Abstract
Turnip yellow mosaic virus (TYMV), a positive-strand RNA virus in the alphavirus-like superfamily, encodes two replication proteins, 140K and 66K, both being required for its RNA genome replication. The 140K protein contains domains indicative of methyltransferase, proteinase, and NTPase/helicase, and the 66K protein encompasses the RNA-dependent RNA polymerase domain. During viral infection, the 66K protein localizes to virus-induced chloroplastic membrane vesicles, which are closely associated with TYMV RNA replication. To investigate the determinants of its subcellular localization, the 66K protein was expressed in plant protoplasts from separate plasmids. Green fluorescent protein (GFP) fusion and immunofluorescence experiments demonstrated that the 66K protein displayed a cytoplasmic distribution when expressed individually but that it was relocated to the chloroplast periphery under conditions in which viral replication occurred. The 66K protein produced from an expression vector was functional in viral replication since it could transcomplement a defective replication template. Targeting of the 66K protein to the chloroplast envelope in the course of the viral infection appeared to be solely dependent on the expression of the 140K protein. Analysis of the subcellular localization of the 140K protein fused to GFP demonstrated that it is targeted to the chloroplast envelope in the absence of other viral factors and that it induces the clumping of the chloroplasts, one of the typical cytological effects of TYMV infection. These results suggests that the 140K protein is a key organizer of the assembly of the TYMV replication complexes and a major determinant for their chloroplastic localization and retention.
A universal feature of eukaryotic positive-strand RNA viruses is that replication of their genomes is closely associated with intracellular membranes (reviewed in reference 8). Most purified viral RNA replication complexes copurify with membrane extracts from infected cells (reviewed in reference 10) and, although in some cases RNA synthesis activity can be solubilized (24, 67), in vivo and in vitro studies suggest that the presence of membranes and/or phospholipids is essential for at least some steps of RNA replication (37, 41, 67). It was proposed that these membranes can play both a structural and a functional role in the replication complex.
Electron microscopy observations of infected cells revealed that many positive-stranded RNA viruses induce proliferation and/or reorganization of the intracellular membranes of their host to create a membrane compartment in which RNA replication takes place. Depending on the virus, a variety of membrane systems can be concerned, including the early and late endomembrane systems (52, 59), the nuclear envelope (13), the vacuole (64), the endosomes and lysosomes (17, 59), the peroxisomes (56), chloroplasts (35), and mitochondria (14, 40). The fact that distinct types of membranes are involved in the replication of different viruses suggests the establishment of specific interactions between such host membranes and virus-encoded proteins. A number of viral proteins that target replication complexes to intracellular membranes have been identified (9, 48, 55, 63). Membrane interaction of host-encoded factors that are part of the viral replication complex has also been reported (22, 68).
Despite this universal association of positive-strand RNA virus replication complexes with intracellular membranes, little is known about the mechanisms by which the viral replication complexes are targeted to and assembled on specific membrane sites. Characterizing these structures and the mechanisms of their localization may help to identify general principles in positive-strand RNA virus replication. We address here this question by studying the assembly of the replication complex of Turnip yellow mosaic virus (TYMV), the type member of the tymovirus group. TYMV shares viral replication features with positive-strand RNA viruses from other members of the alphavirus-like supergroup of viruses and has proven useful in investigating fundamental aspects of viral multiplication (3, 65).
TYMV is a small spherical plant virus that infects members of the Cruciferae (reviewed in reference 39). Upon infection, TYMV triggers the development of typical cytological abnormalities that appear to be confined to the chloroplasts (39). These include the swelling and clumping of the chloroplasts and the appearance of peripheral structures consisting of membrane vesicles, 50 to 100 nm in diameter, that are likely to result from the invagination of the chloroplast envelope into the organelle (23). These small vesicles are closely associated with TYMV RNA replication, as revealed by previous in vivo RNA labeling observations (35) and immunocytochemical experiments with antibodies raised against the viral replication complex (19). Consistent with these results, membrane-associated extracts that selectively synthesize TYMV negative-strand RNAs have been isolated from TYMV-infected plant cells (11, 44).
The TYMV genome is composed of a monopartite, positive-sense RNA genome of 6,718 nucleotides (nt) (Fig. 1). It is capped and directs the expression of two extensively overlapping nonstructural proteins: 69-kDa (69K) protein and 206K protein (42, 66). A third open reading frame (ORF) encodes the 20-kDa coat protein (CP), which is expressed from a subgenomic RNA. The 206K protein is the only viral protein required for TYMV RNA replication (66). It shows considerable amino acid sequence similarities with nonstructural putative replication proteins of several positive-strand RNA viruses and domains indicative of methyltransferase, NTPase/helicase, and RNA-dependent RNA polymerase (RdRp) activities have been highlighted in its sequence (20, 31, 53). This large nonstructural protein also contains a papain-like cysteine proteinase domain located between the methyltransferase and the NTPase/helicase domains (54) that is responsible for the cotranslational proteolytic cleavage of the 206K protein in vitro (6, 43). Mapping of the cleavage site (between A1259 and T1260) revealed that the resulting N-terminal protein product of 140 kDa (140K) contains the methyltransferase, proteinase, and NTPase/helicase motifs, whereas the C-terminal 66K protein encompasses the RdRp domain (5, 30). This cleavage was also demonstrated to be functional in vivo (50), and both the 140K and 66K viral proteins appear to be essential for the replication of TYMV RNA genome (65). We showed previously that, during viral infection, the TYMV 66K protein localizes to the virus-induced membrane vesicles present at the chloroplast envelope (50). It is not known, however, whether the 66K protein carries the determinants for its own localization.
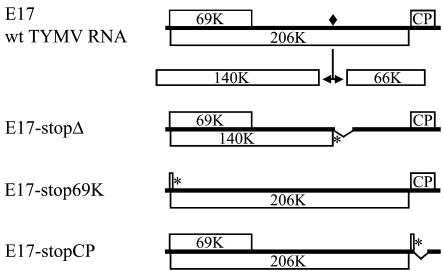
FIG. 1.
Schematic representation of the genomic organization of wild-type TYMV RNA and mutant genomes used in the present study. Open bars denote viral ORFs. The encoded 206K protein is proteolytically processed at a peptide bond signified by a filled square. Deletions are indicated by broken bars, and introduced stop codons are indicated by asterisks.
In the present study, we used the ability to express the TYMV-encoded replication proteins in plant protoplasts from separate plasmids to investigate the determinants of their localization. Green fluorescent protein (GFP) fusion and immunofluorescence experiments provided the opportunity to trace the 66K protein within transfected or infected protoplasts. We demonstrate that the 66K protein displays a cytoplasmic distribution when expressed individually and that its targeting to the chloroplast envelope in the course of the viral infection is solely dependent on the expression of the 140K protein. Our studies also demonstrate that the 140K protein is targeted to the chloroplast envelope in the absence of other viral factors and that it induces the clumping of the chloroplasts, one of the typical cytological effects of TYMV infection.
MATERIALS AND METHODS
Plasmid constructions.
All DNA manipulations were performed by using standard techniques (2, 57). The sequences of PCR-generated DNA fragments were confirmed by DNA sequencing, and the overall structures of all plasmids were confirmed by restriction analysis. Laboratory designations for plasmids are given in parentheses.
Intermediate plasmids.
A DNA fragment corresponding to the 140K ORF was amplified by PCR with Pfu DNA polymerase (Promega) and the primers 5′-CCGGAATTCGAACTAGTCATGGCCTTCCAATTAGCATTGGACGCCC-3′and 5′-GGAAGCTTGAATTCAGGCCCCGTTGAGTTTGGGGCCGCG-3′ with E17 (15) as a template. After digestion with SpeI and EcoRI, the fragment was cloned in the similarly restricted pGadGH vector (Clontech) to create pGad-140K.
A DNA fragment corresponding to the 66K ORF was amplified by PCR with E17 as a template, Pfu DNA polymerase, and primers 5′-CCGGAATTCGAACTAGTCACCCCCAGCGCATCCCCCACCCACCGTTCG-3′ and 5′-GGCCGCTCGAGGATCCTATTGGACGTAGTGAAGCAATTCAGACTC-3′. The resulting PCR product was digested with SpeI and BamHI and cloned into the SpeI-BglII-restricted pGadGH vector to create pGad-66K. Cloning of the AvrII-SacII DNA fragment of E17 into the SpeI-SacII-restricted pGad-66K created pGad-JN115.
The NcoI-PstI fragment of p17AE corresponding to the N terminus of the 66K ORF (25) was cloned into the similarly restricted vector pBS-Nco+, a derivative of pBlueScript II KS(+) in which a unique NcoI restriction site replaced the SmaI site, to create pBS-66-Nterm. The C terminus of the 66K ORF was then excised from pGad-66K by digestion with PstI and SmaI and recloned into the PstI-EcoRV-restricted pBS-66-Nterm to create the plasmid pBS-66K.
A similar strategy was used to create pBS-His66K, except that plasmid p8AF (25) was used to provide the N-terminal His6-tagged version of the 66K ORF.
In order to obtain the clone pBS-66KHis in which the His6 tag is fused to the C-terminal of the 66K ORF, a DNA fragment was amplified by PCR with E17 as a template, Pfu DNA polymerase, and the primers 5′-ACTCGCCACGACTGGCCATCCG-3′ and 5′-GCGAATTCAAGCTTAATGGTGATGGTGATGGTGTTGGACGTAGTGAAGCAATTCAGACTCAGCG-3′. The resulting PCR product was digested with PstI and HindIII and cloned into the similarly restricted pBS-66-Nterm to create pBS-66KHis.
Full-length cDNA constructs and derivatives.
Plasmid E17, from which genomic length infectious transcripts corresponding to the entire genome of TYMV can be obtained, was described previously (15).
Plasmid E17-stopΔ (dpA3) was constructed in two steps. Frist, E17 was digested with SalI and SacII, followed by insertion of the adaptor 5′-TCGACTCTAGAACCGC-3′. The resulting plasmid E17Δ contains an in-frame deletion within the 66K ORF (i.e., amino acids 74 to 165 missing). Then, the BamHI-SalI fragment of pGad-140K was cloned in the similarly restricted E17Δ to create E17-stopΔ, in which a stop codon truncates the 206K ORF at the cleavage site (amino acid 1259) and in which nt 3872 to 4366 of the TYMV genome are deleted (amino acids 1 to 165 of the 66K ORF).
To obtain the plasmid E17-stop69K (gdA2), a DNA fragment was amplified by PCR with Pfu DNA polymerase and primers 5′-GGAAACAGCTATGACCATG-3′ and 5′-CCTTCGGAATGGACCATGGGTAGGTCTGTATCGACGATCGAATTGAATCAACTGTGGATTCGAGAATC-3′ with E17 as a template. After digestion with HindIII and NcoI, the fragment was cloned in the similarly restricted E17 to create E17-stop69K, in which a stop codon truncates the 69K ORF at amino acid 30 without modification of the 206K ORF.
Plasmid E17-stopCP (C4+mcs) was constructed in two steps. First, a DNA fragment was amplified by PCR with Pfu DNA polymerase and primers 5′-GACCCTTCTACCGGCCTCCATC-3′ and 5′-CACGTGCCCGGGACGCGTCTGGTAGGACGGTGGCGACGGTGAC-3′ with E17 as a template. After digestion with SstII and XmaI, the fragment was cloned in the similarly restricted E17. The resulting plasmid C4 contains a deletion within the CP ORF (i.e., nt 5708 to 6058 of the TYMV genome missing). Then, the adaptor oligonucleotide 5′-GTGAATAGGCCTCGAGGGCCTTCGAACTAGTACGTATCTAGAGCGGCCGCCC-3′ was inserted in the SmaI restriction site to create a stop codon interrupting the CP ORF at amino acid 26.
Expression vectors.
All plant expression vectors derive from pDH51, a pUC-based plant expression cassette that includes the cauliflower mosaic virus (CaMV) 35S promoter and terminator (49). To facilitate future cloning of the genes of interest and enhance their translation, the original polylinker was replaced by the Ω adaptor 5′-TATTTTTACAACAATTACCAACAACAACAAACAACAAACAACATTACAATTACTATTTACAATTACCATGGTACTAGTCCCGGGATCCT-3′. It contains the 5′-untranslated leader of tobacco mosaic virus (TMV) (the omega element; underlined), a sequence that functions as a translational enhancer (18) and several restriction sites. The corresponding plasmid was designated pΩ and was further used for cloning the TYMV genes.
To construct the expression vector encoding the 66K protein and its histidine-tagged derivatives, the 66K ORFs were excised from pBS-66K, pBS-His66K, and pBS-66KHis by digestion with NcoI and XhoI and recloned into the NcoI-SalI-restricted pΩ vector to create pΩ-66K, pΩ-His66K, and pΩ-66KHis, respectively.
To obtain the expression plasmid encoding the 140K protein, the 140K ORF was excised from pGad-140K by digestion with SpeI and SalI and recloned into the similarly restricted pΩ vector to create pΩ-140K.
To obtain the expression plasmid encoding the 206K protein, a SmaI site was first inserted in pΩ-140K in place of the SalI site with the use of the adaptor oligonucleotide 5′-TCGAGCCCGGGC-3′. The SacI-SmaI DNA fragment of pGad-JN115 was then excised and recloned into the similarly restricted pΩ-140K to create pΩ-206K (dpB16).
To construct the expression vector pΩ-CP encoding the TYMV CP, a DNA fragment was amplified by PCR with Pfu DNA polymerase and primers 5′-GGACTAGTCCATGGAAATCGACAAAGAACTCGC-3′ and 5′-GCTCTAGAGGATCCTTAGGTGGAAGTGTCCGT-3′ with E17 as a template. The resulting PCR product was digested with SpeI and XbaI and cloned into the similarly restricted pΩ expression vector.
A similar strategy was used to construct PΩ-69K (gdG3) encoding the TYMV 69K protein with a His6 tag at the C terminus, except that the plasmid E17-206K-stop (15) was used as a template for the PCR. This clone contains three point mutations aiming at preventing the expression of the overlapping 206K protein without introducing any modification of the 69K ORF. The primers used for PCR were 5′-TAATACGACTCACTATAGGGCTCGAGACTAGTCATGAGTAACGGCCTTCCAATAAGCATTGGACGC-3′ and 5′-GTCGACTCTAGAGCATGCTCAGTGATGGTGATGGTGATGATCGGTGTCGGGGGCGCTGCCGTAGTC-3′. The resulting DNA fragment was digested with SpeI and SphI and cloned into the similarly restricted pΩ expression vector.
To construct GFP gene fusions, the polylinker of pΩ was first replaced by the adaptor oligonucleotide 5′-CTAGTCGCGAGCTCTGCAGATCTCTAGATCCATGGGATCCGTACGCGGCCGCCCTAGGTACGTAGTCGACTCGAGCATG-3′ to create pΩ-adaptGFP (dpC4). The DNA fragment corresponding to the enhanced green fluorescent protein (EGFP) gene (Clontech) was then excised from pCK-EGFP (16) by digestion with NcoI and BsrGI and cloned into the NcoI-BsiWI-restricted pΩ-adaptGFP to create the expression vector pΩ-EGFP (dpD1) encoding free EGFP.
Cloning of the SpeI-XhoI fragment of pGad-66K into the AvrII-XhoI-restricted pΩ-EGFP created pΩ-EGFP-66K (dpE7).
Similarly, cloning of the SpeI-SalI fragment of pGad-140K into the AvrII-SalI restricted pΩ-EGFP created pΩ-EGFP-140K (ajB2).
In order to obtain the plasmid pΩ-66K-EGFP (dpG40) in which the EGFP gene is fused at the C terminus of the 66K protein coding sequence, the SpeI-PstI fragment of pBS-66-Nterm was first cloned into the similarly restricted pΩ-EGFP to create pΩ-66-Nterm-EGFP (dpF30). Then, a DNA fragment was amplified by PCR with Pfu DNA polymerase and primers 5′-ACTCGCCACGACTGGCCATCCG-3′ and 5′-GCTCTAGATTGGACGTAGTGAAGCAATTCAGAC-3′ with E17 as a template. The resulting PCR product was digested with PstI and XbaI and cloned into the similarly restricted pΩ-66-Nterm-EGFP vector to create pΩ-66K-EGFP.
In vitro transcription and translation.
Capped in vitro transcripts were obtained from AgeI-linearized DNA templates by using the Message Machine Transcription system (Ambion) according to the supplier's instructions. In vitro translation reactions were carried out in micrococcal nuclease-treated reticulocyte lysate as described previously (15).
Preparation and infection of protoplasts.
Protoplasts of Arabidopsis thaliana ecotype Columbia were prepared from a cell suspension culture as described previously (50). A total of 106 protoplasts were transfected with 5 μg of (i) viral RNA, (ii) in vitro transcript, (iii) plasmid expression vector, or (iv) a mixture of both in cotransfection experiments. Transfected protoplasts were incubated at 24°C in the dark for the indicated period of time as described previously (50). Under these conditions, routine transfection levels were ca. 10% of transfected protoplasts, and the efficiency of cotransfection was estimated to be at least 70% of the transfected protoplasts.
Plant inoculations.
For plant inoculations, aliquots of 4 × 105 protoplasts were collected at 48 h posttransfection, lysed in 50 μl of inoculation buffer (20 mM sodium phosphate [pH 7.5]), and rubbed on two young leaves of 5- to 6-week-old Chinese cabbage (Brassica pekinensis cv. Granaat) plants, using celite as an abrasive. The plants were grown at 20 to 25°C with a 16-h daylength. At 4 weeks postinoculation, both the inoculated and uninoculated young developing leaves were collected, and detection of the viral CP in the samples was performed as described previously (50).
SDS-PAGE and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis with polyclonal anti-66K or anti-CP antibodies were performed as previously described (50), except that the anti-CP antibody was diluted 10,000-fold.
Immunofluorescent labeling, GFP observation, and epifluorescence microscopy.
TYMV-infected or plasmid-transfected protoplasts (∼106 cells) were harvested for immunofluorescence staining at 18 to 48 h posttransfection and were processed essentially as described previously (50), except that goat anti-rabbit immunoglobulin G conjugated to Alexafluor-488 (Molecular Probes) diluted 2,000-fold in phosphate-buffered saline-1% bovine serum albumin was used as a secondary antibody. To observe GFP fluorescence in living cells, ∼105 cells were harvested, immobilized between microscope slides and coverslips, and examined immediately.
Fluorescence microscopy and image acquisitions were performed as previously described (50) by using a Leica DMR epifluorescence microscope. Standard filters for fluorescein isothiocyanate were used to monitor and record Alexafluor-488 and EGFP fluorescence. Chloroplasts were identified in fixed cells by DAPI (4′,6′-diamidino-2-phenylindole) staining of the chloroplastic DNA as described previously (50) or in living cells by the chlorophyll autofluorescence that was monitored and recorded by using the rhodamine channel.
RNA extraction and Northern blot hybridization.
The protoplasts were collected by centrifugation at 80 × g for 5 min at room temperature, and nucleic acids were extracted as described previously (29). Total RNA (2.5 μg) was loaded onto 1% agarose-formaldehyde gels and, after they were blotted onto charged nylon membranes (Roche), the membrane was hybridized with a digoxigenin (DIG)-labeled RNA probe according to the supplier's instructions (Roche). After stringent washes (in 0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-SDS 0.1% at 68°C), the blot was incubated with anti-DIG antibody conjugated to alkaline phosphatase and then visualized with CDP-Star as a substrate according to the supplier's instructions (Roche).
For the production of the DIG-labeled probe, plasmid 1AH was constructed by subcloning the 3,044-bp XhoI-EcoRI fragment of E17 in the similarly restricted pBluescript II KS(+). The probe was prepared by in vitro transcription with T7 RNA polymerase of BglII-restricted 1AH, in the presence of DIG-UTP (Roche) according to the supplier's instructions. The 338-nt riboprobe hybridized to nt 5984 to 6318 of the viral genomic RNA and was suitable for detection of the viral genomic plus-strand RNA and the subgenomic RNA.
RESULTS
Construction of plasmids expressing the 66K protein and its derivatives in plant cells.
In order to study the localization of the viral proteins independently of the viral infection, we first constructed the expression plasmid vector pΩ, a derivative of pDH51 (49) that allows the expression of cloned genes in plant cells under the transcriptional control of the CaMV 35S promoter. The TMV omega element, which functions as a translational enhancer (18), was inserted downstream of the promoter sequence. Cloning of the TYMV 66K protein coding sequence into pΩ resulted in pΩ-66K, whereas pΩ-His66K and pΩ-66KHis encode derivatives of the 66K protein with in frame His6 tags at the N or C terminus, respectively.
The use of GFP, which allows direct fluorescence microscopy of living cells, was also explored. EGFP, a fluorescence-enhanced version of GFP (Clontech), was fused in frame to the N or C terminus of 66K protein to create two fusions, EGFP-66K and 66K-EGFP, that were encoded by the plasmids pΩ-EGFP-66K and pΩ-66K-EGFP, respectively.
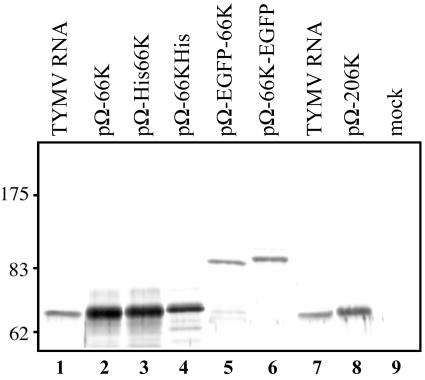
The expression in Arabidopsis cells of the 66K protein and its derivatives was analyzed by transfection of each of the plasmid constructs in protoplasts, followed by immunoblotting with anti-66K specific antibodies (Fig. 2). As shown in lane 2, in cells transfected with pΩ-66K the anti-66K antibodies recognized a major protein that comigrated with the 66K protein produced in the course of the viral infection (lane 1). In cells expressing the His-tagged derivatives (lanes 3 and 4) or the GFP-fused versions of the 66K protein (lanes 5 and 6), the anti-66K antibody-reactive band shifted to a higher position that was consistent with the expected molecular mass of the fusion proteins.
FIG. 2.
Expression of the TYMV 66K protein and its derivatives in plant cells. Arabidopsis protoplasts were transfected with TYMV RNA (lanes 1 and 7) or plasmids pΩ-66K (lane 2), pΩ-His66K (lane 3), pΩ-66KHis (lane 4), pΩ-EGFP-66K (lane 5), pΩ-66K-EGFP (lane 6), pΩ-206K (lane 8), and water (lane 9). The cells were harvested at 27 h posttransfection, and total proteins were extracted and subjected to SDS-8% PAGE and immunoblot analysis with anti-66K polyclonal antibodies. The position of molecular weight markers (Biolabs) is indicated.
The 66K protein and its derivatives have a cytoplasmic distribution when expressed individually.
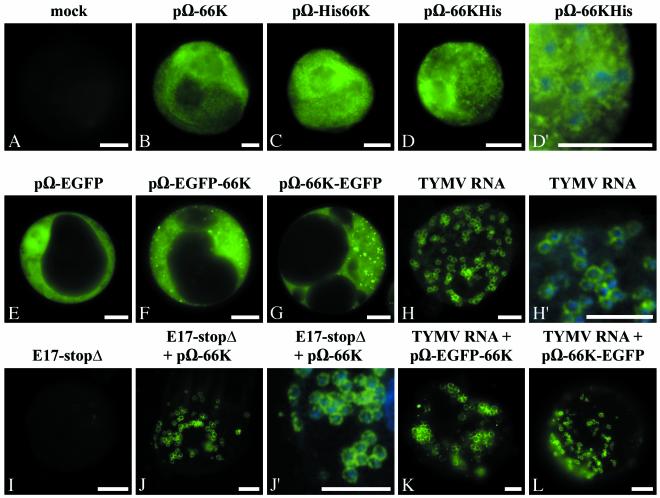
In order to determine the intracellular sites of accumulation of the TYMV 66K protein in the absence of other virus-encoded proteins, Arabidopsis protoplasts were transfected with the expression vectors pΩ-66K, pΩ-His66K, or pΩ-66KHis. At 46 h posttransfection, the transfected cells were fixed, and indirect immunofluorescence microscopy was carried out by using anti-66K antibodies and secondary antibodies conjugated to Alexafluor-488 fluorochrome. Chloroplasts were identified in fixed cells by DAPI staining of the chloroplastic DNA. Figure 3B to D′ show representative epifluorescence micrographs of such cells. Transfected cells displayed a bright fluorescent staining of the 66K protein (green) throughout the cytoplasm. The protein was excluded from subcellular compartments such as the nucleus, the vacuole, or chloroplasts, as evidenced by confocal microscopy (data not shown). Mock-infected cells processed and imaged in parallel showed negligible fluorescence (Fig. 3A).
FIG. 3.
The 66K protein and its derivatives have a cytoplasmic distribution when expressed individually but relocalize to the chloroplast envelope in infected cells. Arabidopsis protoplasts were transfected with water (A); with the expression plasmids pΩ-66K (B), pΩ-His66K (C), pΩ-66KHis (D and D′), pΩ-EGFP (E), pΩ-EGFP-66K (F), or pΩ-66K-EGFP (G); and with the viral RNA (H and H′). Transfections were also performed with transcript E17-stopΔ alone (I), or together with the plasmid pΩ-66K (J and J′), or with plasmid pΩ-EGFP-66K (K) or pΩ-66K-EGFP (L), together with viral RNA. The protoplasts were collected at 22 h posttransfection (H) or 46 h posttransfection (A to G and I to L), and they were either observed directly for GFP fluorescence (E to G, K, and L) or processed for indirect immunofluorescence labeling by using anti-66K antiserum followed by secondary antibodies coupled to Alexafluor-488 (A to D′ and H to J′). The cells were observed by epifluorescence microscopy. Scale bars, 10 μm. Panels D′, H′ and J′ are enlargements of panels D, H, and J, respectively, in which the DAPI staining (blue) of the chloroplastic DNA was acquired and superimposed to the fluorescence signal of the viral protein (green) to visualize the location of chloroplasts.
The cytoplasmic distribution of the 66K protein was confirmed by protoplast transfection experiments with the expression vectors pΩ-EGFP-66K or pΩ-66K-EGFP. In these cases, the subcellular distribution of the fusion proteins can be directly observed by fluorescence microscopy in living cells due to the green fluorescence of the GFP moiety. When expressed from pΩ-EGFP-66K or pΩ-66K-EGFP, the GFP-fused 66K proteins appeared distributed diffusely in the cytoplasm (Fig. 3F and G). In some cases, the fluorescent proteins were also observed as punctate structures of variable size and intensity distributed throughout the cytoplasm. These foci do not correspond to chloroplasts, as evidenced by the simultaneous observation of chlorophyll red autofluorescence in living cells, or staining of the chloroplastic DNA with DAPI in fixed cells. When EGFP was produced in a free form from pΩ-EGFP, diffuse fluorescence characteristic of cytosoluble GFP was observed throughout the cell (Fig. 3E).
These studies demonstrate that the 66K protein, when expressed individually, is not targeted to the chloroplast but that it accumulates in the cytoplasm. These data therefore suggest that targeting of the 66K protein to the chloroplast envelope in the course of the viral infection, as can be seen in Fig. 3H and H′ and as evidenced previously (50), is not an intrinsic property of the 66K protein but rather a process that is dependent upon virus infection.
The 66K protein and some of its derivatives can support RNA replication in trans.
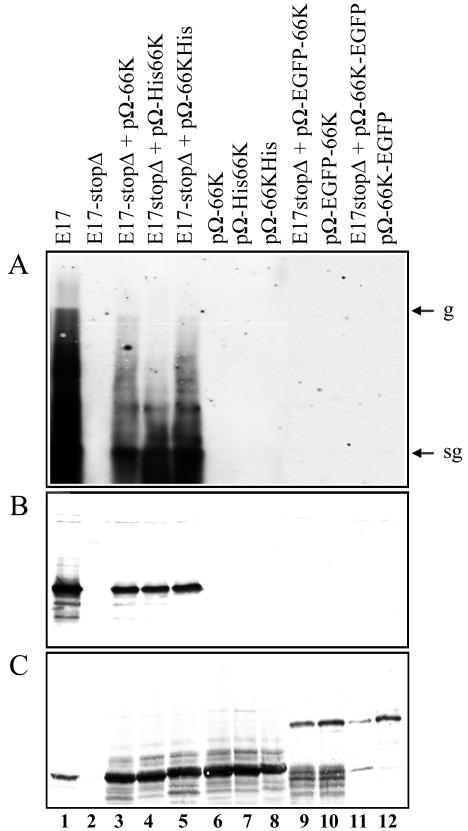
To test whether the 66K protein, and its fused derivatives, expressed in trans from expression plasmids can support TYMV RNA replication and subgenomic mRNA synthesis, the expression vectors pΩ-66K, pΩ-His66K and pΩ-66KHis, pΩ-EGFP-66K, and pΩ-66K-EGFP expressing wild-type 66K, His-tagged 66K derivatives, and GFP-fused 66K proteins, respectively, were introduced into plant cells, together with a defective TYMV-derived replication template. The variant RNA to be used as a template for replication, named E17-stopΔ, contains two alterations aimed at preventing the synthesis of a functional 66K protein (Fig. 1). First, a stop codon was introduced at amino acid 1259 of the 206K ORF, i.e., at the C terminus of the 140K protein (5, 30). This stop codon was followed by a 494-nt deletion (nt 3872 to 4366 of the TYMV genome), the region that formerly encoded the first 165 amino acids of the 66K protein. As expected, upon in vitro translation in rabbit reticulocyte lysate of the corresponding transcript, the 66K protein was not produced, but wild-type amounts of the 69K and the 140K proteins were synthesized (data not shown). Upon transfection of Arabidopsis protoplasts with E17-stopΔ transcript, neither accumulation of viral progeny was detectable by Northern blot hybridization (Fig. 4A, lane 2), nor accumulation of the CP was detectable by Western blot (Fig. 4B, lane 2). These observations are consistent with the essential role played by the 66K protein within the replication process and its debilitation in the E17-stopΔ transcript.
FIG. 4.
The 66K protein and some of its derivatives can support TYMV RNA replication in trans. Arabidopsis protoplasts were transfected with transcript E17 (lane 1), transcript E17-stopΔ (lane 2), or combinations of transcript E17-stopΔ with the plasmids pΩ-66K (lane 3), pΩ-His66K (lane 4), pΩ-66KHis (lane 5), pΩ-EGFP-66K (lane 9), and pΩ-66K-EGFP (lane 11). Each of the expression plasmids was also transfected individually (lanes 6 to 8 and lanes 10 and 12, respectively). The transfected protoplasts were collected at 46 h posttransfection and used both for RNA and protein analyses. (A) Equivalent amounts of total RNA prepared from the transfected protoplasts were analyzed by Northern blotting with a single-stranded, DIG-labeled RNA probe complementary to positive-strand TYMV RNA. The migration positions of genomic (g) and subgenomic (sg) RNAs are indicated. (B and C) Equivalent amounts of proteins were analyzed by SDS-PAGE on 15% (B) or 8% (C) polyacrylamide gels. The gels were electroblotted onto a nitrocellulose filter, and proteins were revealed by Western blotting with anti-CP (B) or anti-66K (C) antiserum.
In contrast, in cells cotransfected by this defective replication template and the plasmid construct pΩ-66K expressing wild-type 66K protein, both genomic and subgenomic RNA (Fig. 4A, lane 3) and a large amount of CP (Fig. 4B, lane 3) could be detected. This demonstrates that the 66K protein produced in trans from an expression vector is able to complement the E17-stopΔ defective RNA. Similar data were observed when the expression vectors pΩ-His66K (Fig. 4A and B, lanes 4) and pΩ-66KHis (Fig. 4A and B, lanes 5) were used, demonstrating that the presence of the His6 tag at one or the other terminus of the protein did not interfere with its functionality. On the other hand, it appeared that the GFP fusion proteins did not support genomic RNA replication and CP synthesis, as shown by the absence of viral RNA progeny and CP in cells that were cotransfected with E17-stopΔ transcript and the plasmid constructs pΩ-EGFP-66K or pΩ-66K-EGFP (Fig. 4A and B, lanes 9 and 11). Thus, the GFP moiety is apparently deleterious for the proper functioning of the 66K protein in the replication process. Western blotting experiments with the anti-66K antibodies were used as controls to verify that all of the constructs used were properly expressed (Fig. 4C).
Given the alterations introduced in E17-stopΔ, it appears extremely unlikely that the replication observed in the cotransfection experiments is due to reversion of the defective replication template or to recombination with the expression vector. Nevertheless, experiments were carried out to verify that wild-type RNA was absent from the progeny of the complementation experiments. For that purpose, a fraction of the protoplasts used in the experiment was collected 46 h posttransfection, lysed and used to inoculate Chinese cabbage plants. Those were inspected 4 weeks postinoculation for disease symptoms, and the presence of CP in the inoculated and young expanding leaves was assayed by Western blotting. This potent assay (65) failed to detect any wild-type RNA molecule in the progeny (data not shown), thereby demonstrating that the 66K protein produced from an expression vector can transcomplement a transcript in which the copy of the 66K gene has been disabled.
The 66K protein and its derivatives relocalize to the chloroplast envelope in infected cells.
Because the above data suggested that viral infection is required for proper targeting of the 66K protein to the chloroplast envelope, it was of interest to investigate the subcellular distribution of the 66K protein and its derivatives within cells during the infection process. For this purpose, Arabidopsis protoplasts were cotransfected with the expression vector pΩ-66K, together with E17-stopΔ transcript. As shown above, under these conditions transcomplementation allows the viral replication to take place. Indirect immunofluorescence microscopy was then carried out on the transfected cells by using anti-66K antibodies. Figure 3J and J′ show a representative epifluorescence micrograph of such cells. In cells where replication occurs, the 66K protein encoded by pΩ-66K appeared targeted to the periphery of chloroplasts, whereas the cytoplasm was almost completely depleted of any fluorescence. Cells transfected with E17-stopΔ transcript alone showed negligible fluorescence (Fig. 3I). Similar experiments were also performed with the expression vector pΩ-EGFP-66K or pΩ-66K-EGFP that were transfected together with wild-type TYMV RNA to provide a functional 66K protein. Localization of the GFP-fused 66K proteins was observed by fluorescence microscopy of the living cells. As shown by representative epifluorescence micrographs of such cells, both EGFP-66K (Fig. 3K) and 66K-EGFP (Fig. 3L) fusion proteins appeared targeted to the chloroplast envelope in cells where replication occurs. Similar observations were made on fixed cells, although GFP fluorescence was reduced relative to unfixed cells (data not shown). In control experiments, cotransfection of TYMV viral RNA with pΩ-EGFP demonstrated that viral infection had no influence on the distribution of free EGFP, which presented a typical cytosoluble protein distribution pattern (data not shown).
Within cotransfected cells, the subcellular distribution of the plasmid-encoded 66K protein and its derivatives, in the shape of rings around the chloroplasts, therefore matched perfectly that of wild-type 66K produced during TYMV viral infection (Fig. 3H and H′). We conclude that during virus multiplication the 66K protein and its derivatives are relocated within the cell and that targeting of the 66K protein to the chloroplast envelope where the replication takes place is dependent upon virus infection. This leads to the assumption that proteins produced during the viral infection (e.g., 69K, CP, 206K, and/or 140K) are necessary to target and/or anchor the 66K protein to the chloroplast envelope.
Targeting of the 66K protein to the chloroplast envelope is not dependent on the 69K or CP protein.
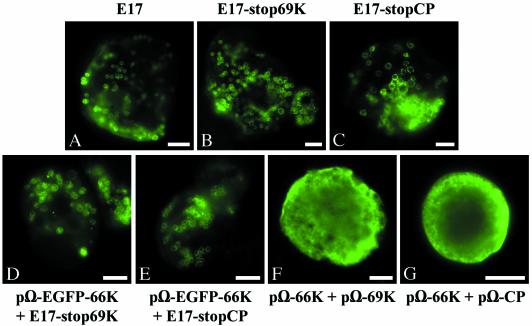
In order to evaluate further the role played by each of the TYMV-encoded proteins on the targeting of the 66K protein to the chloroplast envelope, we examined the effect of disrupting either the 69K movement protein or the 20K CP in the viral genome. For that purpose, two mutant plasmids were obtained (Fig. 1): E17-stop69K, in which a stop codon truncates the 69K ORF at amino acid 30 without modification of the overlapping 140K ORF, and E17-stopCP, in which the CP gene is interrupted at amino acid 26. In vitro translation experiments in rabbit reticulocyte lysate of the corresponding transcripts revealed the expected profile of expression (data not shown). Arabidopsis protoplasts were transfected with each of the mutant transcripts, and indirect immunofluorescence microscopy was then carried out on the transfected cells by using anti-66K antibodies. As shown by representative epifluorescence micrographs of such cells in Fig. 5B and C, the 66K protein encoded by both transcripts was properly targeted to the chloroplast envelope. No major differences in terms of protein distribution were observed compared to wild-type E17 transcripts transfected in parallel (Fig. 5A). Similar results were obtained in experiments involving the expression vector pΩ-EGFP-66K, which was cotransfected together with each of the mutant transcripts. In both cases, the GFP-66K protein, observed by fluorescence microscopy of the living cells was efficiently targeted to the chloroplast envelope (Fig. 5D and E).
FIG. 5.
Targeting of the 66K protein to the chloroplast envelope is not dependent on the 69K movement protein or CP. Arabidopsis protoplasts were transfected with transcript E17 (A), with transcript E17-stop69K alone (B) or together with plasmid pΩ-EGFP-66K (D), with transcript E17-stopCP alone (C) or together with plasmid pΩ-EGFP-66K (E), or with plasmid pΩ-66K together with plasmid pΩ-69K (F) or pΩ-CP (G). The protoplasts were collected at 25 h (A to C) or 46 h (D to G) posttransfection, and they were either observed directly for GFP fluorescence (D to E) or they were processed for indirect immunofluorescence labeling by using anti-66K antiserum, followed by secondary antibodies coupled to Alexafluor-488 (A to C and F to G). The cells were observed by epifluorescence microscopy. Scale bars, 10 μm.
As a complementary approach, we also investigated whether the localization of the 66K protein was affected by the simultaneous expression of the 69K protein or CP. For this purpose, expression vectors encoding the 69K protein or the CP (pΩ-69K and pΩ-CP, respectively) were constructed and Arabidopsis protoplasts were transfected with these plasmids together with the construct pΩ-66K. Indirect immunofluorescence microscopy was then carried out on the cotransfected cells by using anti-66K antibodies. Examination of hundreds of cells revealed that, in both cases, the 66K protein remained in the cytoplasm and was not targeted to the chloroplast envelope. Representative epifluorescence micrographs of such cells are shown in Fig. 5F and G.
These experiments clearly demonstrate that the 69K movement protein and CP are not required for the proper targeting of the 66K protein to the chloroplast envelope.
Targeting of the 66K protein to the chloroplast envelope is dependent on the 140K protein.
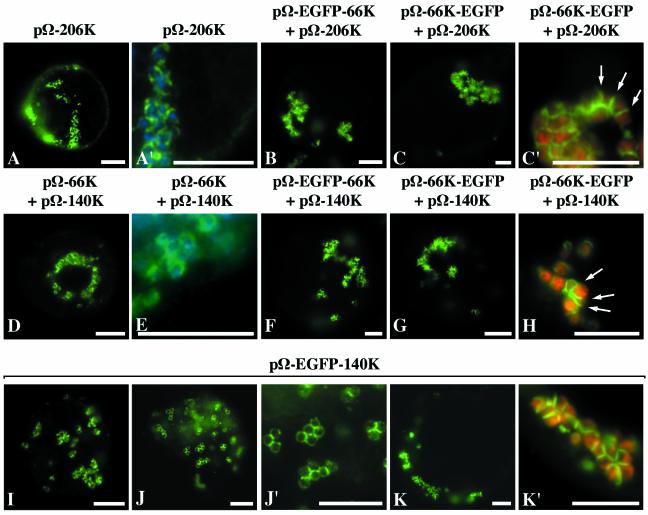
To further assess the role played by the other virus-encoded proteins, the expression vector pΩ-206K encoding the 206K protein was constructed. Upon its transfection into Arabidopsis protoplasts, analysis of protein extracts revealed that the 66K protein was easily detected by immunoblotting with anti-66K antiserum (Fig. 2, lane 8), therefore demonstrating that the 206K protein produced in this system is properly processed. The inability to detect the uncleaved 206K product is consistent with protoplast infection experiments with viral RNA (Fig. 2, lanes 1 and 7) (50) and is most likely due to the fast and efficient processing of the 206K protein into its 140K and 66K protein products in plant cells. Arabidopsis protoplasts were transfected with the expression vector pΩ-206K, and indirect immunofluorescence microscopy was then carried out on the transfected cells by using anti-66K antibodies. Figure 6A and A′ show representative epifluorescence micrographs of such cells in which the periphery of chloroplasts displayed a bright fluorescent staining. These results demonstrate that expression of the 206K protein allows targeting of the 66K protein to the chloroplast envelope. Similar experiments were also performed by using the expression vectors pΩ-EGFP-66K or pΩ-66K-EGFP that were transfected together with pΩ-206K. Localization of the GFP-fused 66K proteins was observed by fluorescence microscopy of the living cells. As shown by representative epifluorescence micrographs of such cells, both EGFP-66K (Fig. 6B) and 66K-EGFP (Fig. 6C and C′) fusion proteins were targeted to the chloroplast envelope.
FIG. 6.
Targeting of the 66K protein to the chloroplast envelope is dependent on the 140K protein, which localizes to the chloroplast envelope in the absence of other viral components. Arabidopsis protoplasts were transfected with the plasmid pΩ-206K alone (A and A′) or pΩ-206K together with the plasmids pΩ-EGFP-66K (B) or pΩ-66K-EGFP (C and C′) or with the plasmid pΩ-140K together with the plasmids pΩ-66K (D and E), pΩ-EGFP-66K (F), or pΩ-66K-EGFP (G and H), or with the plasmid pΩ-EGFP-140K (I to K′). The protoplasts were collected at 22 h (A and A′) or 46 h (B to K′) posttransfection, and they were either observed directly for GFP fluorescence (B to C′ and F to K′) or they were processed for indirect immunofluorescence labeling by using anti-66K antiserum, followed by secondary antibodies coupled to Alexafluor-488 (A, A′, D, and E). The cells were observed by epifluorescence microscopy. Scale bars, 10 μm. Panels A′, C′, J′, and K′ are enlargements of panels A, C, J, and K, respectively. In order to visualize the location of chloroplasts, the DAPI staining (blue) of the chloroplastic DNA was acquired and superimposed onto the fluorescence signal of the viral protein (green) in panels A′ and E, whereas in panels C′, H, and K′ the chlorophyll autofluorescence (red) was acquired and superimposed onto the GFP fluorescence (green).
Despite the proper targeting of the 66K protein and its GFP fusion derivatives to the chloroplast envelope, it should be noted that the pattern of localization observed was somehow different from the one observed during the viral infection process. In particular, the fluorescent signal corresponding to the 66K protein often formed partial rings or arcs, as opposed to the images of circles typically seen during viral infection. These arcs appeared to localize preferentially at the junctions between clumped chloroplasts (see arrows in Fig. 6C′). In addition, the clumping of chloroplasts within cells transfected with pΩ-206K was much more severe than the one that typically appears during TYMV infection (Fig. 3H), leading to the formation of crowded masses of chloroplasts.
Precise data concerning the timing of processing of the 206K protein are currently lacking. However, the fact that the unprocessed 206K protein was not detected in protoplasts transfected with pΩ-206K (Fig. 2, lane 8) and the observation that targeting of the 66K protein to the chloroplasts could be achieved during transcomplementation experiments in which only the 140K and 66K processed products are provided (Fig. 3), led to the hypothesis that the 140K cleavage product rather than the unprocessed 206K polyprotein may be involved in mediating chloroplast targeting of the 66K protein.
To investigate more precisely the role played by the 140K protein in this process, pΩ-140K, an expression vector encoding the 140K protein was constructed and Arabidopsis protoplasts were cotransfected with the expression vector pΩ-66K together with the pΩ-140K construct. Indirect immunofluorescence microscopy was then carried out on the transfected cells by using anti-66K antibodies. As seen in Fig. 6D and E (representative epifluorescence micrographs of such cells), when the 66K protein was expressed in the presence of the 140K protein, it appeared distributed at the periphery of chloroplasts. Similar experiments were also performed by cotransfection of the expression vectors pΩ-EGFP-66K or pΩ-66K-EGFP together with the expression vector pΩ-140K. Observation by fluorescence microscopy of the living cells confirmed that, in both cases, the GFP-fused 66K proteins were efficiently targeted to the chloroplast envelope by the simultaneous expression of the 140K protein (Fig. 6F to H). In some cases, the distribution of the 66K protein was in the shape of rings (Fig. 6D and F), but in others the labeling formed arcs that seem to be present preferentially at the junctions between clumped chloroplasts (arrows in Fig. 6H).
Altogether, these results demonstrate that relocalization of the 66K protein to the chloroplast envelope during viral replication is solely dependent on the expression of the 140K protein.
The 140K protein localizes to the chloroplast envelope in the absence of other viral components.
To explore the independent localization properties of the 140K protein, the expression vector pΩ-EGFP-140K was constructed, encoding the 140K protein fused to EGFP. Arabidopsis protoplasts were transfected with this plasmid, and the intracellular distribution of the EGFP-140K protein was observed by fluorescence microscopy of the living cells. Representative epifluorescence micrographs of such cells are shown in Fig. 6I to K′. Expression of EGFP-140K induced a clumped chloroplast distribution, demonstrating that the 140K viral protein is involved in that process. The EGFP-140K fusion protein displayed a perfect localization at the chloroplast envelope, in the shape of partial or complete rings, with a stronger labeling at the junctions between adjacent chloroplasts (Fig. 6J′ and K′).
These data therefore demonstrate that the 140K protein is targeted to the chloroplast envelope independently of any other TYMV-encoded proteins or RNA replication and is able to induce the clumping of the chloroplasts, one of the typical cytological effects of TYMV infection.
DISCUSSION
A universal feature of eukaryotic positive-strand RNA viruses is that replication of their genomes is closely associated with intracellular membranes (reviewed in reference 8). In the case of TYMV, replication complexes colocalize with virus-induced membrane vesicles that are thought to result from the invagination of the chloroplast envelope (23). We showed previously by immunodetection experiments and electron microscopy observations performed on infected plants and protoplasts that the TYMV 66K protein localizes to these membrane vesicles during viral infection (50). We investigated here the intracellular localization and organellar targeting of TYMV 66K protein in Arabidopsis cells with the aim of defining the determinants for this subcellular localization.
We were interested in locating the 66K protein when it is expressed individually in transfected cells, and the effectiveness of noninvasive techniques, such as GFP tagging, had already been documented for this purpose (9, 58). However, since GFP fusions may debilitate 66K protein function or affect its location in cells, the 66K protein was also expressed with His6 tags or as an unfused version, which could be detected by immunofluorescence. The 66K protein and its derivatives were properly expressed and were readily detectable in transfected cells either by Western blotting (Fig. 2) or by fluorescence microscopy (Fig. 3). In all cases, the 66K protein displayed a cytoplasmic distribution and the presence of the His tag or the GFP moiety had no apparent effect on its subcellular localization. In some cases, discrete spots were observed that did not colocalize with chloroplasts but whose nature was not investigated in details. These foci may result from the aggregation of the 66K protein, since it has been previously reported that the 66K protein formed aggregates when produced in large amounts in insect cells (25) and that similar tendancies to aggregation or oligomerization have previously been reported for other viral RdRps, including Brome mosaic virus (BMV) 2a (9, 62) and poliovirus 3D proteins (47).
Remarkably, the 66K protein produced from the expression vector pΩ-66K was functional since it had the potential to transcomplement a replication-defective RNA (Fig. 4). The His-tagged versions behaved comparably, demonstrating that the presence of the His6 tag at one or the other terminus of the protein did not interfere with its functionality. It is clear that the rescued mutant produced less positive-strand RNA than did the wild-type viral RNA, but these amounts were not quantified. The low complementation efficiency obtained in the rescue experiment may be due to the cis-preferential replication of TYMV that has been previously reported (65). In contrast, fusion of EGFP to the N or C terminus of the 66K protein abolished its ability to support viral replication, possibly because the GFP moiety interfered with the proper folding of the protein or its binding to other components of the replication complex.
Under conditions where viral replication occurs, i.e., in the transcomplementation experiments, the 66K protein encoded by pΩ-66K was relocated to the chloroplast envelope (Fig. 3). This suggests that the proper targeting of the 66K protein to the replication complexes depends on the expression of other virus-encoded proteins. Interestingly, although the GFP-fused versions of the 66K protein were not functional in replication (Fig. 4), their distribution was indistinguishable from that of the functional proteins in coinfection experiments involving wild-type viral RNA (Fig. 3). This suggests that the ability of the GFP fusions to establish the proper interactions needed for chloroplast localization is not impaired and that they might constitute suitable markers for studying the determinants of 66K protein localization, as long as a functional 66K protein is provided simultaneously to allow the viral replication to proceed. These data also indicate that the GFP fusion proteins did not interfere in a dominant manner with the proper targeting of the replication complexes to the chloroplast envelope.
The results obtained with the various TYMV-derived mutants, as well as in the coexpression experiments, clearly demonstrated that neither CP nor the 69K movement protein is involved in the chloroplastic localization of the 66K protein (Fig. 5). These data are consistent with previous results demonstrating that these two viral proteins are not required for viral replication (7, 66). In contrast, expression of the 140K protein, either in the form of the 206K polyprotein precursor or as an independent protein, allowed the relocation of the 66K protein to the chloroplast envelope (Fig. 6). Our studies also demonstrate (Fig. 6) that the 140K protein is targeted to the chloroplast envelope in the absence of other viral factors and that it induces the clumping of the chloroplasts, a typical cytopathic effect of TYMV infection (39). Altogether, these findings reveal a novel role of the 140K protein as a key organizer of the assembly of the TYMV replication complexes and a major determinant for their chloroplastic localization and retention.
The results presented here address several important aspects of TYMV replication complex formation and reveal multiple parallels between membrane localization of RNA replication complexes by TYMV and other alphavirus-like viruses, such as BMV or Semliki forest virus (SFV). Like BMV 1a and SFV nsP1 (48, 51), the TYMV 140K protein is able to interact with cellular membranes in the absence of other viral factors and to relocate the viral protein encompassing the RdRp domain (BMV 2a, SFV nsP4, and TYMV 66K, respectively) that otherwise is cytoplasmic (9, 34). This observation is consistent with the amino acid sequence similarities existing among these proteins (33) and the fact that the 140K protein encompasses the methyltransferase domain, which has been identified as the main membrane association determinant of BMV 1a and SFV nsP1 (1, 12). In the case of BMV, the recruitment of 2a to intracellular membranes mediated by 1a correlates with their ability to interact, as evidenced in vitro or in two-hybrid experiments (32, 45). It is thus expected that the TYMV 140K and 66K proteins would also interact with each other, and experiments are currently under way to test this hypothesis.
Interestingly, although the assembly of multiprotein replication complexes on intracellular membranes appears to be a general aspect of genome replication by all positive-strand RNA viruses, unknown variations in virus-host interaction lead different viruses to direct replication complex assembly to different membrane sites. For instance, the replication complexes of BMV, SFV, and TYMV target the endoplasmic reticulum, the endosomal or lysosomal membranes, and the chloroplast envelope, respectively (17, 35, 52). Although a detailed basis for this specificity remains to be determined, it has been proposed that tethering of the replication complexes to particular membranes can be controlled by a direct interaction between a helical peptide region and phospholipids (1, 12, 36) and/or by specific interactions with membrane-bound host proteins (22, 68). Further experiments are needed to identify the membrane association determinants of the TYMV 140K protein, as well as its eventual chloroplast partner(s). This might be facilitated by the fact that preparative procedures to isolate chloroplast envelopes as well as methods for the detailed analysis of their lipid and protein composition have already been described (28, 61).
The viral replication process often results in proliferation, vesiculation, and severe rearrangement of the cellular membrane compartments. The importance of these rearrangements in virus replication is not yet clear, but it has been suggested that this process may provide a means of increasing the key lipid constituents and/or the membrane surface area used as a scaffold to assemble the replicative machinery. The compartmentalization resulting from the membrane vesiculation may also be critical for viral replication by diverting the viral RNA from the translation machinery (27), by increasing the local concentration of replication complexes and their associated components (58), or by avoiding host defense responses directed against double-stranded RNA replicative intermediates (4, 26). The cytological abnormalities induced by TYMV infection appear to be confined to the chloroplasts. In infected cells, the chloroplasts are swollen, rounded, and clumped together (39). The data presented here demonstrate that the expression of the sole 140K protein is sufficient to promote the clumping of chloroplasts. This process is therefore unrelated to the production of CP or to the accumulation of TYMV particles between adjacent chloroplasts as previously suggested (38). Although the molecular mechanisms enabling the 140K protein to alter the intracellular distribution of chloroplasts still remain to be defined, it is noteworthy that its protein counterparts in TMV and BMV (proteins 126K and 1a, respectively) can form multimers that result in a higher-order protein structure (21, 46). Therefore, we propose a model in which the 140K protein would interact peripherically with the chloroplast envelope membrane and in which intermolecular self-interactions between multiple 140K proteins would result in the formation and stabilization of chloroplast aggregates. It will be interesting to investigate this model in the future.
Another conspicuous cytopathic effect of TYMV infection is the occurrence of characteristic double membrane vesicles at the chloroplast periphery (39) that are thought to result from invaginations of the chloroplast envelope into the organelle (23) and that serve as sites for viral RNA replication (19, 35). Information concerning the origin and biogenesis of these TYMV-induced vesicles is still missing, but it is tempting to draw an analogy with membrane vesiculation in BMV-infected cells, which was reported to be induced by the expression of the sole viral protein 1a (60). Further ultrastructural studies on cells expressing the TYMV 140K protein will be necessary to clarify whether this individual viral protein also triggers membrane vesiculation processes similar to those occurring during viral infection. This may help to identify the intracellular structures induced by TYMV replication in plant cells and give new insights about the specific virus-host interactions that ultimately determine a successful viral infection.
Acknowledgments
D.P. and A.J. contributed equally to this work.
D.P. and A.J. were the recipients of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT). This work was supported in part by grants from CNRS (Programme Jeunes Equipes) and MENRT (Action Concertée Incitative “Jeunes Chercheurs”) to I.J.
We thank Leonie van Dinten, Julien Notaise, and Mildred Noizet for constructing some of the intermediate plasmids used in the present study; Thierry Candresse for providing anti-CP antibodies; Marie Aronson for the gift of plasmid pDH51; Patrice Dunoyer for the gift of plasmid pCK-EGFP; and members of the lab for comments on the manuscript. The kind help of Leonie van Dinten, Mildred Noizet, and Stéphanie Pflieger for the maintenance of the Arabidopsis cell suspension is also greatly acknowledged.
REFERENCES
- 1.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley, New York, N.Y.
- 3.Barends, S., H. H. Bink, S. H. van den Worm, C. W. Pleij, and B. Kraal. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112:123-129. [DOI] [PubMed] [Google Scholar]
- 4.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 5.Bransom, K. L., S. E. Wallace, and T. W. Dreher. 1996. Identification of the cleavage site recognized by the turnip yellow mosaic virus protease. Virology 217:404-406. [DOI] [PubMed] [Google Scholar]
- 6.Bransom, K. L., J. J. Weiland, and T. W. Dreher. 1991. Proteolytic maturation of the 206-kDa nonstructural protein encoded by turnip yellow mosaic virus RNA. Virology 184:351-358. [DOI] [PubMed] [Google Scholar]
- 7.Bransom, K. L., J. J. Weiland, C. H. Tsai, and T. W. Dreher. 1995. Coding density of the turnip yellow mosaic virus genome: roles of the overlapping coat protein and p206-readthrough coding regions. Virology 206:403-412. [DOI] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Graaf, M., and E. M. J. Jaspars. 1994. Plant viral RNA synthesis in cell-free systems. Annu. Rev. Phytopathol. 32:311-335. [Google Scholar]
- 11.Deiman, B. A., K. Seron, E. M. Jaspars, and C. W. Pleij. 1997. Efficient transcription of the tRNA-like structure of turnip yellow mosaic virus by a template-dependent and specific viral RNA polymerase obtained by a new procedure. J. Virol. Methods 64:181-195. [DOI] [PubMed] [Google Scholar]
- 12.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Zoeten, G. A., G. Gaard, and F. B. Diez. 1972. Nuclear vesiculation associated with pea enation mosaic virus-infected plant tissue. Virology 48:638-647. [DOI] [PubMed] [Google Scholar]
- 14.Di Franco, A., M. Russo, and G. P. Martelli. 1984. Ultrastructure and origin of multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233-1237. [Google Scholar]
- 15.Drugeon, G., and I. Jupin. 2002. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83:3187-3197. [DOI] [PubMed] [Google Scholar]
- 16.Dunoyer, P., E. Herzog, O. Hemmer, C. Ritzenthaler, and C. Fritsch. 2001. Peanut clump virus RNA-1-encoded P15 regulates viral RNA accumulation but is not abundant at viral RNA replication sites. J. Virol. 75:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallie, D. R., D. E. Sleat, J. W. Watts, P. C. Turner, and T. M. A. Wilson. 1987. A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 15:8693-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier, M., T. Candresse, and J. M. Bové. 1986. Immunocytochemical localization of TYMV-coded structural and nonstructural proteins by the protein A-gold technique. Virology 151:100-109. [DOI] [PubMed] [Google Scholar]
- 20.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair, and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagiwara, Y., K. Komoda, T. Yamanaka, A. Tamai, T. Meshi, R. Funada, T. Tsuchiya, S. Naito, and M. Ishikawa. 2003. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22:344-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatta, T., S. Bullivant, and R. E. Matthews. 1973. Fine structure of vesicles induced in chloroplasts of Chinese cabbage leaves by infection with turnip yellow mosaic virus. J. Gen. Virol. 20:37-50. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 25.Héricourt, F., S. Blanc, V. Redeker, and I. Jupin. 2000. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 349:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 27.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyard, J., and R. Douce. 1982. The chloroplast envelope: structure and biochemical properties. Prog. Clin. Biol. Res. 102:77-89. [PubMed] [Google Scholar]
- 29.Jupin, I., K. Richards, G. Jonard, H. Guilley, and C. W. Pleij. 1990. Mapping sequences required for productive replication of beet necrotic yellow vein virus RNA 3. Virology 178:273-280. [DOI] [PubMed] [Google Scholar]
- 30.Kadaré, G., M. Rozanov, and A. L. Haenni. 1995. Expression of the turnip yellow mosaic virus proteinase in Escherichia coli and determination of the cleavage site within the 206-kDa protein. J. Gen. Virol. 76:2853-2857. [DOI] [PubMed] [Google Scholar]
- 31.Kamer, G., and P. Argos. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, C. C., R. Quadt, R. P. Hershberger, and P. Ahlquist. 1992. Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J. Virol. 66:6322-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 34.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laflèche, D., C. Bové, G. Dupont, C. Mouchès, T. Astier, M. Garnier, and J. M. Bové. 1972. Site of viral replication in the cells of higher plants: TYMV-RNA synthesis on the chloroplast outer membrane system, p. 43-71. In Proceedings of the 8th FEBS Meeting: RNA viruses/ribosomes. Elsevier/North Holland, Amsterdam, The Netherlands.
- 36.Lampio, A., I. Kilpelainen, S. Pesonen, K. Karhi, P. Auvinen, P. Somerharju, and L. Kaariainen. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 275:37853-37859. [DOI] [PubMed] [Google Scholar]
- 37.Lee, W.-M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martelli, G. P., S. Sabanadzovic, N. Abou-Ghanem Sabanadzovic, M. C. Edwards, and T. Dreher. 2002. The family Tymoviridae. Arch. Virol. 147:1837-1846. [DOI] [PubMed] [Google Scholar]
- 39.Matthews, R. E. F. 1973. Induction of disease by viruses, with special reference to turnip yellow mosaic virus. Annu. Rev. Phytopathol. 11:147-170. [Google Scholar]
- 40.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molla, A., A. V. Paul, and E. Wimmer. 1993. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 67:5932-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morch, M. D., J. C. Boyer, and A. L. Haenni. 1988. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 16:6157-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morch, M. D., G. Drugeon, P. Szafranski, and A. L. Haenni. 1989. Proteolytic origin of the 150-kilodalton protein encoded by turnip yellow mosaic virus genomic RNA. J. Virol. 63:5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouchès, C., T. Candresse, and J. M. Bové. 1984. Turnip yellow mosaic virus RNA-replicase contains hosts and virus-encoded subunits. Virology 134:78-90. [DOI] [PubMed] [Google Scholar]
- 45.O'Reilly, E. K., J. D. Paul, and C. C. Kao. 1997. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol. 71:7526-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Reilly, E. K., Z. Wang, R. French, and C. C. Kao. 1998. Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol. 72:7160-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 48.Peranen, J., P. Laakkonen, M. Hyvonen, and L. Kaariainen. 1995. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology 208:610-620. [DOI] [PubMed] [Google Scholar]
- 49.Pietrzak, M., R. D. Shillito, T. Hohn, and I. Potrykus. 1986. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 14:5857-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prod'homme, D., S. Le Panse, G. Drugeon, and I. Jupin. 2001. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281:88-101. [DOI] [PubMed] [Google Scholar]
- 51.Restrepo-Hartwig, M. A., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozanov, M., E. Koonin, and A. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 73:2129-2134. [DOI] [PubMed] [Google Scholar]
- 54.Rozanov, M. N., G. Drugeon, and A.-L. Haenni. 1995. Papain-like proteinase of turnip yellow mosaic virus: a prototype of a new viral proteinase group. Arch. Virol. 140:4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 56.Russo, M., A. Di Franco, and G. P. Martelli. 1983. The fine structure of Cymbidium ringspot virus infections in host tissues. III. Role of peroxisomes in the genesis of multivesicular bodies. J. Ultrastruct. Res. 82:52-63. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 61.Seigneurin-Berny, D., N. Rolland, J. Garin, and J. Joyard. 1999. Technical advance: differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J. 19:217-228. [DOI] [PubMed] [Google Scholar]
- 62.Tomita, Y., T. Mizuno, J. Díez, S. Naito, P. Ahlquist, and M. Ishikawa. 2003. Mutation of host dnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 77:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 64.Van Der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiland, J. J., and T. W. Dreher. 1993. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. USA 90:6095-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiland, J. J., and T. W. Dreher. 1989. Infectious TYMV RNA from cloned cDNA: effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 17:4675-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]