = 0.001) was ≤ 1.96 × 10−7. Although Bonferroni's correction can be overly conservative for certain analyses (81), its use here is consistent with the primary goal of determining with high confidence whether or not latent HSV impacts gene expression in the host ganglion, as opposed to a goal of generating a list of hypothetically differentially expressed genes with high false-positive rates.
In situ hybridization
Methods for ganglion tissue preparation and in situ hybridization have been described previously (23). Plasmids used as probes for cellular gene expression included cDNAs encoding Chrm1, Ptprs, and Kcnc1. These genes were selected on the basis of their known neuronal specificity and the availability of specific reagents at the time the tests were conducted. Plasmid pIPH, used as a probe for HSV LATs, was included with each analysis to confirm latent or mock infection in ganglia and to verify the neuronal localization of HSV. DNA probes were generated by radiolabeling plasmids with α-35S-dATP and α-35S-dCTP (Amersham, Arlington Heights, Ill.) via nick translation. Two to six mock-infected or latently HSV infected ganglia were serially cryosectioned (thickness, 6 μm). Sequential sections from at least four different locations in each ganglion were hybridized with the various probes described above.
Real-time RT-PCR.
Real-time reverse transcriptase PCR (RT-PCR) was performed on the ABI Prism 7700 Sequence Detection system (Applied Biosystems, Foster City, Calif.). Primers were designed according to the manufacturer's guidelines. Aliquots of RNA from each sample of pooled ganglia were treated extensively with RQ1 DNase, purified by using a MasterPure RNA Purification kit (Epicentre Technologies, Madison, Wis.), and reverse transcribed with a mixture of gene-specific downstream PCR primers. PCR primers were based on clone sequences and homology data retrieved from the Mouse Genome Database (6). We used SYBR Green fluorescence detection for all assays (2× PCR Master Mix; Applied Biosystems). The specificity of each assay was verified by electrophoretic separation of exponential-phase PCR products on polyacrylamide gels stained with a SYBR Gold nucleic acid gel stain (Molecular Probes, Eugene, Oreg.). Each set of samples analyzed included cDNAs derived from mock- and HSV-infected samples from a given experiment, RT-negative controls, and a panel of primer pairs that always included those for mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and LAT. Gapdh was used as the normalization standard after it was determined that the amounts of mouse Gapdh RNA per total RNA measured spectrophotometrically were comparable in the latently infected and mock-infected samples. LAT was included to confirm the identities of latently versus mock-infected samples.
Threshold cycle numbers (CT) for each specific and reference (Gapdh) cDNA within a given sample were measured at least twice in duplicate or triplicate from each of two cDNA preparations of a given RNA sample. Replicate CTs were within 1 CT of each other. CTs were averaged (AvCT). CT is inversely proportional to the amount of starting material and represents a power of 2; the amount is calculated as 1/(2f)−CT, where f is amplification efficiency. Based on the assumption that each sequence was amplified with comparable efficiency in all mock- and HSV-infected pairs of samples, the difference between the AvCT of the reference gene and that of the specific gene (ΔCT) [AvCT(Gapdh) − AvCT(specific gene) = ΔCT(specific gene)] inversely correlates with the relative amount of the specific gene in a given sample. To compare genes in mock- and HSV-infected samples within each of the three experimental sets, the difference between their ΔCTs was determined [ΔCT(gene in mock-infected sample) − ΔCT(gene in HSV-infected sample) = ΔCT(infection)]. The ratio of gene expression in HSV-infected samples to that in mock-infected samples is calculated as 2ΔCT(infection) for each experimental set.
RESULTS
Microarray analysis of host gene expression in latently infected ganglia.
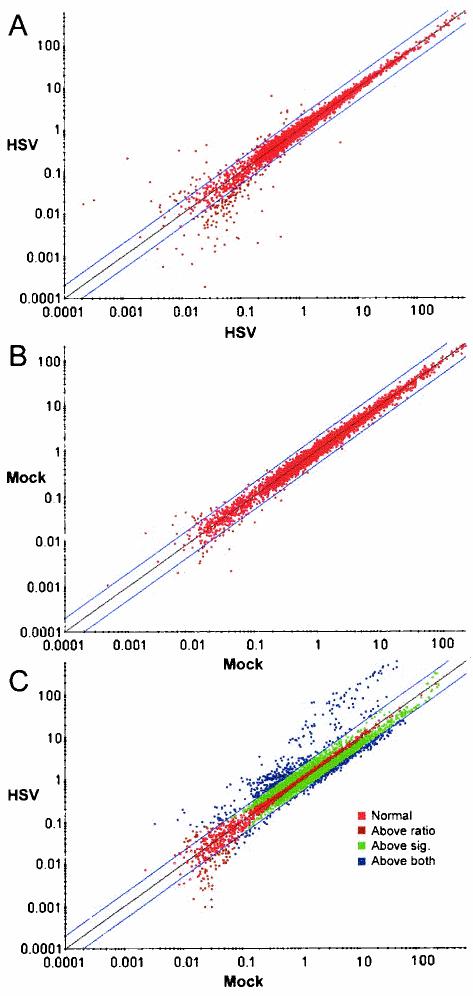
We hypothesized that latent HSV infection alters neuronal gene expression in ways that might affect neuronal physiology and viral gene expression. To investigate whether latent HSV infection alters neuronal gene expression, we conducted transcript profiling using microarray technology. We used a well-characterized mouse model in which HSV-1 strain KOS is inoculated onto a scarified mouse cornea, undergoes productive infection in the corneal epithelium and trigeminal ganglion for several days, and then establishes a latent infection in trigeminal ganglia by day 30 postinoculation (48). RNA was prepared from pooled ganglia (10 to 16 for each sample) harvested as described previously (41) at day 30 postinfection (p.i.). RNA samples from latently and mock-infected mice were used to generate radiolabeled cDNA probes. Probes were hybridized to duplicate nylon filters, with each printed in duplicate with 2,556 PCR-amplified, mostly full length mouse cDNA clones plus quality and normalization control clones. Scatterplots of duplicate hybridizations demonstrated the filter-to-filter consistency of the arrays (Fig. 1A and B), while scatterplots of latently infected versus mock-infected ganglia revealed numerous differences consistent with increases and decreases in host transcript levels (Fig. 1C). Thus, differences in gene expression between the experimental and control groups were detected by microarray analysis.
FIG. 1.
Scatterplots comparing 33P signal intensities in microarrays after background subtraction and normalization. Each point represents the log10 phosphorimager intensity value normalized to the median intensity of the microarray for each filter. (A and B) Results from duplicate array hybridizations. The x and y axes each represent one of the two duplicate arrays. HSV, one representative probe generated from pooled ganglia 30 days after infection. Mock, one representative probe generated from pooled ganglia 30 days after mock infection, and from the same experiment as the HSV probe shown. (C) Results for HSV-infected versus mock-infected ganglia. y axis, one of the two duplicate HSV arrays; x axis, one of the duplicate Mock arrays.
Several factors could potentially limit the confidence with which one can interpret microarray results. First, there is experimental variability of microarray measurements, as has been observed by others (69). Second, only a small fraction of cells in trigeminal ganglia harbor latent HSV, so that even large differences in gene expression in individual infected cells might give rise to very small differences in expression on a whole-ganglion basis. Thus, we wished to evaluate relatively small differences in gene expression. For these reasons, we performed three independent experiments that entailed three separate infections of mice and preparation of cDNA probes to increase the statistical power of the analysis. Microarray sets used for each experiment came from separate printings. Each probe was hybridized on replicate arrays such that each clone was represented at least four times. Differences between latently HSV infected and mock-infected samples were evaluated statistically in each experiment by using a variant of the Student t test in which standard deviations were regularized by an independent estimate of measurement errors. The P values from each experiment were combined, and a correction was applied to account for the total number of genes tested. Only genes whose expression differed in the same direction in all three experiments were included. Differences between latently and mock-infected samples were statistically significant for 51 genes, and 8 of these differences were highly significant (Tables 1 and 2). (The entire annotated list of genes represented in the data set, along with averaged, normalized values and ratios of gene expression in HSV-infected samples to that in mock-infected samples, is available for viewing at http://coen.med.harvard.edu.)
TABLE 1.
Changes in mRNA levels during latent HSV infection following peak expression during acute infectiona
| Geneb | Gene product | MGI accession no. | Functional category | Change (n-fold)c at day 30 p.i.
|
Changes (n-fold) at days 3 and 10 p.i. (peak day p.i.f) | |
|---|---|---|---|---|---|---|
| Arrayd | RT-PCRe | |||||
| H2-D1 | Histocompatibility 2, D region locus 1 | 95896 | Immune response | 4.1 | 9.2, 6.0 (3) | |
| Psmb8 | Proteasome subunit, beta type 8 | 1346527 | Immune response | 3.7* | 20.5, 15.6 (3) | |
| Psmb9 | Proteasome subunit, beta type 9 | 1346526 | Immune response | 2.4 | 9.6, 7.8 (3) | |
| Stat1 | Signal transducer and activator of transcription | 103063 | Gene expression | 2.4 | 7.5 | 14.2, 9.2 (3) |
| C2 | Complement component 2 (within H-2S) | 88226 | Immune response | 2.0* | 7.0, 3.7 (3) | |
| Prss12 | Protease, serine, 12 neurotrypsin | 1100881 | Axonal remodeling | 2.0 | 6.0, 4.8 (3) | |
| Flt1 | FMS-like tyrosine kinase 1 | 95558 | Immune response | 1.9 | 9.4, 3.3 (3) | |
| C1r | Complement component 1, r subcomponent | 1355313 | Immune response | 1.6 | 4.8, 2.5 (3) | |
| Cxcr6 | Chemokine (C-X-C) receptor 6 | 1934582 | Immune response | 4.8 | 1.4, 5.7 (10) | |
| B2m | β2 microglobulin | 88127 | Immune response | 4.5* | 8.3, 13.4 (10) | |
| Ii | Ia-associated invariant chain | 96534 | Immune response | 4.3* | 1.8, 14.5 (10) | |
| C3 | Complement component 3 | 88227 | Immune response | 3.2 | 12.9, 13.3 (10) | |
| Itgb2 | Integrin beta 2 | 96611 | Immune response | 3.1 | 3.1, 9.1 (10) | |
| Cd3g | CD3 antigen, gamma polypeptide | 88333 | Immune response | 3.1* | 0.68, 3.1 (10) | |
| Hipk2 | Homeodomain-interacting protein kinase 2 | 1314872 | Gene expression | 3.0* | 5.0 | 1.6, 9.1 (10) |
| Man2b1 | Mannosidase 2, alpha B1 | 107286 | Turnover | 2.3 | 1.0, 3.0 (10) | |
| Ptprc | Protein tyrosine phosphatase, receptor, C | 97810 | Immune response | 2.2* | 5.6, 10.2 (10) | |
| Cd68 | CD68 antigen | 88342 | Immune response | 2.1 | 1.3, 8.4 (10) | |
| Lck | Lymphocyte protein tyrosine kinase | 96756 | Immune response | 1.9 | 1.7, 3.3 (10) | |
| Cd3d | CD3 antigen, delta polypeptide | 88331 | Immune response | 1.9 | 0.83, 3.0 (10) | |
| Axl | AXL receptor tyrosine kinase | 1347244 | Immune response | 1.9 | 1.9, 3.4 (10) | |
| Lyn | Yamaguchi sarcoma viral oncogene homolog | 96892 | Immune response | 1.9* | 2.7, 4.2 (10) | |
| Apobec1 | Apolipoprotein B editing complex 1 | 103298 | Gene expression | 1.9 | 1.8 | 2.4, 5.1 (10) |
| Ctsd | Cathepsin D | 88562 | Turnover | 1.9 | 0.84, 2.8 (10) | |
| Fcgr3 | Fc receptor, IgG, low affinity III | 95500 | Immune response | 1.8 | 1.6, 6, 1 (10) | |
| I12rg | Interleukin 2 receptor, gamma chain | 96551 | Immune response | 1.8 | 3.2, 5.5 (10) | |
| Lgals3 | Lectin, galactose binding, soluble 3 | 96778 | Immune response | 1.7 | 2.1, 11.0 (10) | |
| Grn | Granulin | 95832 | Immune response | 1.6 | 3.8, 4.3 (10) | |
| Aebp1 | AE binding protein 1 | 1197012 | Gene expression | 1.6 | 0.98, 1.9 (10) | |
Peak changes were observed at day 3 or 10 p.i.
Identified as top BLAST hit of microarray clone sequence. Gene designations, products, and accession numbers are those in the Mouse Genome Informatics (MGI) database.
Asterisks indicate highly significant differences between mock- and HSV-infected samples in three experiments. For all other values, differences are significant.
Average microarray ratio derived from three experiments.
Average RT-PCR ratio from three experiments.
Results from one experiment.
TABLE 2.
Changes in mRNA levels associated with latent HSV infectiona
| Geneb | Gene product | MGI accession no. | Functional category | Change (n fold) at day 30 p.i.
|
|
|---|---|---|---|---|---|
| Arrayc | RT-PCRd | ||||
| Traf3 | TNF receptor-associated factor 3 | 108041 | Immune response | 8.2 | |
| Gprc1g | G protein-coupled receptor, family C, 1, G | 1351344 | Neurotransmission | 8.1 | 2.3 |
| Trc8 | Translocated in renal cancer 8 | Signaling | 7.7 | ||
| Clk2 | CDC-like kinase2 | 1098669 | Gene expression | 7.3 | 4.3 |
| Chrm1 | Cholinergic receptor, muscarinic 1, central nervous system | 88396 | Neurotransmission | 4.9 | 6.3 |
| Ptprs | Protein tyrosine phosphatase, receptor S | 97815 | Signaling | 3.9 | |
| Gabbr1 | γ-Aminobutyric acid receptor, 1 | 1860139 | Neurotransmission | 2.8 | |
| Kcncl | Potassium voltage-gated channel, Shaw 1 | 96667 | Ion channel | 2.6 | 2.4 |
| Scyb14 | Small inducible cytokine subfamily B, 14 | 1888514 | Immune response | 2.2 | |
| Ulk1 | Unc-51-like kinase 1 | 1270126 | Axonal remodeling | 2.1 | |
| Adrbk1 | Adrenergic receptor kinase, β1 | 87940 | Signaling | 1.9 | |
| Kcnab2 | Potassium voltage-gated channel, shaker β2 | 109239 | Ion channel | 1.8 | |
| Col15al | Procollagen, type XV | 88449 | Axonal remodeling | 1.8 | |
| Gpr56 | G protein-coupled receptor 56 | 1340051 | Signaling | 1.7 | 1.5 |
| Adam23 | A disintegrin and metalloprotease domain 23 | 345162 | Axonal remodeling | 1.5 | |
| Ppp3ca | Protein phosphatase 3, catalytic subunit | 107164 | Signaling | 0.7 | |
| Usp20 | Ubiquitin specific protease 20 | 1921520 | Turnover | 0.7 | |
| Chn1 | n-Chimaerin | 1923974 | Axonal remodeling | 0.7 | 0.6 |
| Rgs4 | Regulator of G-protein signaling 4 | 108409 | Signaling | 0.6 | 0.6 |
| Rab27b | RAB27b, member of Ras oncogene family | 1931295 | Signaling | 0.6 | |
| Ubelx | Ubiquitin-activating enzyme E1, Chr X | 98890 | Turnover | 0.6 | |
| Acox2 | Acyl-coenzyme A oxidase 2, branched chain | 97381 | Turnover | 0.5 | |
| Gh | Growth hormone | 95707 | Signaling | 0.3 | 0.3 |
Peak changes were observed at day 30 p.i.
Identified as top BLAST hit of microarray clone sequence. Gene designations, products, and accession numbers are those in the Mouse Genome Informatics (MGI) database. There is no MBI accession no. for Trc8. The accession no. for human Trc8 is BC021571.
Average microarray ratio derived from three experiments.
Average RT-PCR ratio from three experiments.
Altered expression of immune response genes.
Infiltration of inflammatory cells in latently infected ganglionic tissue has been observed (53, 78). Thus, it was not surprising that about half of the 51 genes in Tables 1 and 2 are involved in the immune response and that all of these immune response genes exhibited increased expression in latently infected versus mock-infected ganglia. This group included genes expressed specifically by immune cells, such as the CD3 subunit gene Cd3g (27), and/or in target cells involved in antigen presentation or in responding to immune stimuli, such as the major histocompatibility complex class I genes H2-D1 and β2 microglobulin, which can be expressed in the nervous system (19, 39, 63). These results confirm and extend previous observations of a persistent immune response in latently infected ganglia (7, 9, 28, 53, 78), thus validating the array results.
Alteration of neuronal gene expression.
Strikingly, a surprising number of the genes in Tables 1 and 2 (mostly in Table 2) are known to be expressed in nervous system tissue and/or neurons. Twenty-one genes of this class exhibited increased expression in latently infected versus mock-infected ganglia, and eight exhibited decreased expression. Many of the genes whose expression was altered play definite roles in neuronal physiology. These include genes encoding neurotransmitter receptors, such as Gprc1g (75), Chrm1 (30), and Gabbr1 (16); voltage-gated ion channels, such as Kcnc1 (57, 82) and Kcnab (13, 58); proteins involved in signaling, including Trc8 (8, 24), Ptprs (21, 32), Adrbk1 (2, 5), Gpr56 (51), Ppp3Ca (95), Rgs4 (64, 65), Rab27b (11, 70, 94), and Gh (4, 18); proteins involved in neurite extension, axonal elongation, and cell-matrix interactions (“axonal remodeling”), such as Prss12 (26), Ulk1 (66, 88), Col15a1 (56, 76), Adam23 (37, 74), and n-chimaerin (29, 50); and proteins involved in catabolic functions, such as Man2b1 (84), Ctsd (1), Usp20 (49), Ube1x (89), and Acox2 (35, 40). Several genes, such as Stat1 (17, 55), Hipk2 (38, 67), Apobec1 (61, 80), and Clk2 (20, 59), are involved in gene expression. One gene, Aebp1, encodes a protein described variously as being involved in either gene expression or axonal remodeling or both (93).
In situ hybridization analysis was performed on a few genes that appeared significantly altered in all statistical analyses in an attempt to determine the cell types that expressed the cellular transcripts. Neuronal expression of Chrm1 was observed in sensory neurons in mock-infected (data not shown) and latently infected (Fig. 2A) ganglia. Neuronal expression of Ptprs was also observed in mock-infected (data not shown) and latently infected (Fig. 2B) ganglia. Finally, neuronal expression of Kcnc 1 was observed in mock-infected and latently infected ganglia (data not shown). We confirmed that these ganglia were latently infected by in situ hybridization with a LAT probe (data not shown). Specific neuronal expression of another gene, Stat1, in mouse trigeminal ganglia has recently been shown by immunohistochemistry (43). Although expression of Gh in neurons has not been documented to our knowledge, the expression of Gh receptors and target genes has been demonstrated (31).
FIG. 2.
Neuronal expression of host genes confirmed by in situ hybridization. Mouse trigeminal ganglia latently infected with HSV at 30 days p.i. hybridized with radiolabeled DNA probes for cellular genes. Neurons were identified morphologically by their large size and staining pattern. (A) Muscarinic acetylcholine receptor gene (Chrm1). Grains indicate nuclear and cytoplasmic distribution of the mRNA in neurons (magnification, ×200). (B) Protein tyrosine phosphatase sigma gene (Ptprs). Grains cluster over neurons (magnification, ×400).
Confirmation by real-time RT-PCR.
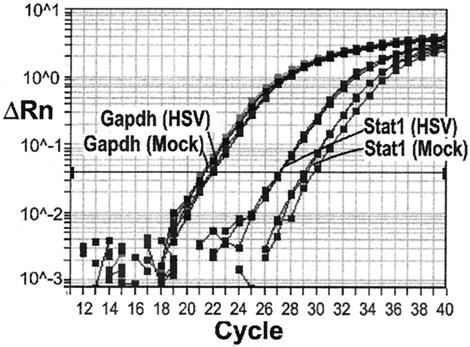
To verify these changes in gene expression by an alternate method, we quantified the expression of 11 genes (Tables 1 and 2) representing various neuronal functions by RT-PCR using real-time technology. Mouse Gapdh was assayed as a normalization control. As shown in Fig. 3, Stat1 transcripts, which were more abundant in latently infected than in mock-infected ganglia in the three array analyses, were also reproducibly more abundant in latently infected than in mock-infected ganglia relative to the abundance of Gapdh transcripts in the RT-PCR assay. The mean ratios of expression in latently infected ganglia to expression in mock-infected ganglia were of similar magnitude by the two assays. Thus, we verified changes in expression of specific neuronal genes in latently infected ganglia by RT-PCR.
FIG. 3.
Quantitative real-time RNA PCR. This example illustrates that the level of Stat1 RNA normalized to Gapdh RNA is higher in latently infected ganglia (HSV) than in mock-infected ganglia (Mock). ΔRn, fluorescence difference from background determined by using Sequence Detection System software (Applied Biosystems). Each assay was performed in triplicate. The relative level of Stat1 was determined by the difference in average cycle number at a constant threshold ΔRn. Gapdh, mouse Gapdh mRNA; Stat1, mouse Stat1 mRNA. View is expanded around exponential phase.
Time course of host gene expression.
The altered levels of specific mouse RNAs in latently infected ganglia on day 30 p.i. could reflect changes in cellular gene expression specific to the latent state or changes induced initially during acute viral replication. To distinguish between these possibilities, in one of the three experiments we harvested trigeminal ganglia from HSV- and mock-infected mice on days 3 and 10 p.i., and we assayed the RNA by using the same arrays used for the 30-day samples. Limiting the analysis just to the 52 genes in Table 1, those whose expression was greater in latently infected ganglia formed two expression profile clusters: one, of special interest, comprising 15 genes showing maximal expression on day 30 p.i. (Fig. 4 and Table 2) and the other showing maximal expression on either day 3 or day 10 p.i. (Fig. 4 and Table 1). As expected, most of the host immune response genes exhibited peak expression on day 3 or 10 p.i., with peak levels of lymphocyte markers such as Cd3d, Cd3g, Cxcr6, Lyn, Axl, Il2rg, Lck, and Ptprc at day 10 p.i. (Many other genes known to be involved in classical host immune responses exhibited increased expression at days 3 and 10, as expected. Only those whose expression was significantly increased at day 30 are examined here.) The peak expression of Stat1 on day 3 p.i. was consistent with high expression of cytokines at this time (7, 9, 28, 53). In contrast, most of the genes that are known to be expressed relatively abundantly in nervous tissue and/or neurons, including those shown by in situ hybridization to be specifically expressed in neurons of latently infected trigeminal ganglia, were expressed in greatest abundance on day 30 p.i., well after cessation of acute viral replication in ganglia. Additionally, the eight genes whose expression was lower in latently infected ganglia all exhibited the greatest decrease relative to levels in mock-infected ganglia at day 30 p.i. (Table 2). Thus, taken together, these data show that latent infection rather than acute infection was associated with altered expression of these neuronal genes.
FIG. 4.
Kinetic expression profiles of selected genes whose up-regulation at day 30 p.i. correlated significantly with HSV latency. HSV/Mock, ratios of mean normalized signal intensities in HSV-infected samples to those in mock-infected samples.. Each bar for day 3 or 10 p.i. represents an average ratio from one experiment, and each bar for day 30 p.i. represents average ratios from all three experiments.
DISCUSSION
An important problem in virology is to understand the interaction of latent viral infections with the host cell and to identify host responses to virus latency. In these studies, we observed up-regulation or down-regulation during latent infection of the levels of a number of host genes, including several thought to be expressed specifically in neurons. Our confidence that levels of expression of the genes listed in Tables 1 and 2 are truly altered is based on stringent statistical criteria applied to the microarray hybridization data coupled with validation by quantitative real-time RT-PCR. Evidence that expression of the genes listed in Table 2 is altered only during latent infection and that these changes are not remnants of acute infection is provided by the results in Fig. 4. In contrast to our results, Hill and colleagues (34) did not identify changes in gene expression between uninfected and latently infected ganglia. One reason for the differing results could be that they looked at a total of only 149 mouse genes in a “stress/toxicology” set, whereas our data set included 2,500 genes in a diverse set.
Potential effects of latent HSV infection on host gene expression.
Our microarray data reflect changes in gene expression in the entire trigeminal ganglion, which is a heterogeneous tissue composed of many cell types. The alterations in expression of neuron-specific genes observed in these studies could represent a direct effect of HSV on cellular gene expression in latently infected cells. Alternatively, the virus may act indirectly on cellular gene expression, for example, by inducing neuronal injury or by inducing a persistent immune response to latent infection that includes expression of cytokines known to alter host gene expression. The increased expression of Stat1 is consistent with the latter mechanism. Although it is easier to explain the decreased expression of certain genes by such an indirect mechanism, to date, little or no precedent exists for immune modulation of neuronal gene expression. Direct and indirect effects of latent HSV infection on cellular gene expression are not necessarily mutually exclusive. These two mechanisms make specific predictions about the levels of expression of particular cellular genes in latently infected versus uninfected neurons. Preliminary studies using in situ hybridization to examine this issue have been inconclusive due to the qualitative nature of this technique and the apparent variability of expression from cell to cell. Thus, PCR-based methods to examine individual cells may be necessary to test these models.
Potential effects of changes in host gene expression on viral gene expression.
Alternatively, changes in cellular gene expression may represent a host response that contributes to the repression of productive-cycle HSV gene expression characteristic of latent infection. Of particular interest are cellular genes encoding proteins that regulate gene expression. Aebp1 has been reported to be a transcriptional repressor, and Hipk2 has been reported to be a corepressor (38, 72). A relative of Clk2, Clk1, has been reported to induce neuronal differentiation of PC12 cells in a manner akin to nerve growth factor (62), which helps to maintain HSV latency in both in vitro and in vivo models (33, 92). The increased expression of Apobec1, an RNA-editing enzyme, is especially intriguing, given the abundant expression of LAT, an RNA of uncertain coding capacity, in latently infected neurons. Altered expression of genes may provide clues to changes in signaling pathways that regulate gene expression. The increased expression of Chrm1 is interesting in that it is ordinarily down-regulated by ciliary neurotrophic factor (CNTF), which has been reported to promote HSV reactivation in humans (42). Certain cellular genes may maintain the virus in a state of readiness for reactivation. Stat1 may also be in this category, as it is activated by CNTF. Other neuronal genes whose expression is altered could also influence viral gene expression less directly. For example, Chrm1 couples to G proteins that can activate mitogen-activated protein kinase cascades that lead to changes in gene expression (30). These various hypotheses make specific, testable predictions regarding the effects of knocking out or overexpressing these genes in latently infected ganglia. Cellular proteins that help to maintain latency or promote reactivation could potentially serve as targets for therapeutic intervention.
Potential implications for HSV infections in humans.
Given that the expression of several genes with known roles in neurotransmission and signaling is altered in latently infected ganglia, our results, using a mouse model and a virus strain with which no spontaneous reactivation has ever been observed (9), raise the possibility that latent HSV can affect sensation. Moreover, several of the genes whose expression is altered, including Gprc1g, Gabbr1, Kcnab2, and Kcnc1, affect neuronal excitability in an interconnected manner (13, 16, 36, 57, 58, 71, 75, 82), which suggests a mechanism by which changes in neuronal physiology are induced by latent virus and could alter sensation. This predicts that neurons in latently infected ganglia would exhibit alterations in ion channel function such as have been observed in neurons in vitro (86). Consistent with this idea, postherpetic allodynia and hyperalgesia have been observed in mice latently infected with HSV for at least 40 days p.i. (87). Interestingly, there are reports of patients with recurrent HSV with symptoms of abnormal sensation, including pain, both before and even long after clinically evident disease (14, 25, 45, 46, 77). Although these symptoms are often ascribed to reactivating HSV (91), or could be due to sensory nerve or central nervous system injury, our results provoke the speculation that these clinical phenomena and perhaps more subtle changes in sensation may be due to effects of latent HSV infection on sensory neurons. These studies demonstrate that latent herpes simplex virus infection can alter neuronal gene expression and might provide a new mechanism for how persistent viral infection can cause chronic disease.
Acknowledgments
This work was supported by NIH grant PO1 NS 35138, a grant from Millennium Pharmaceuticals, and an institutional grant from the Howard Hughes Medical Institute Biomedical Research Support Program for Medical Schools.
We thank M. Chen for initial work on this project; A. Marshall for assistance with RT-PCR; and M. Greenberg, P. Schaffer, T. Peng, and members of the Knipe and Coen laboratories for helpful discussions. We also acknowledge the contributions of numerous colleagues at Millennium Pharmaceuticals, particularly T. Burwell, M. Cloutier, L. Rudolf-Owen, F. Spaltmann, R. Walker, and T. Wilson.
REFERENCES
- 1.Adamec, E., P. S. Mohan, A. M. Cataldo, J. P. Vonsattel, and R. A. Nixon. 2000. Up-regulation of the lysosomal system in experimental models of neuronal injury: implications for Alzheimer's disease. Neuroscience 100:663-675. [DOI] [PubMed] [Google Scholar]
- 2.Arriza, J. L., T. M. Dawson, R. B. Simerly, L. J. Martin, M. G. Caron, S. H. Snyder, and R. J. Lefkowitz. 1992. The G-protein-coupled receptor kinases beta ARK1 and beta ARK2 are widely distributed at synapses in rat brain. J. Neurosci. 12:4045-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. [DOI] [PubMed] [Google Scholar]
- 4.Bartke, A., V. Chandrashekar, D. Turyn, R. W. Steger, L. Debeljuk, T. A. Winters, J. A. Mattison, N. A. Danilovich, W. Croson, D. R. Wernsing, and J. J. Kopchick. 1999. Effects of growth hormone overexpression and growth hormone resistance on neuroendocrine and reproductive functions in transgenic and knock-out mice. Proc. Soc. Exp. Biol. Med. 222:113-123. [DOI] [PubMed] [Google Scholar]
- 5.Benovic, J. L., J. J. Onorato, J. L. Arriza, W. C. Stone, M. Lohse, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, M. G. Caron, and R. J. Lefkowitz. 1991. Cloning, expression, and chromosomal localization of beta-adrenergic receptor kinase 2. A new member of the receptor kinase family. J. Biol. Chem. 266:14939-14946. [PubMed] [Google Scholar]
- 6.Blake, J. A., J. E. Richardson, C. J. Bult, J. A. Kadin, J. T. Eppig, and the Mouse Genome Database Group. 2002. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res. 30:113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantin, E. M., D. R. Hinton, J. Chen, and H. Openshaw. 1995. Gamma interferon expression during acute and latent nervous system infection by herpes simplex virus type 1. J. Virol. 69:4898-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charytoniuk, D., B. Porcel, J. R. Gomez, H. Faure, M. Ruat, and E. Traiffort. 2002. Sonic hedgehog signalling in the developing and adult brain. J. Physiol. (Paris) 96:9-16. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S. H., D. A. Garber, P. A. Schaffer, D. M. Knipe, and D. M. Coen. 2000. Persistent elevated expression of cytokine transcripts in ganglia latently infected with herpes simplex virus in the absence of ganglionic replication or reactivation. Virology 278:207-216. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S. H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y., P. Samaraweera, T. T. Sun, G. Kreibich, and S. J. Orlow. 2002. Rab27b association with melanosomes: dominant negative mutants disrupt melanosomal movement. J. Investig. Dermatol. 118:933-940. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, L. W., J. M. Grenier, L. Ettwiller, L. P. Jenkins, D. Ficenec, J. Martin, F. Jin, P. S. DiStefano, and A. Wood. 2001. An orchestrated gene expression component of neuronal programmed cell death revealed by cDNA array analysis. Proc. Natl. Acad. Sci. USA 98:2814-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, J. A., M. Arai, E. L. Prak, S. A. Brooks, L. H. Young, and M. B. Prystowsky. 1992. Characterization of a novel mRNA expressed by neurons in mature brain. J. Neurosci. Res. 31:273-284. [DOI] [PubMed] [Google Scholar]
- 14.Constantine, V. S., R. D. Francis, and L. F. Montes. 1968. Association of recurrent herpes simplex with neuralgia. JAMA 205:181-183. [PubMed] [Google Scholar]
- 15.Cummings, C. A., and D. A. Relman. 2000. Using DNA microarrays to study host-microbe interactions. Genomics 6:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, A. M., T. R. Henion, and S. A. Tobet. 2002. Gamma-aminobutyric acid B receptors and the development of the ventromedial nucleus of the hypothalamus. J. Comp. Neurol. 449:270-280. [DOI] [PubMed] [Google Scholar]
- 17.De-Fraja, C., L. Conti, S. Govoni, F. Battaini, and E. Cattaneo. 2000. STAT signalling in the mature and aging brain. Int. J. Dev. Neurosci. 18:439-446. [DOI] [PubMed] [Google Scholar]
- 18.delaTorre, J. C., and M. B. Oldstone. 1992. Selective disruption of growth hormone transcription machinery by viral infection. Proc. Natl. Acad. Sci. USA 89:9939-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorries, R. J. 2001. The role of T-cell-mediated mechanisms in virus infections of the nervous system. Curr. Top. Microbiol. Immunol. 253:219-245. [DOI] [PubMed] [Google Scholar]
- 20.Duncan, P. I., D. F. Stojdl, R. M. Marius, K. H. Scheit, and J. C. Bell. 1998. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res. 241:300-308. [DOI] [PubMed] [Google Scholar]
- 21.Elchebly, M., J. Wagner, T. E. Kennedy, C. Lanctot, E. Michaliszyn, A. Itie, J. Drouin, and M. L. Tremblay. 1999. Neuroendocrine dysplasia in mice lacking protein tyrosine phosphatase sigma. Nat. Genet. 21:330-333. [DOI] [PubMed] [Google Scholar]
- 22.Feldman, L. T., A. R. Ellison, C. C. Voytek, L. Yang, P. Krause, and T. P. Margolis. 2002. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 99:978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gemmill, R. M., J. D. West, F. Boldog, N. Tanaka, L. J. Robinson, D. I. Smith, F. Li, and H. A. Drabkin. 1998. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl. Acad. Sci. USA 95:9572-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzales, G. R. 1992. Postherpes simplex type 1 neuralgia simulating postherpetic neuralgia. J. Pain Symptom Manage. 7:320-323. [DOI] [PubMed] [Google Scholar]
- 26.Gschwend, T. P., S. R. Krueger, S. V. Kozlov, D. P. Wolfer, and P. Sonderegger. 1997. Neurotrypsin, a novel multidomain serine protease expressed in the nervous system. Mol. Cell Neurosci. 9:207-219. [DOI] [PubMed] [Google Scholar]
- 27.Haks, M. C., P. Krimpenfort, J. Borst, and A. M. Kruisbeek. 1998. The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J. 17:1871-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halford, W. P., B. M. Gebhardt, and D. J. Carr. 1996. Persistent cytokine expression in trigeminal ganglion latently infected with herpes simplex virus type 1. J. Immunol. 157:3542-3549. [PubMed] [Google Scholar]
- 29.Hall, C., G. J. Michael, N. Cann, G. Ferrari, M. Teo, T. Jacobs, C. Monfries, and L. Lim. 2001. α2-Chimaerin, a Cdc42/Rac1 regulator, is selectively expressed in the rat embryonic nervous system and is involved in neuritogenesis in N1E-115 neuroblastoma cells. J. Neurosci. 21:5191-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton, S. E., and N. M. Nathanson. 2001. The M1 receptor is required for muscarinic activation of mitogen-activated protein (MAP) kinase in murine cerebral cortical neurons. J. Biol. Chem. 276:15850-15853. [DOI] [PubMed] [Google Scholar]
- 31.Harvey, S., I. Lavelin, and M. Pines. 2002. Growth hormone (GH) action in the brain: neural expression of a GH-response gene. J. Mol. Neurosci. 18:89-95. [DOI] [PubMed] [Google Scholar]
- 32.Hendriks, W., J. Schepens, C. Brugman, P. Zeeuwen, and B. Wieringa. 1995. A novel receptor-type protein tyrosine phosphatase with a single catalytic domain is specifically expressed in mouse brain. Biochem. J. 305:499-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hill, J. M., H. H. Garza, Jr., M. F. Helmy, S. D. Cook, P. A. Osborne, E. M. Johnson, Jr., H. W. Thompson, L. C. Green, R. J. O'Callaghan, and B. M. Gebhardt. 1997. Nerve growth factor antibody stimulates reactivation of ocular herpes simplex virus type 1 in latently infected rabbits. J. Neurovirol. 3:206-211. [DOI] [PubMed] [Google Scholar]
- 34.Hill, J. M., W. J. Lukiw, B. M. Gebhardt, S. Higaki, J. M. Loutsch, M. E. Myles, H. W. Thompson, B. S. Kwon, N. G. Bazan, and H. E. Kaufman. 2001. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes 23:273-280. [DOI] [PubMed] [Google Scholar]
- 35.Huyghe, S., M. Casteels, A. Janssen, L. Meulders, G. P. Mannaerts, P. E. Declercq, P. P. Van Veldhoven, and M. Baes. 2001. Prenatal and postnatal development of peroxisomal lipid-metabolizing pathways in the mouse. Biochem. J. 353:673-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang, J. H., and T. L. Yaksh. 1997. The effect of spinal GABA receptor agonists on tactile allodynia in a surgically-induced neuropathic pain model in the rat. Pain 70:15-22. [DOI] [PubMed] [Google Scholar]
- 37.Karkkainen, I., E. Rybnikova, M. Pelto-Huikko, and A. P. Huovila. 2000. Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Mol. Cell Neurosci. 15:547-560. [DOI] [PubMed] [Google Scholar]
- 38.Kim, Y. H., C. Y. Choi, S. J. Lee, M. A. Conti, and Y. Kim. 1998. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 273:25875-25879. [DOI] [PubMed] [Google Scholar]
- 39.Kimura, T., and D. E. Griffin. 2000. The role of CD8+ T cells and major histocompatibility complex class I expression in the central nervous system of mice infected with neurovirulent Sindbis virus. J. Virol. 74:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoll, A., J. Salles, F. Sargueil, C. Cassagne, and B. Garbay. 2000. Peroxisomal beta-oxidation enzyme gene expression in the developing mouse brain. Neurosci. Lett. 285:201-204. [DOI] [PubMed] [Google Scholar]
- 41.Kramer, M. F., and D. M. Coen. 1995. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 69:1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriesel, J. D., B. Araneo, J. P. Petajan, S. L. Spruance, and S. Stromatt. 1994. Herpes labialis associated with recombinant human ciliary neurotrophic factor. J. Infect. Dis. 170:1046. [DOI] [PubMed] [Google Scholar]
- 43.Kriesel, J. D., B. B. Jones, I. P. Hwang, K. M. Dahms, and S. L. Spruance. 2001. Signal transducers and activators of transcription (Stat) are detectable in mouse trigeminal ganglion neurons. J. Interferon Cytokine Res. 21:445-450. [DOI] [PubMed] [Google Scholar]
- 44.Kristie, T. M., J. L. Vogel, and A. E. Sears. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc. Natl. Acad. Sci. USA 96:1229-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krohel, G. B., J. R. Richardson, and D. F. Farrell. 1976. Herpes simplex neuropathy. Neurology 26:596-597. [DOI] [PubMed] [Google Scholar]
- 46.Layzer, R. B., and M. A. Conant. 1974. Neuralgia in recurrent herpes simplex. Arch. Neurol. 31:233-237. [DOI] [PubMed] [Google Scholar]
- 47.Lazarov, N. E. 2002. Comparative analysis of the chemical neuroanatomy of the mammalian trigeminal ganglion and mesencephalic nucleus. Prog. Neurobiol. 66:19-59. [DOI] [PubMed] [Google Scholar]
- 48.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, Z., D. Wang, X. Na, S. R. Schoen, E. M. Messing, and G. Wu. 2002. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem. Biophys. Res. Commun. 294:700-709. [DOI] [PubMed] [Google Scholar]
- 50.Lim, H. H., G. J. Michael, P. Smith, L. Lim, and C. Hall. 1992. Developmental regulation and neuronal expression of the mRNA of rat n-chimaerin, a p21rac GAP:cDNA sequence. Biochem. J. 287:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, M., R. M. Parker, K. Darby, H. J. Eyre, N. G. Copeland, J. Crawford, D. J. Gilbert, G. R. Sutherland, N. A. Jenkins, and H. Herzog. 1999. GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics 55:296-305. [DOI] [PubMed] [Google Scholar]
- 52.Liu, T., K. M. Khanna, X. Chen, D. J. Fink, and R. L. Hendricks. 2000. CD8+ T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. J. Exp. Med. 191:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, T., Q. Tang, and R. L. Hendricks. 1996. Inflammatory infiltration of the trigeminal ganglion after herpes simplex virus type 1 corneal infection. J. Virol. 70:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 55.Maier, J., C. Kincaid, A. Pagenstecher, and I. L. Campbell. 2002. Regulation of signal transducer and activator of transcription and suppressor of cytokine-signaling gene expression in the brain of mice with astrocyte-targeted production of interleukin-12 or experimental autoimmune encephalomyelitis. Am. J. Pathol. 160:271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marneros, A. G., and B. R. Olsen. 2001. The role of collagen-derived proteolytic fragments in angiogenesis. Matrix Biol. 20:337-345. [DOI] [PubMed] [Google Scholar]
- 57.Massengill, J. L., M. A. Smith, D. I. Son, and D. K. O'Dowd. 1997. Differential expression of K4-AP currents and Kv3.1 potassium channel transcripts in cortical neurons that develop distinct firing phenotypes. J. Neurosci. 17:3136-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCormack, K., J. Connor, L. Zhou, L. Ho, B. Ganetzky, S. Chiu, and A. Messing. 2002. Genetic analysis of the mammalian K+ channel beta subunit Kvβ2 (Kcnab2). J. Biol. Chem. 277:13219-13228. [DOI] [PubMed] [Google Scholar]
- 59.Moeslein, F. M., M. P. Myers, and G. E. Landreth. 1999. The CLK family kinases, CLK1 and CLK2, phosphorylate and activate the tyrosine phosphatase, PTP-1B. J. Biol. Chem. 274:26697-26704. [DOI] [PubMed] [Google Scholar]
- 60.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukhopadhyay, D., S. Anant, R. M. Lee, S. Kennedy, D. Viskochil, and N. O. Davidson. 2002. C→U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am. J. Hum. Genet. 70:38-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Myers, M. P., M. B. Murphy, and G. Landreth. 1994. The dual-specificity CLK kinase induces neuronal differentiation of PC12 cells. Mol. Cell. Biol. 14:6954-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neumann, H., H. Schmidt, A. Cavalie, D. Jenne, and H. Wekerle. 1997. Major histocompatibility complex (MHC) class I gene expression in single neurons of the central nervous system: differential regulation by interferon (IFN)-gamma and tumor necrosis factor (TNF)-alpha. J. Exp. Med. 185:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni, Y. G., S. J. Gold, P. A. Iredale, R. Z. Terwilliger, R. S. Duman, and E. J. Nestler. 1999. Region-specific regulation of RGS4 (regulator of G-protein-signaling protein type 4) in brain by stress and glucocorticoids: in vivo and in vitro studies. J. Neurosci. 19:3674-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nomoto, S., K. Adachi, L. X. Yang, Y. Hirata, S. Muraguchi, and K. Kiuchi. 1997. Distribution of RGS4 mRNA in mouse brain shown by in situ hybridization. Biochem. Biophys. Res. Commun. 241:281-287. [DOI] [PubMed] [Google Scholar]
- 66.Okazaki, N., J. Yan, S. Yuasa, T. Ueno, E. Kominami, Y. Masuho, H. Koga, and M. Muramatsu. 2000. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 85:1-12. [DOI] [PubMed] [Google Scholar]
- 67.Pierantoni, G. M., A. Bulfone, F. Pentimalli, M. Fedele, R. Iuliano, M. Santoro, L. Chiariotti, A. Ballabio, and A. Fusco. 2002. The homeodomain-interacting protein kinase 2 gene is expressed late in embryogenesis and preferentially in retina, muscle, and neural tissues. Biochem. Biophys. Res. Commun. 290:942-947. [DOI] [PubMed] [Google Scholar]
- 68.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 69.Pritchard, C. C., L. Hsu, J. Delrow, and P. S. Nelson. 2001. Project normal: defining normal variance in mouse gene expression. Proc. Natl. Acad. Sci. USA 98:13266-13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramalho, J. S., T. Tolmachova, A. N. Hume, A. McGuigan, C. Y. Gregory-Evans, C. Huxley, and M. C. Seabra. 2001. Chromosomal mapping, gene structure and characterization of the human and murine RAB27B gene. BMC Genet. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasband, M. N., E. W. Park, T. W. Vanderah, J. Lai, F. Porreca, and J. S. Trimmer. 2001. Distinct potassium channels on pain-sensing neurons. Proc. Natl. Acad. Sci. USA 98:13373-13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ro, H. S., S. W. Kim, D. Wu, C. Webber, and T. E. Nicholson. 2001. Gene structure and expression of the mouse adipocyte enhancer-binding protein. Gene 280:123-133. [DOI] [PubMed] [Google Scholar]
- 73.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 74.Sagane, K., K. Yamazaki, Y. Mizui, and I. Tanaka. 1999. Cloning and chromosomal mapping of mouse ADAM11, ADAM22 and ADAM23. Gene 236:79-86. [DOI] [PubMed] [Google Scholar]
- 75.Sansig, G., T. J. Bushell, V. R. Clarke, A. Rozov, N. Burnashev, C. Portet, F. Gasparini, M. Schmutz, K. Klebs, R. Shigemoto, P. J. Flor, R. Kuhn, T. Knoepfel, M. Schroeder, D. R. Hampson, V. J. Collett, C. Zhang, R. M. Duvoisin, G. L. Collingridge, and H. van Der Putten. 2001. Increased seizure susceptibility in mice lacking metabotropic glutamate receptor 7. J. Neurosci. 21:8734-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sasaki, T., H. Larsson, D. Tisi, L. Claesson-Welsh, E. Hohenester, and R. Timpl. 2000. Endostatins derived from collagens XV and XVIII differ in structural and binding properties, tissue distribution and anti-angiogenic activity. J. Mol. Biol. 301:1179-1190. [DOI] [PubMed] [Google Scholar]
- 77.Selmanowitz, V. J. 1971. Neurocutaneous herpes simplex. Int. J. Dermatol. 10:227-232. [DOI] [PubMed] [Google Scholar]
- 78.Shimeld, C., J. L. Whiteland, S. M. Nicholls, E. Grinfeld, D. L. Easty, H. Gao, and T. J. Hill. 1995. Immune cell infiltration and persistence in the mouse trigeminal ganglion after infection of the cornea with herpes simplex virus type 1. J. Neuroimmunol. 61:7-16. [DOI] [PubMed] [Google Scholar]
- 79.Simmons, A., D. Tscharke, and P. Speck. 1992. The role of immune mechanisms in control of herpes simplex virus infection of the peripheral nervous system. Curr. Top. Microbiol. Immunol. 179:31-56. [DOI] [PubMed]
- 80.Skuse, G. R., A. J. Cappione, M. Sowden, L. J. Metheny, and H. C. Smith. 1996. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 24:478-485. [DOI] [PMC free article] [PubMed]
- 81.Sokal, R. R., and F. J. Rohlf. 1995. Biometry: the principles and practice of statistics in biological research, 3rd ed. W. H. Freeman, New York, N.Y.
- 82.Sontheimer, H. 1994. Voltage-dependent ion channels in glial cells. Glia 11:156-172. [DOI] [PubMed]
- 83.Stamm, W. E., H. H. Handsfield, A. M. Rompalo, R. L. Ashley, P. L. Roberts, and L. Corey. 1988. The association between genital ulcer disease and acquisition of HIV infection in homosexual men. JAMA 260:1429-1433. [PubMed] [Google Scholar]
- 84.Stinchi, S., R. Lullmann-Rauch, D. Hartmann, R. Coenen, T. Beccari, A. Orlacchio, K. von Figura, and P. Saftig. 1999. Targeted disruption of the lysosomal alpha-mannosidase gene results in mice resembling a mild form of human alpha-mannosidosis. Hum. Mol. Genet. 8:1365-1372. [DOI] [PubMed] [Google Scholar]
- 85.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Storey, N., D. Latchman, and S. Bevan. 2002. Selective internalization of sodium channels in rat dorsal root ganglion neurons infected with herpes simplex-1. J. Cell Biol. 158:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takasaki, I., A. Sasaki, T. Andoh, H. Nojima, K. Shiraki, and Y. Kuraishi. 2002. Effects of analgesics on delayed postherpetic pain in mice. Anesthesiology 96:1168-1174. [DOI] [PubMed] [Google Scholar]
- 88.Tomoda, T., R. S. Bhatt, H. Kuroyanagi, T. Shirasawa, and M. E. Hatten. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24:833-846. [DOI] [PubMed] [Google Scholar]
- 89.van Kerkhof, P., R. Govers, C. M. Alves dos Santos, and G. J. Strous. 2000. Endocytosis and degradation of the growth hormone receptor are proteasome-dependent. J. Biol. Chem. 275:1575-1580. [DOI] [PubMed] [Google Scholar]
- 90.Wald, A., and K. Link. 2001. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J. Infect. Dis. 185:45-52. [DOI] [PubMed] [Google Scholar]
- 91.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 92.Wilcox, C. L., and E. M. Johnson. 1987. Nerve growth factor deprivation results in the reactivation of latent herpes simplex virus in vitro. J. Virol. 61:2311-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xin, X., R. Day, W. Dong, Y. Lei, and L. D. Fricker. 1998. Cloning, sequence analysis, and distribution of rat metallocarboxypeptidase Z. DNA Cell Biol. 17:311-319. [DOI] [PubMed] [Google Scholar]
- 94.Zhao, S., S. Torii, H. Yokota-Hashimoto, T. Takeuchi, and T. Izumi. 2002. Involvement of Rab27b in the regulated secretion of pituitary hormones. Endocrinology 143:1817-1824. [DOI] [PubMed] [Google Scholar]
- 95.Zhuo, M., W. Zhang, H. Son, I. Mansuy, R. A. Sobel, J. Seidman, and E. R. Kandel. 1999. A selective role of calcineurin Aα in synaptic depotentiation in hippocampus. Proc. Natl. Acad. Sci. USA 96:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]