Abstract
Toroviruses (family Coronaviridae, order Nidovirales) are enveloped, positive-stranded RNA viruses that have been implicated in enteric disease in cattle and possibly in humans. Despite their potential veterinary and clinical relevance, little is known about torovirus epidemiology and molecular genetics. Here, we present the first study into the diversity among toroviruses currently present in European swine and cattle herds. Comparative sequence analysis was performed focusing on the genes for the structural proteins S, M, HE, and N, with fecal specimens serving as sources of viral RNA. Sequence data published for animal and human torovirus variants were included. Four genotypes, displaying 30 to 40% divergence, were readily distinguished, exemplified by bovine torovirus (BToV) Breda, porcine torovirus (PToV) Markelo, equine torovirus Berne, and the putative human torovirus. The ungulate toroviruses apparently display host species preference. In phylogenetic analyses, all PToV variants clustered, while the recent European BToVs mostly resembled the New World BToV variant Breda, identified 19 years ago. However, we found ample evidence for recurring intertypic recombination. All newly characterized BToV variants seem to have arisen from a genetic exchange, during which the 3′ end of the HE gene, the N gene, and the 3′ nontranslated region of a Breda virus-like parent had been swapped for those of PToV. Moreover, some PToV and BToV variants carried chimeric HE genes, which apparently resulted from recombination events involving hitherto unknown toroviruses. From these observations, the existence of two additional torovirus genotypes can be inferred. Toroviruses may be even more promiscuous than their closest relatives, the coronaviruses and arteriviruses.
Toroviruses, coronaviruses, arteriviruses, and roniviruses (order Nidovirales) are evolutionary related enveloped positive-stranded RNA viruses of vertebrates and invertebrates. Their common ancestry is evident from sequence identity in their replicase proteins and from similarities in genome organization, gene order, and replication strategy (6, 11, 51). There are, however, marked differences in genome size, host range, and virion architecture (6, 11). Toroviruses are morphologically unique and typically occur as a collection of discoidal, kidney-, and rod-shaped particles, with a tubular nucleocapsid enveloped by a membrane carrying large spikes (59).
Whereas coronaviruses and arteriviruses have been studied in detail, little is known about toroviruses. One important reason for this gap is that toroviruses have not been propagated in cell culture, with the sole exception so far of the Swiss equine isolate Berne virus (BEV). Early seroepidemiological surveys based upon BEV cross-neutralization assays and enzyme-linked immunosorbent assay (ELISA) indicated that toroviruses occur in a wide variety of ungulate hosts (cattle, sheep, goats, and swine) (4, 5, 28, 33, 60; for a review, see reference 26).
Toroviruses (ToVs; acronyms used are according to the recommendations of the ICTV Coronavirus Study Group) seem to be associated with asymptomatic enteric infections in swine (33, 42) but can cause serious, at times fatal, diarrheal disease in cattle (25, 61-63). Berne virus-related bovine toroviruses (BToVs), designated Breda virus (BRV), were first seen in Breda, Iowa, during an outbreak of neonatal calf diarrhea, in which 15% of the affected animals died (62). Both gnotobiotic and conventionally reared calves developed mild to severe diarrhea upon experimental BRV infection (47, 62, 63).
Toroviruses have also been implicated in human gastroenteritis. In fecal samples from children and adults with diarrhea, torovirus-like particles cross-reactive with BEV- and BRV-specific antisera were detected by immunoelectron microscopy (1, 2, 12, 32), and torovirus antigens were detected by ELISA (27, 30). In addition, torovirus RNA was detected in stool specimens of pediatric patients by reverse transcription (RT)-PCR with primers designed after the 3′ nontranslated region (3′-NTR) of the Berne virus genome (12).
Recently, we completed the nucleotide sequence of the Berne virus genomic RNA (S. L. Smits and R. J. de Groot, unpublished data). With a genome length of 28 kb, toroviruses are among the largest RNA viruses, rivaled in size only by the coronaviruses. The 5′-most two-thirds of the genome are taken up by two huge overlapping open reading frames (ORFs), 1a and 1b, encoding polyproteins from which the various subunits of the viral replicase/transcriptase are derived (49; S. L. Smits and R. J. de Groot, unpublished data). Downstream of ORF1b, there are four cistrons of 5 kb, 0.7 kb, 1.2 kb, and 0.5 kb (as ordered from 5′ to 3′). These encode the structural proteins, the spike (S), membrane (M), hemagglutinin-esterase (HE), and nucleocapsid (N) proteins, respectively (51). The structural proteins are translated from a 3′-coterminal nested set of subgenomic mRNAs (52), produced by discontinuous and nondiscontinuous RNA synthesis (54).
Sequence data for toroviruses other than Berne virus are limited to the S, M, HE, and N genes of the bovine torovirus BRV (9, 14), the N gene of porcine torovirus (PToV) field variant Markelo (33), and the HE gene from a putative human torovirus variant (13). Clearly, many questions regarding the genetic diversity of toroviruses need to be addressed. For instance, to what extent do the toroviruses currently present in the field resemble the early BEV and BRV? These viruses were isolated more than two decades ago from different hosts in different geographic locations (59, 62). Are toroviruses host specific, or does transmission occur between vertebrate species? What is the geographic distribution of torovirus genotypes?
To increase our insight into torovirus phylogeny and evolution, we performed comparative sequence analysis of selected European porcine and bovine variants, focusing on the genes for the structural proteins. The toroviruses of cattle and swine display host species preference and belong to distinct genotypic lineages. However, we also provide evidence for multiple intertypic recombination events which involved BToV Breda- and PToV Markelo-like ancestors as well as hitherto unknown toroviruses.
MATERIALS AND METHODS
Specimens.
Pooled fecal samples (20 to 60 animals per specimen) from 75 Dutch veal calf farms with 38 to 930 calves, aged 1 to 52 weeks (53), were kindly provided by W. van der Poel (National Institute of Public Health and the Environment, Bilthoven, The Netherlands). Bovine and porcine fecal samples, taken from diarrheic animals in Italian herds, were screened for the presence of toroviruses by (immuno) electron microscopy (38). Specimens containing torovirus RNA were identified by diagnostic RT-PCR, targeted to the 3′-NTR of the torovirus genome. A fecal sample from a naturally infected pig containing the Dutch PToV variant Markelo (P-MAR) was described previously (33), as were two torovirus-positive specimens obtained during an RT-PCR survey of Hungarian swine and cattle herds (42). All samples selected for further genetic analysis are listed in Table 1.
TABLE 1.
Virus variants and characteristics
| Virus | Host | Designationa | Origin
|
Pooled (n)b | EMc | Reference | |
|---|---|---|---|---|---|---|---|
| Country | Yr | ||||||
| PToV | Swine | P-MAR | The Netherlands | 1995 | − (1) | + | 33 |
| P4 | Italy | 1990 | + (3) | + | 38 | ||
| P9 | Italy | 1996 | + (7) | + | 38 | ||
| P10 | Italy | 1999 | − (1) | + | |||
| P78 | Hungary | 2000 | + (2) | ND | 42 | ||
| BToV | Cattle | B6 | Italy | 1990 | + (2) | + | |
| B1314 | Hungary | 2000 | + (2) | ND | 42 | ||
| B145 | The Netherlands | 1998 | + | ND | 53 | ||
| B150 | The Netherlands | 1998 | + | ND | 53 | ||
| B155 | The Netherlands | 1998 | + | ND | 53 | ||
| B156 | The Netherlands | 1998 | + | ND | 53 | ||
Designation for variant used in text.
n, number of animals per sample.
EM, electron microscopy performed. ND, not determined.
RNA purification, cDNA synthesis, and PCR amplification.
RNA was extracted from fecal samples and concentrated according to the guanidinium isothiocyanate silica protocol of Boom et al. (3) with minor modifications (18). RT-PCR amplification of torovirus sequences was performed with RNase H-free Moloney murine leukemia virus reverse transcriptase (Superscript II) and Taq polymerase (Gibco-BRL, Life Technologies) or with the Expand long-template PCR system (Roche Diagnostics GmbH), according to the instructions of the manufacturers. Precautions to avoid carryover of amplification products included physical separation of the pre- and post-PCR procedures. In each experiment, multiple water controls were taken along. Amplicons were gel purified and cloned into vector pGEM-T Easy (Promega Corp., Madison, Wis.).
Sequence reactions were performed commercially (BaseClear Labservices) with the ABI Prism BigDye Terminators version 3.0 cycle sequencing kit (Applied Biosystems) and an ABI Prism 3100 genetic analyzer. The nucleotide sequences determined for each RT-PCR product on at least two independent clones and, with few exceptions, in both orientations were analyzed and deposited in the EMBL database. Primers used for RT-PCR and sequence analysis are listed in Table 2.
TABLE 2.
Oligonucleotide primers used for RT-PCR
| Primer | Nucleotide sequence (5′ to 3′) | Polarity | Positionsa | Gene | RNA template(s) used in RT-PCR |
|---|---|---|---|---|---|
| 293 | AGCAACCTTGTGGTTGGTCTGT | + | 27805-27826 | 3′-NTR | All |
| 294 | CTTACATGGAGACACTCAACCA | − | 27920-27941 | 3′-NTR | All |
| 1344 | GAGAAAGAGCCAAGATGAATT | + | 27299-27319 | IGR N | All |
| 692 | GGATTAAGCATAGAATTCAT | − | 27313-27322 | N | P-MAR |
| 1383 | TAGCATTTGGATTAAGCATAGA | − | 27319-27340 | N | All |
| 1433 | AAGTTTGAGTAGCCACTTATC | + | 26773-26793 | IGRe M/HE | B6, B145, B150, B155, B156, B1314, P78 |
| 1345 | GAGACACTATCTTTAG | + | 27284-27299 | IGR N | P-MAR, P4, P9, P10 |
| 795 | GGATGACAGACACCACTCATC | − | 660-680b | HE | P-MAR |
| 1434 | CATCTTCTAAAGATAAGTGG | − | 26785-27804 | IGR M/HE | P4, P9, P10, B6, B145, P-MAR |
| 1435 | TCTTTGAAGATTGCCAAAA | + | 26061-26083 | IGR S/M | P4, P9, P10, B6, B145 |
| 1493 | AAGAATATTAAACTCAGCAT | − | 1-21c | HE | B150 |
| 1494 | AAGTTAGTCACTTTCTTTAGT | + | 26048-26068 | IGR S/M | B150, B155 |
| 1478 | GTGTTTAGTACTAGTTTAAGTT | + | 21259-21280 | ORF1b | P-MAR, B145 |
| 1542 | GCTGTACAGAGCATTTGGTCA | + | 853-873c | HE | B6, B145, B150 |
| 1543 | GCTGTTCAGAGTATTTGGTCA | + | 877-897b | HE | B6 |
| 1544 | GTGGTGAAAAATTCACACC | − | 289-307c | M | B150 |
| 1546 | GTGCTGCAAGGTGTTGCAGA | + | 26731-26750 | M | B150 |
| 1833 | CTAGGAAAATATGGATTATTA | − | 123-143d | HE | B155 |
Positions with respect to the Berne virus genome, unless indicated otherwise.
Position as counted from the iniation codon of the PToV Markelo HE gene.
Position as counted from the start codon of the BToV BRV HE gene.
Position as counted from the initiation codon of the B155 HE gene.
IGR, intergenic region.
Phylogenetic analysis.
Torovirus sequences were aligned with the DNAStar program MegAlign (ClustalV method; DNAStar, Inc.) to determine pairwise differences. Multiple sequence alignments were generated with the program Multalin (10) with the Dayhoff symbol comparison table. The sequences of the genes coding for structural proteins of BToV Breda 1, EToV Berne, and HToV were extracted from the NCBI database (accession numbers D00563, X52506, X52505, AF076621, and AF159585).
Phylogenetic and molecular evolutionary analyses were conducted with Mega version 2.1 (34). To detect sites of recombination, the program PhylPro was used (58).
The distribution of synonymous substitutions (Dss) was determined by pairwise comparison of protein-coding nucleotide sequences via sliding-window analysis. Gaps were excluded from the alignments. The number of synonymous substitutions per synonymous site (Ks) was estimated for overlapping 240-nucleotide gene segments with a 60-nucleotide step size with the program K-estimator 4.5 (7, 8). Dss profiles were generated by plotting the calculated Ks values against nucleotide position.
Nucleotide sequence accession numbers.
The newly determined sequences have been deposited in the EMBL database under accession numbers AJ575358 through AJ575389.
RESULTS
Identification of new bovine and porcine torovirus field variants.
During routine electron microscopic analysis of clinical samples, torovirus-like particles were occasionally observed in the feces of diarrheic Italian cattle and swine (Fig. 1). A diagnostic RT-PCR, targeted to the 3′-NTR of the viral genome (33), detected torovirus RNA in one out of four bovine (B6) and in three out of four porcine (P4, P9, and P10) samples. Four additional bovine specimens containing torovirus RNA (B145, B150, B155, and B156) were identified upon RT-PCR screening of a library of pooled fecal samples from 75 Dutch cattle farms, collected during the winter of 1998 (53). Also included in our studies were fecal samples containing the Dutch PToV variant Markelo (P-MAR) (33) and two specimens (B1314 and P78) identified as torovirus positive in a previous RT-PCR survey of Hungarian swine and cattle herds (42). All samples, with date, location, and host of origin, are listed in Table 1.
FIG. 1.
Electron microscopic detection of toroviruses in fecal specimens from diarrheic swine (P4 and P9) and cattle (B6). Virions were analyzed directly (B6) or after immunoaggregation with convalescent-phase serum from PToV P4-infected animals (P4 and P9). Negative staining was performed with 2% sodium phosphotungstic acid. Bars, 100 nm.
Viral RNA extracted from feces served as a template for RT-PCR amplification of the genes for the structural proteins (Fig. 2; Table 1). To avoid primer-directed selection of sequence variants, nonbiased primers were designed after elements conserved among BRV, BEV, and PToV, including the 3′-NTR and the intergenic regions (Table 2) (9, 14, 52, 54). For the amplification of S genes, the positive-stranded primer was designed after ORF1b sequences, conserved between BEV and BRV (14, 49). In all cases, RT-PCR amplicons of the anticipated size were obtained (not shown), which were analyzed either directly or after cloning.
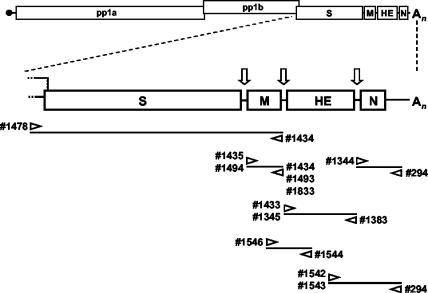
FIG. 2.
Torovirus genome organization and schematic outline of the strategies used for RT-PCR amplification of the genes for the structural proteins. The upper panel shows a schematic representation of the genome. Boxes represent the genes for the polymerase (ORF1a and ORF1b), the spike protein (S), the membrane protein (M), the hemagglutinin-esterase (HE), and the nucleocapsid protein (N). Open arrows indicate intergenic regions and transcription-regulating sequences. Also indicated are the cap structure (black dot) and the poly(A) tail (An). The lower panel shows a schematic outline of the RT-PCR assays employed to amplify S, M, HE, and N sequences. The orientations and positions of the oligonucleotides on the torovirus genome are shown.
Genetic diversity of torovirus field variants.
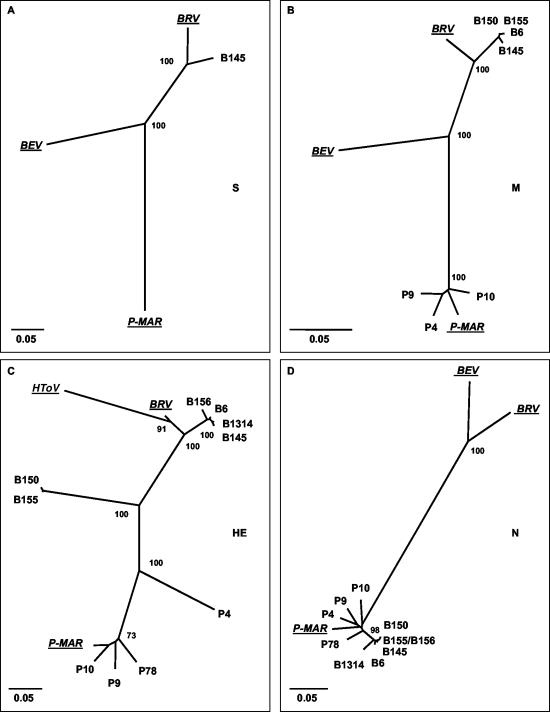
Pairwise comparison of toroviral nucleic acid sequences revealed considerable diversity, with identities ranging from 55.9 to 100% (Tables 3, 4, and 5). Phylogenetic trees were generated for each gene by neighbor-joining (Fig. 3), maximum parsimony, and maximum-likelihood methods (not shown); these methods yielded essentially identical topologies. Three distinct torovirus genotypes with apparent preferences for horse, cattle, and swine could be discerned, represented by BEV, BRV, and P-MAR, respectively; HToV apparently constitutes a fourth genotype (Fig. 3C).
TABLE 3.
Sequence relationships between toroviral M genes
| Virus | % Nucleotide sequence identity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BRV | B6 | B145 | B150 | B155 | P4 | P9 | P10 | P-MAR | |
| BEV | 81.8 | 81.8 | 81.7 | 82 | 82 | 78 | 77.5 | 76.8 | 77.1 |
| P-MAR | 77.6 | 77.5 | 77.8 | 77.8 | 77.8 | 96 | 95.4 | 96.3 | |
| P10 | 78.1 | 77.8 | 78.1 | 78.2 | 78.2 | 96.3 | 95.6 | ||
| P9 | 76.8 | 77.5 | 77.8 | 77.9 | 77.9 | 96.3 | |||
| P4 | 76.7 | 76.9 | 77.2 | 77.4 | 77.4 | ||||
| B155 | 93.9 | 99.6 | 99.1 | 100 | |||||
| B150 | 93.9 | 99.6 | 99.1 | ||||||
| B145 | 93.7 | 99 | |||||||
| B6 | 93.4 | ||||||||
TABLE 4.
Sequence relationships between toroviral HE genes
| Virus | % Nucleotide sequence identity
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRV | B6 | B145 | B156 | B1314 | B150 | B155 | P4 | P9 | P10 | P78 | P-MAR | |
| HToV | 83.4 | 73.6 | 73.2 | 73.4 | 72.9 | 59.6 | 59.7 | 56.4 | 55.9 | 56.4 | 56.9 | 56 |
| P-MAR | 61.8 | 66.6 | 65.9 | 65.8 | 65.5 | 67.8 | 68.1 | 76.8 | 92.6 | 94 | 94 | |
| P78 | 62.4 | 67.7 | 67.3 | 66.9 | 66.8 | 68.6 | 68.6 | 77 | 93.3 | 92.7 | ||
| P10 | 62.2 | 66.6 | 66.1 | 66.3 | 65.8 | 67.9 | 68.1 | 77 | 92.9 | |||
| P9 | 61.8 | 65.6 | 65.4 | 65.2 | 64.9 | 67.6 | 67.7 | 76.6 | ||||
| P4 | 63.4 | 66.7 | 66.7 | 66.6 | 66 | 67.1 | 66.9 | |||||
| B155 | 67.1 | 73.6 | 73.1 | 73.3 | 72.8 | 99 | ||||||
| B150 | 66.7 | 73.6 | 73.2 | 73.2 | 73 | |||||||
| B1314 | 86.8 | 98 | 98.4 | 96.7 | ||||||||
| B156 | 87 | 97.5 | 96.6 | |||||||||
| B145 | 87.1 | 98.3 | ||||||||||
| B6 | 87.7 | |||||||||||
TABLE 5.
Sequence relationships between toroviral N genes
| Virus | % Nucleotide sequence identity
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRV | B6 | B145 | B156 | B1314 | B150 | B155 | P4 | P9 | P10 | P78 | P-MAR | |
| BEV | 86.1 | 69.6 | 69.6 | 69.6 | 69.6 | 69.2 | 69.6 | 68.9 | 70 | 70.2 | 69.8 | 69.8 |
| P-MAR | 71.3 | 93.3 | 93.7 | 93.3 | 92.5 | 93.7 | 93.3 | 94.7 | 92.9 | 93.5 | 93.5 | |
| P78 | 72 | 95.3 | 95.3 | 94.9 | 94.1 | 94.9 | 94.9 | 94.7 | 94.5 | 94.3 | ||
| P10 | 72 | 93.3 | 93.7 | 93.3 | 92.9 | 92.9 | 93.3 | 93.9 | 93.1 | |||
| P9 | 71.7 | 93.5 | 93.5 | 94.1 | 92.7 | 94.1 | 93.9 | 95.1 | ||||
| P4 | 71.8 | 94.3 | 94.3 | 94.1 | 92.8 | 94.5 | 94.1 | |||||
| B155 | 72.6 | 98.8 | 99.2 | 100 | 97.4 | 99.6 | ||||||
| B150 | 72.6 | 98.4 | 98.8 | 99.6 | 97 | |||||||
| B1314 | 71.7 | 97.6 | 97.6 | 97.4 | ||||||||
| B156 | 72.6 | 98.8 | 99.2 | |||||||||
| B145 | 72.2 | 99.2 | ||||||||||
| B6 | 72.2 | |||||||||||
FIG. 3.
Unrooted neighbor-joining trees depicting the phylogenetic relationships among torovirus field variants. Trees were constructed for (A) the S gene, (B) the M gene, (C) the HE gene, and (D) the N gene with the Kimura-2 parameter method. Confidence values calculated by bootstrap analysis (1,000 replicates) are indicated at the major branching points. Branch lengths are drawn to scale; the scale bar represents 0.05 nucleotide substitution per site. The torovirus reference strains BRV, BEV, P-MAR, and HToV are italicized and underlined. The tree shown for the HE gene was based upon an alignment corresponding to residues 1 to 1049 of B145. Note that BEV was not included in this tree; as a result of a large deletion, Berne virus has lost most of its HE gene (51).
While all new PToV strains resembled P-MAR, the BToV strains seemed mostly related to BRV. The S gene of the Dutch BToV variant B145 shared 93% identity with that of the prototypic New World BToV variant Breda virus (14). The B150 S gene was partially sequenced. Within its 3′-most 2,436 nucleotides, the B150 S gene was 98% and 95% identical to those of B145 and BRV, respectively. Sequence identities between the BToV S genes and those of Berne virus (50) and P-MAR were considerably lower (77% and 73%, respectively; Fig. 3A).
In trees constructed for the M genes, the new BToV variants clustered with BRV (93 to 94% identity); among these new variants, sequence identity in the M gene was more than 99% (Table 3 and Fig. 3B). Remarkably, however, in the HE gene, only a subset of the BToV variants (B6, B145, B156, and B1314) grouped with BRV (Fig. 3C). The B150 and B155 HE sequences were 99% identical to each other but quite distinct from those of the other BToVs (67 to 73% identity) and equally far removed from those of the PToV variants (65 to 68% identity; Table 4 and Fig. 3C). Moreover, for all new BToV variants, the N genes did not group with that of BRV but, surprisingly, with those of the PToV strains (Fig. 3D). Note, however, that the new variant BToV N genes still formed a separate cluster.
The PToV strains P-MAR, P9, P10, and P78 displayed 5 to 8% divergence and clustered in trees constructed for M, HE, and N (Fig. 3B, C, and D). However, there was an anomaly in the phylogeny of variant P4: in the N and M genes, P4 grouped with the other PToVs, but in trees based upon HE sequences, it was clearly distinct (Fig. 3C). In the HE gene, P4 is only 77% identical to the other PToVs, versus 92 to 94% sequence identity in the M and N genes (Tables 3 to 5).
Evidence for recombination of torovirus sequences.
Conflicting phylogenetic data, such as obtained for the different regions of the BToV and PToV genomes, are indicative of recombination. For example, the B6, B145, B156, and B1314 sequences seem to have resulted from a genetic exchange between BToV and PToV. In order to identify recombination breakpoints, we employed phylogenetic profile analysis (PhylPro) (58), a computer graphic method based on the premise that phylogenetic relationships derived from different regions of a multiple sequence alignment will be similar when no recombination has occurred. Recombination junction sites create areas of low phylogenetic correlation and in PhylPro graphs are depicted as sharp downward peaks. Within the HE genes of B6, B145, B156, and B1314, single recombination breakpoints were detected, located approximately 1,100 nucleotides from the ATG initiation codon (Fig. 4A). Visual inspection of the alignment suggested a template switch between residues 1070 through 1110: sequences coding for the ectodomain of a BRV-type HE were effectively fused to those encoding a PToV-type transmembrane region and cytoplasmic domain (Fig. 4C).
FIG. 4.
Identification of recombination sites in the M, HE, and N genes of torovirus field variants. (A) Recombination sites were identified with the program PhylPro (58). This phylogenetic profile method introduces the phylogenetic correlation measure, i.e., the principle that phylogenetic relationships in different regions of an aligned sequence will be similar when no recombination has occurred. An alignment of the coding regions of the M, HE, and N genes of P-MAR, P4, P9, P10, BRV, B6, B145, B150, and B155 was generated. For each individual sequence in the alignment, the phylogenetic correlations were computed at every position with a sliding-window technique, with window limits fixed at 15 differences. Shown are the phylogenetic profiles of B6 and B145, with x and y axes indicating the nucleotide positions and the phylogenetic correlation, respectively; as a reference, the genes for M, HE, and N, drawn to scale, are depicted as boxes (top). For clarity, profiles of P4, B150, and B155 were hidden in the graph. (B) Phylogenetic profiles of B150 and B155; for clarity, profiles of P4, B6, and B145 were hidden. Window limits were fixed at 10 differences. (C) Alignments of HE sequences surrounding the recombination sites; arrows and numbers correspond to those in panels A and B. Nucleotide differences with respect to the B145 sequences are shown. Nucleotide positions given are numbered from A1 of the B145 HE gene.
PhylPro analysis of the B150 and B155 sequences revealed three potential recombination sites (Fig. 4B), one located near the 5′ end of the HE gene, approximately 80 nucleotides downstream of the ATG codon. Visual inspection of the alignment suggested a template switch within the region comprising residues 70 through 110 (Fig. 4C). The other two breakpoints were located at the 3′ end of the HE gene between residues 1070 and 1110, one at the exact same position as was found for B6, B145, B156, and B1314 and one immediately upstream of it (Fig. 4A to C).
PhylPro comparison of PToV variant P4 with the other PToV variants yielded inconclusive results, with multiple, weak recombination signals scattered throughout the HE region (not shown). This prompted the question of whether the sequence divergence in the PToV HE genes resulted from recombination or merely from antigenic drift. As an alternative approach to test for recombination, we determined the distribution of synonymous substitutions (Dss). “Silent mutations” are fixed at random locations within viral genomes and at an approximately constant rate, providing a molecular clock (16, 24, 39, 66). The number of synonymous substitutions per synonymous site, Ks (8), thus reflects the evolutionary distance between two related coding sequences. If viruses have diverged from a common ancestor without the occurrence of recombination, pairwise sequence comparison should yield essentially identical Ks values throughout the genomes. Differences in Ks value between genomic regions, however, would be a tell-tale sign of recombination.
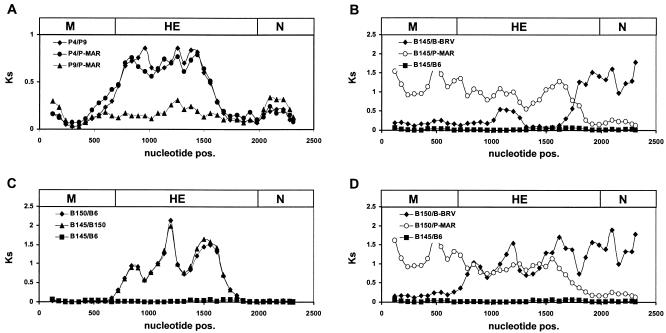
The coding sequences of the various torovirus variants were compared pairwise by sliding-window analysis, during which Ks values were estimated for overlapping 240-nucleotide gene segments with the program K-estimator 4.5 (7, 8). Dss profiles were generated by plotting the calculated Ks values against nucleotide position (Fig. 5). Comparison of P-MAR with the PToV variants P10 and P9 yielded roughly constant Ks values across the M, HE, and N genes (Fig. 5A). Apparently, no intertypic genetic exchanges have occurred in this part of the genome after P9, P10, and P-MAR branched off from their most recent common ancestor. In contrast, pairwise comparison of P4 with the other PToV variants (Fig. 5A and not shown) revealed that within the HE region, comprising nucleotides 20 to 950, the number of synonymous substitutions per synonymous site was four- to fivefold higher than in the flanking HE sequences or in the adjacent M and N genes. The Dss profiles are suggestive of a double recombination event, during which a segment of the PToV HE gene was exchanged for the corresponding HE sequences from an as yet unidentified torovirus. However, the data do not allow a definitive conclusion as to whether the P4 HE sequence is recombinant or, rather, that of the other PToVs.
FIG. 5.
Distribution of synonymous substitutions (Dss) in the M, HE, and N genes of torovirus field variants. Multiple alignments were generated for the protein-coding nucleotide sequences; all gaps were excluded. The sequences were compared pairwise by sliding-window analysis, during which the number of synonymous substitutions per synonymous site (Ks) was estimated for overlapping 240-nucleotide gene segments with a 60-nucleotide step size. Dss profiles were generated by plotting the calculated Ks values against nucleotide positions. As a reference, the genes for M, HE, and N, drawn to scale, are depicted as boxes (top). The Dss profiles shown were produced by pairwise comparison (A) of P4, P9, and P-MAR, (B) of B145 to BRV, P-MAR, or B6, (C) of B145 to B150 or B6 and of B150 to B6, and (D) of B150 to BRV or P-MAR, with the B145-B6 graph providing a baseline.
Dss profiles obtained for the BToV variants were fully consistent with the results of phylogenetic analysis and PhylPro scanning. Comparison of B6 and B145 gave a Dss profile with constant, low Ks values throughout the M, HE, and N coding regions. In contrast, the B145-BRV comparison revealed an abrupt increase in Ks value at the deduced recombination breakpoint within the B145 HE gene, coinciding with a sudden decrease in Ks value in the B145-P-MAR plot (Fig. 5B). Thus, the Dss profiles also indicate a single recombination event in the HE gene, during which BRV- and PToV-type sequences were fused. The Dss profiles of the B150-B6 and B150-B145 pairs showed that, within the region comprising the codons for residues 50 to 1100 of the HE gene, the number of synonymous substitutions per synonymous site was approximately eightfold higher than in the surrounding sequences (Fig. 5C). In profiles comparing B150 to BRV, Ks values were low in the M gene and in the 5′ region of the HE gene, with a sharp eightfold increase in the number of synonymous substitutions per synonymous site around position 40. Conversely, the B150 versus P-MAR profile showed a sixfold drop in Ks value near the 3′ end of the HE gene, around nucleotide 1100 (Fig. 5D). The combined data suggest that the HE sequences amplified for B150 and B155 are a composite derived from a BRV-like parent, a PToV-like parent, and an unknown torovirus.
DISCUSSION
The identification and characterization of new torovirus field variants is arduous. Although seroprevalence data indicate that torovirus infections are common in ungulates (26), naturally infected animals do not display distinctive clinical signs (34). Moreover, fecal or respiratory shedding of virus, detectable by ELISA or RT-PCR, appears to be short-lived (20, 25, 33).
For the present study, we used torovirus-positive fecal specimens from cattle and swine in Italy, the Netherlands, and Hungary which had been either preselected by electron microscopy or prescreened by diagnostic RT-PCR. This allowed the first study into the genetic diversity among toroviruses currently present in European herds. Four genotypes, displaying 30 to 40% sequence divergence, were readily distinguished. These were encountered in horses, cattle, swine, and humans and are represented by BEV, BRV, PToV, and HToV, respectively. Although the ungulate toroviruses may not be species specific (see below), they do display host species preference. In phylogenetic analyses, all PToV variants clustered, whereas the recent European BToVs resembled the New World BToV variant BRV, which was isolated 19 years ago (62). For example, in the S, M, and HE genes (6.7 kb), the European variant B145 shared 93% overall sequence identity with BRV.
Intriguingly, chimeric torovirus sequences were amplified from a number of fecal specimens, suggesting the existence of two additional torovirus genotypes. The detection of chimeric genes would imply intertypic genetic exchange via RNA recombination in natural torovirus populations. However, the fact that pooled fecal samples served as a source of viral RNA poses an obvious caveat. Herds and even individual animals may be subject to coinfection with nonrecombinant toroviruses of different genotypes. Consequently, the chimeric HE sequences could have been generated during RT-PCR, in particular during the reverse transcription step. RNase H-deficient Moloney murine leukemia virus reverse transcriptase was used throughout our experiments to minimize the frequency of template switching, but even under these conditions, recombination in vitro may still have occurred (40). Indeed, in simulation experiments in which we mixed synthetic T7 RNA polymerase transcripts of P-MAR and BRV HE at a 1:1 ratio and performed RT-PCR with nonbiased primers, chimeric amplicons were obtained. However, the in vitro recombination frequency was only 5% (4 out of 76 clones tested), which is in good agreement with the values (3%) reported by Luo and Taylor (40).
Most amplicons (95%) were nonrecombinant and either PToV or BRV specific. Moreover, template switching occurred at seemingly random locations (not shown). In contrast, RT-PCR amplification of HE sequences from the various bovine and porcine specimens gave strictly reproducible results. In independent RT-PCR assays, RNA extracted from each individual sample yielded consistently and exclusively one particular chimeric sequence; parental nonrecombinant HE genes were never detected. Moreover, sequences of identical chimeric structure were obtained with different primer sets and, most notably, were detected in different fecal specimens. The B6, B145, B156, and B1314 HE sequences differed at several nucleotide positions, as would be expected for genes of viruses subject to genetic and/or antigenic drift. Yet they all had identical recombination breakpoints and parents; the same was true for the HE genes of specimens B150 and B155. Based upon the collective evidence, we postulate that the chimeric sequences detected by RT-PCR are not in vitro artifacts but faithfully reflect the genomic organization of torovirus field variants.
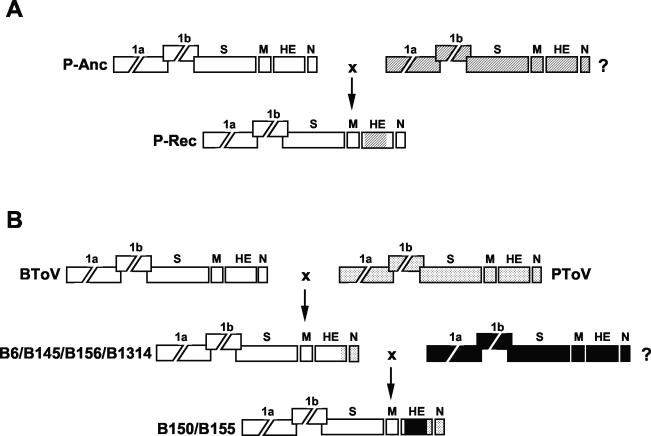
During PToV divergence, a recombination event occurred, apparently involving a donor virus of a hitherto unknown genotype. Thus, the coding sequences for the PToV HE ectodomain were partly replaced, presumably through a double template switch (Fig. 6A). Our data do not allow a conclusion as to whether P4 or the P-MAR-like viruses are the recombinant progeny. As the P4 variant is the oldest field variant, it may well represent the parental PToV type; a definitive answer will have to await identification of the other recombination partner.
FIG. 6.
Hypothetical model for genetic exchanges among torovirus field variants. Torovirus genomes are depicted schematically, with the various genes represented by boxes. (A) Presumptive recombination event during PToV divergence, involving an ancestral PToV strain (P-Anc; genes indicated by white boxes) and an unknown toroviral parent (?; genes indicated by hatched boxes). Exchange of HE sequences resulted in recombinant progeny (P-Rec). (B) Presumptive recombination events during BToV divergence. During an initial single recombination event, the 3′ end of the HE gene and the N gene and the 3′-NTR of a BRV-like BToV variant (indicated in white) were replaced by the corresponding PToV sequences (dotted), giving rise to the B6/B145/B156/B1314 lineage. A subsequent double recombination event, during which BToV HE sequences were replaced by those of an as yet unidentified torovirus (?, indicated in black), resulted in the B150/B155 lineage.
The phylogenetic data for the BToV variants are best explained by two sequential recombination events from which the B150/B155 lineage arose. An initial genetic exchange between BRV- and PToV-like parents gave rise to the B6/B145/B156/B1314 lineage. Apparently, a single template switch occurred at the 3′ end of the HE gene, as a result of which part of the BToV HE gene and the complete BToV N gene were replaced by the corresponding PToV sequences. A B6-type descendant then partook in a second recombination event, this time entailing a double template switch during which the coding sequences for the ectodomain of a BRV-type HE protein were replaced by those of yet another unknown torovirus (Fig. 6B). The occurrence of intertypic recombination implies cross-species transmission. At present, it cannot be established which hosts served as “mixing vessels” to produce the recombinant PToV and BToV variants. Also, the natural hosts of the unknown parental viruses of the PToV and BToV recombinants remain to be identified.
How long ago did the recombination events take place? Our data set is too limited to allow more than an educated guess. From the sequence alignments and the sampling dates of the field variants (1990, 1998, and 2000 for B6, B145, B156, and B1314, respectively), we estimated the rate of synonymous substitutions in BToV to be ≈3 × 10−3 per synonymous site per year. This is somewhat lower than the rates reported for influenzavirus, picornavirus, and human immunodeficiency virus (≈13 × 10−3) (16, 17), but in the same order as the rate estimated for the coronavirus transmissible gastroenteritis virus. Sanchez et al. (48) determined the total mutation fixation rate at ≈0.7 × 10−3 substitutions/site/year. From their data set, we recalculated a rate of ≈1.4 × 10−3 for synonymous substitutions (S. L. Smits and R. J. de Groot, unpublished data).
Under the assumptions of (i) a monophyletic origin of the new BToV strains and (ii) the existence of an approximate molecular clock (16), the B6-type and B150-type lineages would have branched during the mid-1990s. Although this rough estimate should be considered with caution, the data suggest that the extant recombinant BToV variants emerged only recently. The genetic exchanges possibly resulted in antigenic shifts which allowed the recombinant viruses to evade existing herd immunity. It is of note that in two instances the ectodomain of the HE protein was altered as a result of the recombination. HE is a prominent toroviral envelope glycoprotein (9) and hence a potential prime target for the humoral immune response.
Most likely, the recombination mechanism involved similarity-assisted template switching (homologous recombination) (36, 46). In each case, the donor sequence neatly replaced the homologous region of the acceptor sequence, and the crossovers occurred within stretches of 20 to 40 nucleotides that are highly conserved (98 to 100% identity) among currently known torovirus genotypes.
Recombination has been well documented for RNA viruses (64), including other nidoviruses (for reviews, see references 36, 37, and 43). Homologous recombination between related strains was observed for coronaviruses and arteriviruses, both under experimental conditions (31, 41, 55, 57, 65) and in nature (22, 23, 35, 45, 65). The genus Coronavirus consists of three antigenic clusters, which in turn can be subdivided into at least 13 different species on the basis of host specificity and genotype (15). While intraspecies recombination among coronaviruses may be common in the field (22, 35), there are only a few recognized cases of interspecies recombination. The best-known exceptions are the type II feline coronaviruses, which originated from genetic exchanges between canine and type I feline coronavirus strains (18, 44). The type II feline coronaviruses do not form a monophyletic lineage but arose from multiple independent recombination events (19).
The frequency of successful intertypic genetic exchange (those instances in which the recombinant offspring can establish itself in the host population) is determined among others by (i) properties inherent to the viral replicase, (ii) the frequency of cross-species transmission and the odds of double infections (not only of the host, but also of host cells), (iii) the viability of the recombinant progeny, and (iv) the “evolutionary gain,” i.e., the increase in fitness of the recombinant viruses relative to their nonrecombinant parents (64). Coronaviruses and toroviruses are closely related by evolution, occupy similar niches, and possess error-prone replicases that promote RNA recombination. The apparent difference between coronaviruses and toroviruses, with respect to the frequency of natural intertypic recombinant viruses in the field, may therefore reflect differences in viral “life style” and survival strategy.
The evidence that toroviruses cause gastroenteritis in animals and humans ranges between firm and tenuous (2, 12, 21, 25, 29, 30, 56, 61-63), and their veterinary importance and etiological role in human disease remain to be corroborated. Epidemiological studies are hampered by our limited knowledge of toroviral genetic and antigenic diversity. Thus far, most diagnostic assays rely on BEV- or BRV-derived antigens and sequences and are hence of narrow specificity. The data presented in this report not only offer an exciting glimpse into torovirus genetics but also furnish new diagnostic tools, including a set of antigenically distinct HE proteins.
Acknowledgments
We thank Nancy Schuurman and Eva Rendes for technical assistance and Wim van der Poel and Reina van der Heide for providing bovine fecal samples. We are grateful to Ies Nijman for helpful comments and suggestions with respect to phylogenetic analysis and to Jolanda Mijnes and Peter Rottier for critical reading of the manuscript.
REFERENCES
- 1.Beards, G. M., D. W. Brown, J. Green, and T. H. Flewett. 1986. Preliminary characterisation of torovirus-like particles of humans: comparison with Berne virus of horses and Breda virus of calves. J. Med. Virol. 20:67-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beards, G. M., C. Hall, J. Green, T. H. Flewett, F. Lamouliatte, and P. Du Pasquier. 1984. An enveloped virus in stools of children and adults with gastroenteritis that resembles the Breda virus of calves. Lancet i:1050-1052. [DOI] [PMC free article] [PubMed]
- 3.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. W., G. M. Beards, and T. H. Flewett. 1987. Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J. Clin. Microbiol. 25:637-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. W., R. Selvakumar, D. J. Daniel, and V. I. Mathan. 1988. Prevalence of neutralising antibodies to Berne virus in animals and humans in Vellore, South India. Arch. Virol. 98:267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 7.Comeron, J. M. 1999. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15:763-764. [DOI] [PubMed] [Google Scholar]
- 8.Comeron, J. M. 1995. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 41:1152-1159. [DOI] [PubMed] [Google Scholar]
- 9.Cornelissen, L. A., C. M. Wierda, F. J. van der Meer, A. A. Herrewegh, M. C. Horzinek, H. F. Egberink, and R. J. de Groot. 1997. Hemagglutinin-esterase, a novel structural protein of torovirus. J. Virol. 71:5277-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duckmanton, L., B. Luan, J. Devenish, R. Tellier, and M. Petric. 1997. Characterization of torovirus from human fecal specimens. Virology 239:158-168. [DOI] [PubMed] [Google Scholar]
- 13.Duckmanton, L., R. Tellier, C. Richardson, and M. Petric. 1999. The novel hemagglutinin-esterase genes of human torovirus and Breda virus. Virus Res. 64:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Duckmanton, L. M., R. Tellier, P. Liu, and M. Petric. 1998. Bovine torovirus: sequencing of the structural genes and expression of the nucleocapsid protein of Breda virus. Virus Res. 58:83-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. J. M. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Family Coronaviridae, p. 835-849. In M. H. V. van Regenmortel (ed.), Virus taxonomy. Academic Press, London, United Kingdom.
- 16.Gojobori, T., E. N. Moriyama, and M. Kimura. 1990. Molecular clock of viral evolution, and the neutral theory. Proc. Natl. Acad. Sci. USA 87:10015-10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamada, N., K. Masunaga, Y. Ohtsu, H. Kato, K. Tsuji, H. Maeda, M. Shingu, and T. Toyoda. 1998. Nucleotide sequence of the gene encoding the RNA polymerase and the 3′ non-coding region of a bovine enterovirus Japanese isolate: rapid synonymous substitutions between European and Japanese strains. Arch. Virol. 143:815-821. [DOI] [PubMed] [Google Scholar]
- 18.Herrewegh, A. A., R. J. de Groot, A. Cepica, H. F. Egberink, M. C. Horzinek, and P. J. Rottier. 1995. Detection of feline coronavirus RNA in feces, tissues, and body fluids of naturally infected cats by reverse transcriptase PCR. J. Clin. Microbiol. 33:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrewegh, A. A., I. Smeenk, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoet, A. E., K. O. Cho, K. O. Chang, S. C. Loerch, T. E. Wittum, and L. J. Saif. 2002. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 63:342-348. [DOI] [PubMed] [Google Scholar]
- 21.Jamieson, F. B., E. E. Wang, C. Bain, J. Good, L. Duckmanton, and M. Petric. 1998. Hum. torovirus: a new nosocomial gastrointestinal pathogen. J. Infect. Dis. 178:1263-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia, W., K. Karaca, C. R. Parrish, and S. A. Naqi. 1995. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 140:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapur, V., M. R. Elam, T. M. Pawlovich, and M. P. Murtaugh. 1996. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 77:1271-1276. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, M. 1989. The neutral theory of molecular evolution and the world view of the neutralists. Genome 31:24-31. [DOI] [PubMed] [Google Scholar]
- 25.Koopmans, M., H. Cremers, G. Woode, and M. C. Horzinek. 1990. Breda virus (Toroviridae) infection and systemic antibody response in sentinel calves. Am. J. Vet. Res. 51:1443-1448. [PubMed] [Google Scholar]
- 26.Koopmans, M., and M. C. Horzinek. 1994. Toroviruses of animals and humans: a review. Adv. Virus Res. 43:233-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopmans, M., M. Petric, R. Glass, and S. Monroe. 1993. Enzyme-linked immunosorbent assay reactivity of torovirus-like particles in fecal specimens from humans with diarrhea. J. Clin. Microbiol. 31:2738-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koopmans, M., U. van den Boom, G. Woode, and M. C. Horzinek. 1989. Seroepidemiology of Breda virus in cattle with ELISA. Vet. Microbiol. 19:233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koopmans, M., L. van Wuijckhuise-Sjouke, Y. H. Schukken, H. Cremers, and M. C. Horzinek. 1991. Association of diarrhea in cattle with torovirus infections on farms. Am. J. Vet. Res. 52:1769-1773. [PubMed] [Google Scholar]
- 30.Koopmans, M. P., E. S. Goosen, A. A. Lima, I. T. McAuliffe, J. P. Nataro, L. J. Barrett, R. I. Glass, and R. L. Guerrant. 1997. Association of torovirus with acute and persistent diarrhea in children. Pediatr. Infect. Dis. J. 16:504-507. [DOI] [PubMed] [Google Scholar]
- 31.Kottier, S. A., D. Cavanagh, and P. Britton. 1995. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology 213:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnan, T., and T. Naik. 1997. Electronmicroscopic evidence of torovirus like particles in children with diarrhoea. Indian J. Med. Res. 105:108-110. [PubMed] [Google Scholar]
- 33.Kroneman, A., L. A. Cornelissen, M. C. Horzinek, R. J. de Groot, and H. F. Egberink. 1998. Identification and characterization of a porcine torovirus. J. Virol. 72:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 35.Kusters, J. G., E. J. Jager, H. G. Niesters, and B. A. van der Zeijst. 1990. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine 8:605-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, M. M. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavazza, A., P. Candotti, S. Perini, and L. Vezzali. 1996. Electron microscopic identification of torovirus-like particles in post-weaned piglets with enteritis. 14th IPVS Congress, Bologna, Italy.
- 39.Leitner, T., and J. Albert. 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96:10752-10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo, G. X., and J. Taylor. 1990. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 64:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makino, S., J. Keck, S. Stohlman, and M. Lai. 1986. High-frequency RNA recombination of murine coronaviruses. J. Virol. 57:729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matiz, K., S. Kecskemeti, I. Kiss, Z. Adam, J. Tanyi, and B. Nagy. 2002. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 43.Meng, X. J. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet. Microbiol. 74:309-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motokawa, K., T. Hohdatsu, H. Hashimoto, and H. Koyama. 1996. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer, integral membrane and nucleocapsid proteins of feline, canine and porcine coronaviruses. Microbiol. Immunol. 40:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murtaugh, M. P., S. Yuan, and K. S. Faaberg. 2001. Appearance of novel PRRSV isolates by recombination in the natural environment. Adv. Exp. Med. Biol. 494:31-36. [DOI] [PubMed] [Google Scholar]
- 46.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 47.Pohlenz, J., N. Cheville, G. Woode, and A. Mokresh. 1984. Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Vet. Pathol. 21:407-417. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez, C. M., F. Gebauer, C. Sune, A. Mendez, J. Dopazo, and L. Enjuanes. 1992. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 190:92-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., J. A. den Boon, P. J. Bredenbeek, M. C. Horzinek, R. Rijnbrand, and W. J. Spaan. 1990. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 18:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snijder, E. J., J. A. Den Boon, W. J. Spaan, M. Weiss, and M. C. Horzinek. 1990. Primary structure and post-translational processing of the Berne virus peplomer protein. Virology 178:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snijder, E. J., and M. C. Horzinek. 1993. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 74:2305-2316. [DOI] [PubMed] [Google Scholar]
- 52.Snijder, E. J., M. C. Horzinek, and W. J. Spaan. 1990. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J. Virol. 64:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Poel, W. H., J. Vinje, R. van Der Heide, M. I. Herrera, A. Vivo, and M. P. Koopmans. 2000. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 6:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. De Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Vugt, J. J., T. Storgaard, M. B. Oleksiewicz, and A. Botner. 2001. High frequency RNA recombination in porcine reproductive and respiratory syndrome virus occurs preferentially between parental sequences with high similarity. J. Gen. Virol. 82:2615-2620. [DOI] [PubMed] [Google Scholar]
- 56.Vaucher, Y. E., C. G. Ray, L. L. Minnich, C. M. Payne, D. Beck, and P. Lowe. 1982. Pleomorphic, enveloped, virus-like particles associated with gastrointestinal illness in neonates. J. Infect. Dis. 145:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, L., Y. Xu, and E. W. Collisson. 1997. Experimental confirmation of recombination upstream of the S1 hypervariable region of infectious bronchitis virus. Virus Res. 49:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiller, G. F. 1998. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol. Biol. Evol. 15:326-335. [DOI] [PubMed] [Google Scholar]
- 59.Weiss, M., F. Steck, and M. C. Horzinek. 1983. Purification and partial characterization of a new enveloped RNA virus (Berne virus). J. Gen. Virol. 64:1849-1858. [DOI] [PubMed] [Google Scholar]
- 60.Weiss, M., F. Steck, R. Kaderli, and M. C. Horzinek. 1984. Antibodies to Berne virus in horses and other animals. Vet. Microbiol. 9:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woode, G. N., J. F. Pohlenz, N. E. Gourley, and J. A. Fagerland. 1984. Astrovirus and Breda virus infections of dome cell epithelium of bovine ileum. J. Clin. Microbiol. 19:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woode, G. N., D. E. Reed, P. L. Runnels, M. A. Herrig, and H. T. Hill. 1982. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 7:221-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woode, G. N., L. J. Saif, M. Quesada, N. J. Winand, J. F. Pohlenz, and N. K. Gourley. 1985. Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 46:1003-1010. [PubMed] [Google Scholar]
- 64.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 65.Yuan, S., C. J. Nelsen, M. P. Murtaugh, B. J. Schmitt, and K. S. Faaberg. 1999. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 61:87-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, G., D. T. Haydon, N. J. Knowles, and J. W. McCauley. 1999. Molecular evolution of swine vesicular disease virus. J. Gen. Virol. 80:639-651. [DOI] [PubMed] [Google Scholar]