Abstract
Within the large body of research on retroviruses, the distribution and evolution of endemic retroviruses in natural host populations have so far received little attention. In this study, the epidemiology, genetic diversity, and molecular evolution of feline immunodeficiency virus specific to cougars (FIVpco) was examined using blood samples collected over several years from a free-ranging cougar population in the western United States. The virus prevalence was 58% in this population (n = 52) and increased significantly with host age. Based on phylogenetic analysis of fragments of envelope (env) and polymerase (pol) genes, two genetically distinct lineages of FIVpco were found to cooccur in the population but not in the same individuals. Within each of the virus lineages, geographically nearby isolates formed monophyletic clusters of closely related viruses. Sequence diversity for env within a host rarely exceeded 1%, and the evolution of this gene was dominated by purifying selection. For both pol and env, our data indicate mean rates of molecular evolution of 1 to 3% per 10 years. These results support the premise that FIVpco is well adapted to its cougar host and provide a basis for comparing lentivirus evolution in endemic and epidemic infections in natural hosts.
Feline immunodeficiency virus (FIV) is a lentivirus in the family Retroviridae that infects members of the family Felidae exclusively. First described in domestic cats (46), FIV-related lentiviruses have also been documented from a number of wild felid species, including lions (Panthera leo) and leopards (Panthera pardus) in Africa, Pallas' cats (Otocolobus manul) in central Asia, and cougars (Puma concolor) in North and South America (2, 5, 29, 41). While all feline lentiviruses analyzed to date represent a monophyletic group with similar organizations and structures (41), sequence divergence among viruses isolated from different cat species is often high. For example, sequence similarity between the env genes of FIV in the domestic cat and that in the cougar (FIVpco) is only 39% (9).
The clinical manifestation of FIV infection in domestic cats typically involves chronic immune dysfunction, opportunistic infections, and behavioral disorders. In contrast, the limited data available do not indicate that FIV in naturally infected wild cats is associated with overt disease (9, 43). The apparently low virulence coupled with a high genetic divergence among lentiviruses both within and among wild cat species have been interpreted as the outcome of a long history of coevolution between the virus and natural host in the Felidae (7, 9).
The dynamics and natural history of retroviral infections in natural populations remain poorly understood. At the same time, the recognition of human immunodeficiency virus (HIV) infection as a zoonotic disease, which originated from simian immunodeficiency virus (SIV) in wild primates, has highlighted the importance of wildlife species as sources of viral epidemics (18, 19, 22). More generally, wildlife populations are likely to hold many important clues to the maintenance, emergence, and evolution of viral pathogens (13, 64). Not surprisingly, an increasing number of studies in recent years have examined natural lentivirus infections in primate hosts at both the intrahost (4, 10, 12, 20, 36) and the population (23, 37, 49) levels.
Studies addressing the evolution and epidemiology of FIV have been limited to domestic- and feral-cat populations (1, 8, 11, 53, 62), where the virus is reported to have been introduced only within the last few decades (9). No studies have examined the epidemiology and evolution of endemic lentiviruses within a specific, free-ranging population of wild feline hosts. Such a study provides the opportunity to investigate how the virus is transmitted and how it evolves in a natural system where it has putatively persisted for extended periods of time. To this end, phylogenetic analysis of sequence data obtained from naturally infected individuals can be used first to identify routes and rates of virus transmission within a host population. Secondly, obtaining serial samples from the same individuals and from a large number of infected animals from the same population makes it possible to assess the rate and course of molecular evolution in the virus over time. Determining the evolutionary characteristics of endemic viruses is significant, given that strong selective regimes and high rates of nucleotide substitution are factors frequently considered a hallmark of epidemic retroviruses, such as HIV.
The number of analytic tools available to estimate rates of molecular evolution from sequence data has increased rapidly in recent years. Specifically, methods that incorporate time data in a phylogenetic framework and which are particularly suited for the analysis of quickly evolving organisms, such as RNA viruses, have been introduced (14, 15, 52). In these applications, the time of isolation for each virus sequence is taken into account in the reconstruction of phylogenetic relationships, so as to provide an estimate of the expected amount of molecular change occurring per time interval. Therefore, these methods offer a way of comparing the courses of virus evolution both within and among host individuals, and among different viral genes, from sequence data. Moreover, new methods for rate estimation, such as Bayesian inference approaches (16), have recently become available and represent a significant improvement over earlier approaches by not being reliant on a known genealogy for temporally spaced sequences.
Here, we describe the prevalence and molecular evolution of endemic FIVpco in a population of free-ranging cougars. Prevalence in this population was high (58% overall), particularly among older individuals. Using data from two viral genes, pol and env, we document the occurrence of two divergent types of FIVpco that were specific to the study area. In addition, we found that virus sequences were characterized by low intrahost genetic diversity and purifying selection and were evolving at a rate that was considerably lower than estimates for HIV type 1 (HIV-1) over the short term. These results stand in contrast to those observed in symptomatic lentiviral infections and give insights into possible courses and mechanisms of host-virus adaptation in natural host systems.
MATERIALS AND METHODS
Sample collection.
Blood samples were collected from 52 individuals in a population of free-ranging cougars in the Snowy Mountain Range (SR) in southeast Wyoming between 1997 and 2001. This mountain range is separated from other nearby ranges by marginal cougar habitat and thus was likely to represent a demographic unit. Sex and age were recorded for each animal captured. Individuals were classified into the following four age categories: kittens (0 to 12 months), yearlings (13 to 24 months), young adults (25 months to 4.5 years), and old adults (>4.5 years). Several animals were caught and sampled repeatedly, including nine infected cougars that were sampled at intervals ranging from 11 to 49 months. Because the captured animals had been fitted with radio collars and monitored in the field, extensive information on movement and space use was available for most individuals, and maternity was known for all kittens sampled.
DNA isolation.
Genomic DNA was extracted from lymphocytes of captured animals using a Super Quik-Gene isolation kit (AGTC, Denver, Colo.) at Wyoming State Veterinary Laboratory, Laramie. DNA and serum were then sent to the University of Montana, where all remaining work was conducted. For one infected individual (SR627) that died during the course of the study, a second DNA sample was obtained from brain rather than blood.
Serology.
Evidence of FIV infection was determined by using a flow cytometry assay that is based on serological recognition of cells infected with a cougar FIV isolate (M. Poss, unpublished data). Cougar serum was incubated with either a feline thymic lymphoma cell line (3201) or 3201 cells infected with a cougar lentivirus. The cells were then washed twice in phosphate-buffered saline containing 2% fetal calf serum and incubated with cat anti-immunoglobulin G-fluorescein isothiocyanate. The cells were washed and analyzed on a FACSCalibur (Becton Dickinson, San Jose, Calif.), and data were statistically evaluated using Cell Quest software. Serum was considered positive for FIV antibody if there was a significant difference in fluorescence intensity between infected and uninfected 3201 cells that were incubated with the serum. All test results were confirmed by immunoblotting (data not shown).
PCR amplification of pol and env.
Fragments of the proviral pol and env genes were amplified from serial dilutions of DNAs of infected individuals. First, a 462- to 527-bp fragment of pol was amplified by nested PCR. The primers used for the first round were 2479F (5′ TAG AAG CAT TAA CAG AAA TAG TAG AGA 3′) and 3028R (5′ TTG TAA TTT ATC TTC AGG AGT TT 3′) or 1259F (5′ GAA GGA AAG GTA AAA AGA GCA GAT 3′) and 1261R (5′ ATC TTC AGG AGT TTC AAA TCC CCA 3′) (5). If the first round was amplified with 2479F-3028R, the 1259F-1261R pair was used for the second round. In a few cases, instead of 1259F, a third forward primer, 2581F (5′ AAA TCA GGA AAA TGG AGA A 3′), was used with primer 1261R. If the first round had been amplified using 1259F-1261R, the primers for the second round were 2506F (5′ GGT AAA AAG AGC AGA TCC TA 3′) and 3012R (5′ AGT TTC AAA TCC CCA CCA TAG 3′). The conditions for the first round of PCR were as follows: 3 min at 94°C, followed by 30 cycles of 94°C for 30 s, 44°C for 40 s, and 71°C for 50 s, and a final extension for 5 min at 72°C. For the second round of amplification, the conditions were 3 min at 94°C, followed by 30 cycles of 94°C for 30 s, 48°C for 30 s (52°C for 2506F-3012R), and 71°C for 50 s, and a final extension for 5 min at 71°C.
In addition, a 719-bp fragment of env was amplified by nested PCR. The first-round primers were 7175F (5′ ATT GCA TAT TGG GAT TTT A 3′) and 8267R (5′ TAT CTT AGA CAC TCG TTG G 3′), while the second round employed primers 7406dF (5′ CCR TGG GGW GGR AGT AGR T 3′) and 8115R (5′GTG CCA GTG GTT GCT CCT ATC A 3′). The conditions for the first round were 3 min at 94°C, followed by 10 cycles of 94°C for 30 s, 40°C for 30 s, and 72°C for 90 s, followed by 25 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 70 s. The conditions for the second round were 3 min at 94°C, followed by 30 cycles of 94°C for 30 s, 50°C for 40 s, and 71°C for 50 s, and a final extension for 5 min at 72°C.
The PCR products from pol and env amplifications were cloned into a PCR4 vector (Invitrogen, Inc., San Diego, Calif.). One to 12 env clones were sequenced for each time point, with a maximum of four clones originating from the same PCR to reduce the chance of resampling. For pol, multiple clones were obtained only from the nine individuals for whom serial samples were available. All primer sites were removed from the sequences prior to analysis.
A number of precautionary measures were taken to minimize the probability of contamination of samples: DNA extractions took place in Wyoming at a facility where no FIV work is conducted. At the University of Montana, PCRs for FIV gene amplification were set up in a room that was kept free of plasmids and PCR products. Samples from all animals were first assessed by PCR for pol, regardless of the animals' serological status. None of the samples from seronegative animals yielded a PCR product. Samples that were FIV positive were then evaluated by PCR for env. At most, samples from two infected individuals, determined by pol PCR to be infected by unrelated viruses, were amplified at the same time. Sequential samples from the same individuals were analyzed as the samples were received and hence were never subjected to PCR at the same time. Finally, PCR were randomly checked by heteroduplex mobility assay for diversity within the amplification reaction and against the clone generated for an animal to ensure (i) that clones represented the original sample, (ii) that we were not missing diversity within the population of amplified sequences, and (iii) that no contamination from our reference strains or other cloned products had occurred.
To determine Taq polymerase error, a 719-bp cloned cougar lentivirus env sequence was amplified and cloned, and 10 clones were sequenced. A 681-bp section (after primer sequences were excluded) was compared to the original sequence, and the polymerase error rate per nested PCR procedure was defined as the number of nucleotide changes observed per site.
Phylogenetic analysis.
Alignments were conducted in Lasergene99 (version 4.06) from DNASTAR, Inc. (Madison, Wis.), using the CLUSTAL W algorithm, and were adjusted manually. For each of the two gene segments, the most appropriate model of nucleotide substitution was found with the program MODELTEST (50), using the full set of available sequences. In both cases, the best model, based on Akaike's information criterion, was a General Time Reversible + gamma (GTR + Γ) model (Table 1). Unless otherwise indicated, the estimated parameters were used in all further analyses.
TABLE 1.
Parameters of molecular-evolution modelsa
| Gene | Model | Base frequency
|
No. of substitutions
|
No. of invariable sites | Gamma | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | G | T | A-C | A-G | A-T | C-G | C-T | G-T | ||||
| pol | GTR + Γ | 0.388 | 0.175 | 0.178 | 0.259 | 2.836 | 21.384 | 2.170 | 0.180 | 13.625 | 1.000 | 0.00 | 2.044 |
| env | GTR + Γ | 0.346 | 0.210 | 0.194 | 0.250 | 2.537 | 35.073 | 4.018 | 0.745 | 28.915 | 1.000 | 0.00 | 2.588 |
Selected using the program MODEL TEST (50) for a full data set of 231 (pol) and 292 (env) sequences from 28 FIVpco-infected cougars. Models were chosen based on lowest Akaike's information criterion value.
To determine the affiliation of FIVpco from the SR population with sequences from other North American cougar populations, a maximum-likelihood (ML) tree was constructed from the pol data. For this analysis, only one randomly chosen sequence was used per individual. For animals from which sequential samples were available, a sequence from the most recent sampling date was selected. Additional FIVpco pol sequences from North American cougars were obtained from GenBank (accession numbers U53718, U53720, U53721, U53723, U53726, U53727, U53731, U53742, and U53745). As an outgroup, we used a virus sequence from a cougar on Vancouver Island (British Columbia, Canada; accession number AY307116) that represents a distinct clade of FIVpco relative to which all western North American types (except those from California) form a monophyletic group (7).
To examine the ancestral relationships of FIVpco from the SR population, an ML tree was constructed from the entire set of env sequences. As for pol, the sequence from a Vancouver Island cougar was used as an outgroup (accession number AY307115). The large number of sequences and a high number of polytomies found in the tree prevented a full heuristic search, and the search was therefore terminated after 100,000 iterations or 168 h of computing time. While additional trees with higher likelihood probably exist, they are unlikely to differ significantly from the tree obtained. Support for individual clusters was assessed through a bootstrap analysis with 500 replicates, using a neighbor-joining (NJ) tree algorithm and the same likelihood settings used in obtaining the ML tree. Figures for both pol and env trees were generated with the help of the program TreeView (44).
Analysis of env sequences.
The complete alignment of all env sequences obtained from SR cougars (n = 292) was analyzed in several ways for evidence of positive or negative selection. First, the proportions of possible nucleotide substitutions for all sites, as well as for synonymous and nonsynonymous sites (Pamilo-Bianchi-Li method) were calculated in the program MEGA2 (27) based on pairwise comparisons. Second, sequencewide evidence of positive or negative selection was sought with a Z test using MEGA2 based on the number of synonymous substitutions per synonymous site (KS) and the number of nonsynonymous substitutions per nonsynonymous site (KA). This test evaluates whether the null hypothesis KS = KA can be rejected in favor of one of the alternative hypotheses (KS > KA or KS < KA). Third, a codon-based test for selection was also performed using ML to test for selection acting on only a fraction of the codon sites. Two models of codon selection were fitted to the sequence data (38). Model M1 assumes that nonsynonymous substitutions can be either fatally deleterious (dN /dS ratio; ω0 = 0) or neutral (ω1 = 1) to selection. Model M2 extends model M1 by including a third category of otherwise-selected codons, with the nature of the selection indicated by the value of ω2 as positive (ω2 > 1) or negative (ω2 < 1). The parameters of models M1 and M2 were estimated using PAML software (67) from an NJ tree constructed under a GTR model, and selection models were compared using likelihood ratio testing. Different initial values of ω2 were used, as advised by the user documentation, because the software is known to suffer from convergence problems (38). All codon sites were also classified as experiencing purifying, neutral, or some other type of selection using an empirical Bayesian approach also provided in PAML. The analyses were performed twice: (i) after removing those codon sites at which there were gaps or stop codons or sites having ambiguous nucleotides in some sequences (16 of 226 codons) and (ii) after removing those sequences containing gaps, stop codons, or ambiguous nucleotides (2, 35, and 2 of 292 sequences, respectively).
To determine whether nucleotide changes were evenly distributed across the region of env and pol amplified or whether they occurred at particular hot spots, the number of observed nucleotide states per site was calculated using the program MacClade version 3.0 (32a) and plotted as the average number of states per nucleotide found within a sliding window of 10 nucleotides.
Rate estimates of molecular evolution.
Rate estimates for the evolution of env and pol were based on all sequences available for an individual or on a subset of sequences for the population as a whole. Two estimation techniques were employed. First, we used an ML approach implemented in the program TipDate (52). Because this technique requires a known tree topology, NJ trees were first constructed for the respective sets of sequences using PAUP* (62a). To facilitate adequate rooting of the within-individual trees, two or three intermediately distant sequences from other individuals were included as outgroups during tree construction. These taxa (and the branches leading to them) were subsequently removed prior to rate estimation. For the population as a whole, the data set was limited to one randomly selected sequence per individual and time point, to keep the estimation computationally feasible, and the root was found by the midpoint method. Once NJ trees had been obtained, they were entered into the program TipDate version 1.2 (52) together with the respective sequence data and the sampling date for each sequence (to the nearest decimal fraction of a year) to find the ML rate of substitution, as well as an estimated absolute age of the corresponding tree (i.e., time to the most recent common ancestor), including 95% CIs.
For the second estimation approach, we employed a recently developed Monte Carlo Markov chain (MCMC) framework for the Bayesian estimation of evolutionary rates and population parameters (16; http://www.cebl.auckland.ac.nz/mepi/index.html). This technique also uses sets of dated sequences but offers several specific advantages compared to the ML estimation. First, it does not assume that tree topology is known and it is not limited to estimation of evolutionary rates. Instead, it takes account of the uncertainty in any phylogenetic estimate and the error inherent in any sampling scheme by drawing representative samples from the space of plausible population sizes, evolutionary rates, and phylogenies (16). The result is an estimate of the posterior probability distribution of evolutionary rates (marginalized over phylogenies and population size). The method applied uses a coalescent prior on divergence times (26, 55). Under this prior, viral population size within and between hosts was assumed to remain constant over the time spanned by the genealogy. Previous applications have shown that rate estimation using this method is robust to different assumptions of population history (16, 28). An additional value of the Bayesian framework is that it allows specific prior information to be incorporated into the analysis. In our case, since the approximate ages of the cougar hosts were known, we made the assumption that the most recent common viral ancestor within an individual was unlikely to be much older than the infected animal itself or the time since infection. Thus, we used an exponential distribution with expectation equal to the age of the animal (as estimated in the field) or the known time since infection (based on seroconversion) as a prior for the age of the root of the tree (i.e., the age of the most recent common ancestor of all viral sequences in an infected individual). This had the effect of influencing the depth of the within-host viral phylogeny, since the depth of the phylogeny is necessarily constrained to be bounded by the time to infection because it is assumed that either a single virus or a homogenous group of viruses establishes infection in the host. No prior on the age of the root was used for the estimation of evolutionary rates for the SR population as a whole.
Nucleotide sequence accession numbers.
One env sequence per SR individual has been submitted to GenBank under accession numbers AY120787 to AY120810 and AY120812 to AY120815, as well as one pol sequence per individual (AY120816 to AY120839 and AY120841 to AY120844 [see Fig. 2]). Nexus files of the complete set of pol and env sequences used in the analysis can be obtained from http://www.cebl.auckland.ac.nz/FIVpco or from the authors.
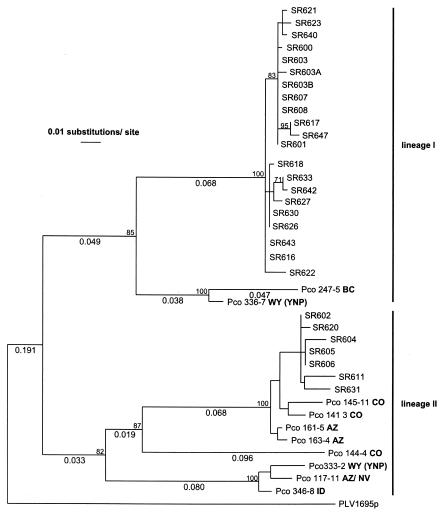
FIG. 2.
ML tree of partial FIVpco pol sequences from free-ranging cougars in the Rocky Mountains (United States and Canada). Taxa designated SR represent animals from the study area. For each SR individual, one sequence was randomly selected from a larger data set. Taxa designated Pco represent previously described FIV pol sequences from other cougar populations in western North America. The state or province where the animal was captured is indicated in boldface letters (YNP, Yellowstone National Park). Branch lengths of >0.02 are shown below the branches. The FIVpco sequence from a Vancouver Island cougar (PLV1695) was used as an outgroup. The values next to tree nodes represent bootstrap values of >70 based on 500 replicates using an NJ tree algorithm and the same model of substitution used for the ML tree.
RESULTS
FIVpco seroprevalence.
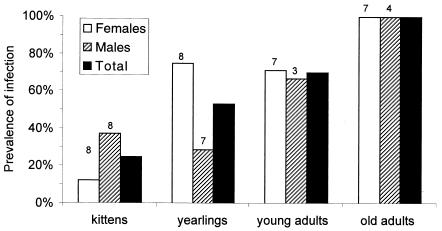
We tested 52 different cougars, of which 30 (58%) showed serological evidence of FIVpco infection. The prevalence increased significantly with age class (Spearman's rank correlation; r = 1.0; P < 0.001) and reached 100% among old adults (Fig. 1). Overall, similar proportions of males and females were infected, but the ratios were more variable among younger age classes (Fig. 1). Of all kittens sampled, 25% were positive. All five young (kittens or yearlings) that had been born to uninfected females tested negative. In contrast, 50% of the young born to infected females (10 of 20) were positive at the time of sampling (when the individuals were between 4 and 16 months old). Two adult females seroconverted over the course of the study. In both cases, the cats had given birth to young near the time of seroconversion, suggesting that they may have become infected during mating.
FIG. 1.
Prevalence of FIVpco infection in a population of free-ranging cougars in southeast Wyoming by sex and age class. Prevalence was determined by serology using a flow cytometric assay and confirmed by PCR. The sample sizes are shown above the bars (n = 52 [total]). For individuals that were sampled repeatedly, only infection status for the most recent sample was considered.
Phylogenetic analysis of pol sequences.
Virus sequences were obtained from all seropositive individuals for which peripheral blood mononuclear cells were available (28 of 30), while none of the seronegative samples yielded a PCR product. An ML tree constructed from pol sequences identified two distinct groups of FIVpco in the SR population that were >20% divergent (corrected distances), with high bootstrap support for the respective nodes (Fig. 2). Previously published pol sequences from other cougar populations all affiliated with these two lineages (7). One lineage (lineage I), which represented 75% of infected cats in the SR population, included sequences from northern Wyoming and British Columbia. The second FIVpco lineage identified (lineage II), which was found in seven SR cougars (25%), contained isolates from Arizona, Colorado, northern Wyoming, and Arizona-Nevada.
Phylogenetic analysis showed that genetic divergence of FIVpco based on pol sequences was high in Rocky Mountain cougars, as documented previously (7). Within lineage I, the corrected divergence among sequences from different areas was 8 to 10%, while differences within lineage II ranged from 2 to 15%. The high diversity found in Rocky Mountain FIVpco contrasted with low degrees of sequence differentiation among pol sequences from within the SR population (Fig. 2). All samples originating from the SR population formed well-supported groups within their respective lineages, with only 0 to 3% sequence divergence within the groups. Together, these data indicate that SR cougars were infected with highly related FIV types that were clearly distinguishable from viruses found in other parts of the Rocky Mountains.
Phylogenetic analysis of env sequences.
The relationship of virus sequences from animals within the SR population was evaluated based on env because env sequences were longer than pol sequences and because we made an initial assumption that the evolutionary rate in env would be higher than that for pol. The general topology of the ML tree of env sequences from the SR population (Fig. 3) was congruent with the clustering of sequences derived from SR animals in the pol tree (Fig. 2). Specifically, pol and env sequences from an individual had the same phylogenetic affiliation relative to those from other individuals. At the same time, the genealogical structure of the env tree was more defined and there was stronger support for substructure in each of the two lineages, possibly due to the larger amount of phylogenetic information contained in the longer env fragment.
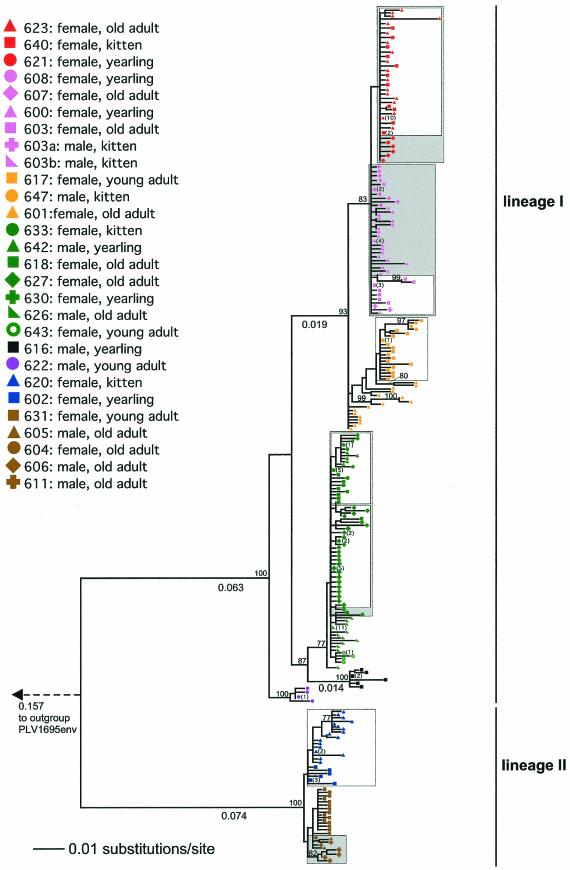
FIG. 3.
ML tree of 292 partial FIV env sequences from 28 infected cougars in a population in southeast Wyoming. Mother-kitten groups are indicated by open boxes, and animals with related viruses and confirmed overlap in home ranges are indicated by shaded boxes. Where identical sequences were identified, their number is indicated in parentheses next to the taxon symbol. Branch lengths of .0.01 are shown below the branches. The ML search was terminated after 100,000 iterations (see the text). See Fig. 2 for further descriptions.
As in the pol analysis, clusters of virus genotypes were often not unique to specific individuals but rather to entire groups (Fig. 3). This resembles the situation described for a domestic-cat population following what appeared to be a singular introduction of FIV (8) and suggests a close epidemiological link among individuals within groups. In most cases, an obvious explanation for such links was apparent. For example, shared or closely related virus genotypes were found in all six known mother-offspring groups, indicating recent vertical-transmission events. Close clustering was also observed for sequences derived from animals for which we lacked familial information but for which field observations demonstrated adjacent or overlapping home ranges (Fig. 3). Observational data on home ranges following capture were unavailable or yielded no evidence for spatial overlap only for the three remaining individuals (SR626, SR631, and SR643) that also had virus genotypes closely related to those found in other SR cougars.
Nucleotide divergence in env.
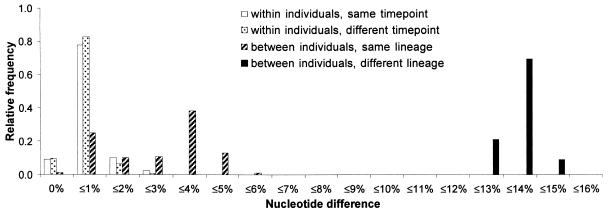
Based on all 292 available clones, uncorrected proportions of possible nucleotide substitutions (p distances) among env sequences derived from the same infected animal varied between 0 and 2% (Fig. 4). This range applied to comparisons both within and among different sampling time points. Different individuals carrying virus from the same phylogenetic lineage exhibited distances ranging from 0.5 to 5.5% but with a bimodal distribution centered around 1 and 4%. The uncorrected average difference among viruses from the two lineages was 14%. It is noteworthy that there were virtually no insertions or deletions among the env sequences.
FIG. 4.
Partitioning of genetic diversity in FIV env sequences from 28 naturally infected cougars based on p distances observed in pairwise comparisons. For each category of comparison (e.g., within individuals, same time point), frequencies sum up to 1.
The error rate of the Taq polymerase was experimentally determined to be 0.015% per site (one substitution observed in 6,810 nucleotides). Because this value is at least an order of magnitude smaller than the average within-host genetic difference among FIV fragments (Fig. 4), it can be assumed that Taq error did not significantly affect our results.
Proportion of synonymous and nonsynonymous changes in env.
Within individuals, the mean proportion of synonymous changes per synonymous site (KS) in env sequences was very similar to the proportion of nonsynonymous changes (KA) (Table 2). Among individuals infected with virus from the same viral lineage, KS was twice times as high as KA, and this increased to a 10-fold difference for comparisons between the two lineages. Concordant with the observation that synonymous changes were more common, a test for possible deviations from neutrality indicated that molecular evolution in env was dominated by purifying selection (Z = 10.41; P < 0.001).
TABLE 2.
Average proportions of synonymous and nonsynonymous sites from pairwise comparisons of FIVpco env sequences
| Site type | Average % of possible substitutions (range)
|
|||
|---|---|---|---|---|
| Within individual
|
Among individuals
|
|||
| Same time point | Different time pointsa | Same lineage | Different lineages | |
| Synonymous | 0.5 (0.0-1.3) | 0.5 (0.2-1.1) | 4.1 (1.3-6.7) | 55.8 (47.8-64.7) |
| Nonsynonymous | 0.5 (0.0-1.6) | 0.5 (0.2-1.2) | 1.9 (0.5-4.4) | 5.6 (4.2-7.9) |
Samples taken 8 to 49 months apart; see Table 4.
For the codon-specific analysis, ML estimates consistently indicated that 60% of sites experienced strong purifying selection regardless of the model and parameters considered. Meaningful estimates of the proportion of sites under neutral or positive selection were precluded by convergence problems (data not shown). PAML routinely returned an estimate of ω2 that matched the seed value (range, 0.7 to 10), suggesting failure to converge. The classification of sites using empirical Bayes' estimates identified 37 to 47% of sites as experiencing purifying selection but, as with the ML estimation, it was inconclusive in respect to the proportion of neutral sites and sites under positive selection (data not shown).
Distribution of molecular changes across env andpol sequences.
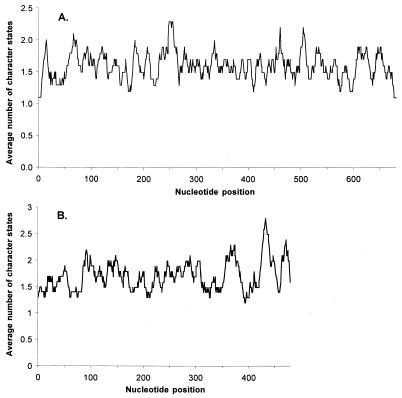
We plotted the average number of different character states observed per nucleotide in a sliding window of 10 nucleotides for both pol and env in order to assess whether the amplified fragments contained both variable and conserved regions (Fig. 5). Both gene fragments were found to contain segments in which nucleotide diversity was elevated, but overall, the distributions of substitutions in both viral genes were uniform.
FIG. 5.
Plots of nucleotide diversity for FIV env (A) and pol (B) gene fragments. Diversity is expressed as the average number of character states found per sliding window of 10 nucleotides.
Evolutionary rates of FIVpco within the SR population.
The two approaches used to estimate FIV evolution within the SR population yielded evolutionary-rate estimates close to 1% per decade for both env and pol (Table 3). Estimates obtained with the MCMC approach were slightly higher (1.54% for both genes) and were associated with smaller variances than the ML estimates. The latter showed a higher rate for env (1.14%) than for pol (0.77%). However, the CIs of all estimates obtained with both ML and MCMC overlapped substantially, indicating no significant difference in evolutionary rates among the different estimation approaches or between the two genes examined. Concordant with the lower mean rate for pol using ML, the estimated time to the most recent common ancestor of all SR sequences (i.e., the ancestor of both viral lineages) was higher for pol (168.6 years) than for env (79.6 years). All estimates were clearly distinguishable from zero, demonstrating sufficient evolutionary signal in the two data sets to identify substitution rates. In comparison, the uncertainty associated with the MCMC estimates was much smaller than for ML estimates and suggested that the two viral lineages found in Rocky Mountain cougars had likely separated 36 to 153 years ago.
TABLE 3.
ML and MCMC estimates of evolutionary rates for FIVpcoa
| Gene | No. of sequences | ML
|
MCMC
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Rate of change (%/site/10 yr)
|
Age of tree (yr)b
|
Rate of change (%/site/10 yr)
|
Age of tree (yr)b
|
||||||
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | ||
| env | 41 | 1.14 | 0.34-1.92 | 79.6 | 49.1-239.6 | 1.54 | 0.89-2.23 | 60.9 | 36.2-91.7 |
| pol | 42 | 0.77 | 0.13-1.49 | 168.6 | 90.1-1,013.9 | 1.54 | 0.63-2.38 | 85.8 | 37.5-152.5 |
Based on sequences from 28 naturally infected hosts (P. concolor) with dated samples collected over a period of 49 months.
Decimals represent fractions of a year.
Evolutionary rates of FIVpco within infected individuals.
Substitution rates were estimated from sequential samples from nine infected individuals (Table 4). Mean rates using ML ranged from 0.39 to 2.35% per 10 years (overall mean, 1.27%) for pol and 0.06 to 3.88% per 10 years for env (overall mean, 1.42%). Thus, evolutionary rates derived for individual animals were similar to the previous estimates of ∼1% per decade for the population as a whole. As before, there was no evidence that the evolutionary rates for the two genes were different. Some problems with uncertainty in the estimation were encountered in that most ML estimates were not significantly different from zero. This suggests that overall, the number of changes in the virus that had occurred within an infected individual was small relative to the time intervals between samples (8 to 49 months). Also, the fact that precision was generally higher for estimates for env, which was based on a fragment that was 220 bp longer than pol, points to the fact that longer sequences were more likely to contain evolutionary signal and thus enhanced the ability to estimate rates. While most estimated times to the most recent common viral ancestor fell within or close to the assumed maximum time since infection, estimated tree ages were much higher (>30 years) in a few cases. Such long time periods, which would span several cougar generations and thus transmission events, appear unrealistic given the small number of viruses presumed to be transmitted during natural retrovirus infections (59, 65). We suspect that unrealistically high tree ages and very low rates were related to difficulties in accurately estimating the tree topology when the signal in the data was weak. However, in light of these results, it remains possible that substantial viral sequence diversity occasionally survives transmission bottlenecks.
TABLE 4.
ML and MCMC estimates of evolutionary rates for two FIVpco genes within naturally infected cougarsa
| Animal | Interval (yr) (no. of samples)b,c | Maximum time since infection (yr)d | Gene | No. of sequences | ML
|
MCMC
|
||
|---|---|---|---|---|---|---|---|---|
| Rate of change (%/site/10 yr) | Tree age (yr)c | Rate of change (%/site/10 yr) | Tree age (yr)c | |||||
| SR600 | 4.1 (3) | 7 | env | 23 | 0.95 (0.00-1.37) | 6.9 (6.1-NEe) | 1.19 (0.34-2.20) | 9.4 (4.9-15.5) |
| pol | 21 | 0.39 (0.00-1.11) | 11.6 (5.4-NE) | 1.38 (0.32-2.58) | 7.3 (4.2-12.5) | |||
| SR617 | 2.9 (2) | 7 | env | 17 | 0.65 (0.00-1.43) | 6.0 (4.1-NE) | 1.27 (0.29-2.51) | 7.7 (3.7-14.0) |
| pol | 25 | 0.50 (0.00-1.09) | 10.6(5.7-NE) | 1.07 (0.30-2.00) | 9.3 (3.9-17.4) | |||
| SR618 | 1.6 (2) | 8 | env | 19 | 2.30 (0.04-3.78) | 1.9 (1.9-52.6) | 1.67 (0.30-3.56) | 6.0 (2.0-13.8) |
| pol | 24 | 0.72 (0.00-1.30) | 4.6 (3.3-NE) | 1.03 (0.18-2.20) | 7.9 (2.3-12.5) | |||
| SR620 | 2.3 (3) | 3 | env | 22 | 3.88 (1.70-6.30) | 2.5 (2.4-3.0) | 3.89 (1.72-6.27) | 3.1 (2.5-4.2) |
| pol | 28 | 2.35 (0.08-3.85) | 8.3 (6.0-192.2) | 5.87 (2.47-9.48) | 5.74 (3.5-8.3) | |||
| SR623 | 1.3 (3) | 2 | env | 28 | 2.57 (0.52-4.07) | 2.2 (1.9-6.3) | 2.48 (0.64-5.63) | 5.4 (1.6-10.0) |
| pol | 23 | 0.52 (0.00-1.25) | 5.9 (2.9-NE) | 6.21 (1.66-11.84) | 1.9 (1.3-2.4) | |||
| SR626 | 0.8 (2) | 4 | env | 23 | 1.54 (0.00-2.47) | 2.1 (1.7-NE) | 3.47 (0.56-3.14) | 2.7 (1.1-5.4) |
| pol | 29 | 1.46 (0.00-2.26) | 1.8 (1.7-NE) | 2.83 (0.36-6.36) | 2.9 (1.0-6.1) | |||
| SR627 | 1.0 (2) | 5 | env | 17 | 0.06 (0.00-1.70) | 88.4 (4.1-NE) | 3.00 (0.73-6.07) | 4.2 (1.6-7.9) |
| pol | 23 | 1.84 (0.00-3.75) | 2.5 (1.6-NE) | 1.85 (0.28-4.06) | 5.4 (1.5-11.9) | |||
| SR630 | 0.7 (2) | 1 | env | 16 | 0.66 (0.00-2.61) | 4.3 (1.3-NE) | 3.03 (0.50-6.82) | 1.7 (0.8-3.1) |
| pol | 18 | 2.28 (0.00-6.02) | 1.5 (1.0-NE) | 4.75 (0.61-10.27) | 1.6 (0.8-2.8) | |||
| SR631 | 2.5 (2) | 6 | env | 11 | 0.13 (0.00-1.14) | 33.0 (4.6-NE) | 1.08 (0.17-2.24) | 7.3 (3.2-14.6) |
| pol | 14 | 1.35 (0.51-2.31) | 2.8 (2.6-5.1) | 1.22 (0.17-3.07) | 9.4 (2.7-21.0) | |||
Based on sequential samples (2 or 3 serial samples per individual) collected 9 to 49 months apart. Shown are means with 95% CIs in parentheses.
Time from earliest to latest sample.
Decimals represent fractions of a year.
Based on estimated age of animal or known time of seroconversion.
NE, upper boundary not estimable.
The described difficulties in generating reasonable and precise rate estimates based on sequence data from individuals could be largely overcome by employing a Bayesian MCMC approach. This method takes the uncertainty of the tree topology into account. In addition, we used prior knowledge about maximum time since infection for each individual to improve estimation. The mean rates obtained ranged from 1.03 to 6.21% per decade for pol (average, 2.91%) and from 1.08 to 3.89% for env (average, 2.34%) (Table 4). These results therefore confirmed the earlier observation that the evolutionary rates of both genes were similar. There was no difference among rate estimates for viruses in lineages I and II. In all cases, the assumed maximum time since infection fell within the 95% CIs of the estimate for the time since the most recent common ancestor.
Whereas evolutionary rates rarely exceeded 2% per decade in most individuals, data for three animals (SR620, SR623, and SR630) were suggestive of higher rates in pol and env in at least three of the four estimates obtained per cat. Notably, these three cougars must have become infected less than a year before the first serial virus sample, because they either were newborn kittens (SR620 and SR630) or had seroconverted since the last sample (SR623). Thus, higher substitution rates appeared to be associated with more recent infections.
DISCUSSION
The overall 58% FIV seroprevalence determined for the SR population is higher than most previous reports for free-ranging cougars in North America (17, 45, 56). The most comprehensive serological survey of North American cougars (n = 225) reported 31% overall FIV prevalence (7). FIV prevalence levels of >30% are also unusual for feral domestic-cat populations (8, 11). Our data suggest that high FIV prevalence in cougars in general may be related to effective transmission of the virus both from mothers to infants or yearlings and among adult individuals. More than half of the young born to infected dams carried the virus at the time of our sampling. Young cougars remain with their mothers for 1 to 2 years. Because our samples were often taken several months before independence, the actual rate of transmission from mother to offspring may be even higher. This high frequency of vertical transmission is in accordance with reports for FIV in domestic cats (39, 40) but markedly higher than that found for lentiviruses in natural primate host populations (23, 37, 49). Vertical transmission is unlikely to be responsible for all infections in cougars, however, given that the prevalence of infection continued to increase with age after reaching maturity. Territorial fights, mating, or other forms of social contact most likely facilitate horizontal transmission of FIVpco in cougars, based on observations of domestic cats (6, 24).
The cooccurrence of several distinct FIV lineages within the same population of hosts has been described previously for free-ranging cougars and African lions (5, 7). While the emergence of regional FIV subtypes is certainly not surprising for an animal with a wide geographic distribution, the intermixing of these distinct types suggests extensive cougar movements, current and/or historic, from or to the SR population. At the same time, SR pol sequences formed monophyletic clusters within their respective lineages, indicating that the two virus strains found within the population had been able to rapidly and effectively spread on a local scale.
The presence of two viral lineages in the SR population also raises the possibility that individuals could become coinfected with both types. Indeed, coinfection with divergent FIVpco genotypes was previously documented in a cougar (7). However, we had no evidence for such coinfection in this population, suggesting that it occurs at a low frequency. Furthermore, coinfection with divergent virus types would have been a prerequisite for the generation of possible recombinant types carrying pol and env from different viral lineages. Such genetic reassortment of viral genotypes has been documented, for example, for HIV both in vivo and in vitro (35, 54). However, the same clusters of infected individuals were identified for both pol and env in our data set, thus providing no evidence for ancestral recombination among lineages. It should be noted, however, that coinfection and recombination involving highly related viruses remain possible. Because of the small sequence differences involved, such events would be very difficult to detect but at the same time would be expected to be inconsequential in respect to the general genealogical patterns.
Phylogenetic analysis of env identified a number of well-supported clusters containing closely related sequences (Fig. 3). Adult female cougars in particular shared highly related viruses with their offspring, supporting the premise of vertical transmission (7). Sequence divergence between viruses in the mother and offspring was usually very small but ranged up to >1%. This was likely due to the fact that samples from mother and kitten were typically obtained at different times, and the interval between sampling and infection was unknown for both. In addition to the close affiliation of FIVpco sequences obtained from females and their offspring, closely related virus sequences were also found in individuals with overlapping or neighboring home ranges (Fig. 3). For example, spatial movement data collected using radio telemetry showed that cougars that had contiguous or overlapping home ranges were infected with viruses that clustered together in the phylogenetic tree (600, 603, 607, and 608 in lineage I and 604, 605, 606, and 611 in lineage II). Adult females frequently share home ranges with offspring from a previous mating, and hence, some of these virus clusters may also be derived from related animals. The relationship of all animals in the population is currently under investigation and will illuminate mechanisms of viral transmission among cougars. However, the bimodal distribution of genetic distances among env sequences from different individuals within the two FIVpco lineages (Fig. 3) suggests that genetic distances of up to 2% among individuals reflect recent infection events. In contrast, distances of 4%, observed among viruses from different individuals, likely represent genotypes separated by more virus generations and potentially more than one transmission event.
In contrast to the common local spread of a particular genotype, the introduction of a new virus into the population appears to be relatively rare. The env sequence most distinguishable from all other SR genotypes was isolated from a yearling male, SR622, which was probably not born in the study area. Unlike female offspring, whose presence close to the mother's territory is often tolerated even after maturity, male yearlings are usually expelled from the natal area and tend to disperse further away from it (31). One plausible scenario is that SR622 had immigrated into the SR population from a neighboring or distant area, where it had become infected with a related, yet distinguishable, variant of FIVpco.
Because retroviral diversity increases with time since infection, we expected to observe larger divergence in env in some individuals. Instead, intrahost env diversity in cougars averaged <1%, which is similar to values for pol in cougars and domestic cats (5) but lower than the 1.1 to 2.5% reported for env in domestic cats (62). This low sequence diversity within hosts is remarkable given that cougars can reach ages of >10 years in the wild and that some of the individuals we examined were known to have harbored infections for several years. Also, we found that within individuals, sequences taken at time points that were 8 to 49 months apart were not more different from each other than sequences derived from the same time point. This may be due, in part, to the fact that we derived our sequence data from proviral DNA, and FIVpco may reside in long-lived cell populations. However, these data suggest that evolution of FIV env within cougar hosts may occur at a lower rate than in epidemically occurring retroviruses. It was also noteworthy that there was a virtual absence of insertions or deletions in env fragments from FIVpco, while such changes are commonly observed in domestic-cat FIV and primate lentiviruses.
Concordant with a low genetic diversity among intrahost isolates and the close relation of sequences from putative family groups, the substitution rates we estimated for cougar FIVpco were low compared to estimates for HIV or SIV. At the same time, it should be noted that evolutionary rates have been obtained using a wide range of techniques and that some of these are not equivalent to our approach, making a comparison with published estimates difficult. The only study we are aware of that estimated evolutionary rates in FIV was done with domestic cats and found no changes in the consensus sequence of pol or gag in a naturally infected cat over a 3-year period (21). For a part of the env gene that included a variable region, a rate of 3.4% per 10 years was estimated in the same study, similar to estimates obtained here. Although results were variable, our estimates suggest an overall rate of 1 to 3% per 10 years for both pol and env. In comparison, studies of HIV evolution within hosts or small groups of hosts reported higher rates of 0.3 to 1.0% per year (30, 36, 47, 60), while rates up to 3% per year have been calculated for SIV (36, 47). Interestingly, HIV-1 evolves considerably more slowly at the population level, where, using sequence data collected over a decade or more, rates of 1 to 2 and 4 to 6% per decade have been estimated for pol (based on all substitutions) and env (V3; based on synonymous changes alone), respectively (32). Whereas FIVpco evolution within some infected cougars in our study appeared to be faster than for the population as a whole, our data do not support rates that would exceed half a percent per year. Comparisons of our estimates to those for primate lentiviruses should be done cautiously, however, because most of the studies focused on particularly variable regions of env and, in the case of SIV, were almost all conducted with experimentally rather than naturally infected individuals (36, 47). More studies of naturally infected primate and feline hosts are needed to determine whether a lower rate of molecular evolution, relative to HIV, could indeed be a characteristic of natural host-virus systems or of feline lentiviruses.
Given the estimated substitution rate of <0.5% per year, the time intervals between our samplings (8 to 49 months) were at the low end of what would yield measurable changes in the virus. Clearly, extending the time frame of virus sampling should improve the precision of rate estimates, as would longer sequence data. For the data collected here, the use of a Bayesian MCMC approach that integrated over all possible tree topologies markedly improved our estimates for evolutionary rates from virus sequences compared to the ML approach. Specifically, incorporating prior information on the probable time since infection made it possible to obtain estimates for virus evolution within individuals where ML rates had not been distinct from zero (Table 4). In almost all cases, the probable time since infection fell within the obtained 95% CIs of both ML and MCMC estimates, indicating that this prior information was in agreement with the data.
No prior for tree age was used for the analysis of virus sequences from all SR individuals combined. The obtained estimates overall suggest that the split between the two FIVpco lineages in Rocky Mountain cougars occurred within the last 200 years (Table 3). This relatively recent time point is consistent with the fact that FIVpco from this region is distinct from virus found in cougars elsewhere in North America (7) and that cougar populations in the western United States are currently recovering from low population levels due to persecution during the 19th century and the first half of the 20th century (3).
Several factors may be involved in a lower rate of molecular evolution in FIVpco than in other lentiviruses, including the fidelity of replication and the length of virus generation time. Reported error rates for the replication enzymes of different retroviruses vary by a factor of 20 (51), raising the possibility that lower mutation rates are due to a higher fidelity of the viral reverse transcriptase. In addition, low evolutionary rates may be related to longer virus generation times. Although in HIV, the generation time is as short as 1 to 3 days, it is thought to be substantially longer for a small percentage of viruses that establish latent infections in long-lived cell types, such as macrophages (48). Therefore, it may be that cell types facilitating slower virus replication comprise the majority of FIVpco-infected cells in cougars, resulting in slow virus population turnover. Although the question of cell tissue tropism in FIVpco remains speculative, the observation that HIV variants targeting long-lived cells outlive fast-replicating variants (42, 63) suggests a potential selective regime that could favor slowly replicating viruses in a coevolutionary process.
Evasion of the host immune system is frequently considered a partial explanation for the high rate of molecular evolution observed in many lentiviruses, especially for the envelope glycoprotein. At the same time, evidence for positive selection has been more or less limited to studies of HIV-1 in humans (33, 34, 58, 66) and is controversial even there (25, 57). Higher proportions of nonsynonymous changes also characterize SIV infection of macaques, which are not a natural host of primate lentiviruses. In African green monkeys, a natural host for SIV, the proportion of synonymous substitutions was higher than that of nonsynonymous changes (12, 61). In the domestic cat, a strong role of positive selection in FIV lacks support based on reported KS/KA ratios that rarely exceed 1 (1, 53, 62). Consistent with the latter results, we found evidence for selection largely against rather than in favor of nonsynonymous changes in cougar FIVpco env, supporting the idea that molecular evolution of this gene occurs under some functional constraint. Results for the identification of sites under positive selection were ambiguous but suggest that the number of such sites is likely to be low. In addition, the low rate of nonsynonymous substitutions relative to synonymous substitutions between hosts argues against any process of host-specific adaptation as the virus moves from one host to the next during virus transmission. This lends some weight to the argument that forms of FIV maintained in wild cats are well adapted to their host species. Still, compared to nonpathogenic lentiviruses in other natural hosts, which are often characterized by rapid accumulation of evolutionary changes resulting from high rates of replication (4, 36), intrahost genetic diversity and substitution rates were low in the cougar FIVpco system. This shows that despite overall similarities, life history strategies and evolutionary characteristics may vary considerably among different lentiviruses in their natural hosts.
Acknowledgments
This work was partially funded by grants from the University of Montana, MONTS, and the Wilburforce Foundation. Funding for capture efforts was provided by the Wyoming Game and Fish Department, the Wyoming Animal Damage Management Board, and the Pope and Young Club.
The cell line infected with a cougar lentivirus was generously donated by S. VandeWoude (Colorado State University, Fort Collins). D. Wroe conducted captures and sampling of cougars, and S. Painter, E. Villegas, and N. Akamine provided technical assistance in the laboratory. We thank three anonymous reviewers for their comments.
REFERENCES
- 1.Bachmann, M. H., C. Mathiason-Dubard, G. H. Learn, A. G. Rodrigo, D. L. Sodora, P. Mazzetti, E. A. Hoover, and J. I. Mullins. 1997. Genetic diversity of feline immunodeficiency virus: dual infection, recombination, and distinct evolutionary rates among envelope sequence clades. J. Virol. 71:4241-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr, M. C., L. Zou, D. L. Holzschu, L. Phillips, F. W. Scott, J. W. Casey, and R. J. Avery. 1995. Isolation of a highly cytopathic lentivirus from a nondomestic cat. J. Virol. 69:7371-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier, P. 1991. Cougar attacks on humans in the United States and Canada. Wildl. Soc. Bull. 19:403-412. [Google Scholar]
- 4.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, and J. S. Allan. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. W., N. Yuhki, C. Packer, and S. J. O'Brien. 1994. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J. Virol. 68:5953-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkhard, M. J., L. A. Obert, L. L. O'Neil, L. J. Diehl, and E. A. Hoover. 1997. Mucosal transmission of cell-associated and cell-free feline immunodeficiency virus. AIDS Res. Hum. Retrovir. 13:347-355. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter, M. A., E. W. Brown, M. Culver, W. E. Johnson, J. Pecon-Slattery, D. Brousset, and S. J. O'Brien. 1996. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J. Virol. 70:6682-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter, M. A., E. W. Brown, D. W. MacDonald, and S. J. O'Brien. 1998. Phylogeographic patterns of feline immunodeficiency virus genetic diversity in the domestic cat. Virology 251:234-243. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, M. A., and S. J. O'Brien. 1995. Coadaptation and immunodeficiency virus: lessons from the Felidae. Curr. Opin. Genet. Dev. 5:739-745. [DOI] [PubMed] [Google Scholar]
- 10.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courchamp, F., N. G. Yoccoz, M. Artois, and D. Pontier. 1998. At-risk individuals in Feline Immunodeficiency Virus epidemiology: evidence from a multivariate approach in a natural population of domestic cats (Felis catus). Epidemiol. Infect. 121:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courgnaud, V., W. Saurin, F. Villinger, and P. Sonigo. 1998. Different evolution of simian immunodeficiency virus in a natural host and a new host. Virology 247:41-50. [DOI] [PubMed] [Google Scholar]
- 13.Daszak, P., A. A. Cunningham, and A. D. Hyatt. 2000. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287:443-449. [DOI] [PubMed] [Google Scholar]
- 14.Drummond, A., R. Forsberg, and A. G. Rodrigo. 2001. The inference of stepwise changes in substitution rates using serial sequence samples. Mol. Biol. Evol. 18:1365-1371. [DOI] [PubMed] [Google Scholar]
- 15.Drummond, A., and A. G. Rodrigo. 2000. Reconstructing genealogies of serial samples under the assumption of a molecular clock using serial-sample UPGMA. Mol. Biol. Evol. 17:1807-1815. [DOI] [PubMed] [Google Scholar]
- 16.Drummond, A. J., G. K. Nicholls, A. G. Rodrigo, and W. Solomon. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evermann, J. F., W. J. Foreyt, B. Hall, and A. J. McKeirnan. 1997. Occurrence of puma lentivirus infection in cougars from Washington. J. Wildl. Dis. 33:316-320. [DOI] [PubMed] [Google Scholar]
- 18.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 19.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene, W. K., J. Meers, G. del Fierro, P. R. Carnegie, and W. F. Robinson. 1993. Extensive sequence variation of feline immunodeficiency virus env genes in isolates from naturally infected cats. Arch. Virol. 133:51-62. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 23.Jolly, C., J. E. Phillips-Conroy, T. R. Turner, S. Broussard, and J. S. Allan. 1996. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops). J. Med. Primatol. 25:78-83. [DOI] [PubMed] [Google Scholar]
- 24.Jordan, H. L., J. G. Howard, J. G. Bucci, J. L. Butterworth, R. English, S. Kennedy-Stoskopf, M. B. Tompkins, and W. A. Tompkins. 1998. Horizontal transmission of feline immunodeficiency virus with semen from seropositive cats. J. Reprod. Immunol. 41:341-357. [DOI] [PubMed] [Google Scholar]
- 25.Kils-Hutten, L., R. Cheynier, S. Wain-Hobson, and A. Meyerhans. 2001. Phylogenetic reconstruction of intrapatient evolution of human immunodeficiency virus type 1: predominance of drift and purifying selection. J. Gen. Virol. 82:1621-1627. [DOI] [PubMed] [Google Scholar]
- 26.Kingman, J. F. C. 1982. On the genealogy of large populations. J. Appl. Probab. 19A:27-43.
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Lambert, D. M., P. A. Ritchie, C. D. Millar, B. Holland, A. J. Drummond, and C. Baroni. 2002. Rates of evolution in ancient DNA from Adelie penguins. Science 295:2270-2273. [DOI] [PubMed] [Google Scholar]
- 29.Langley, R. J., V. M. Hirsch, S. J. O'Brien, D. Adger-Johnson, R. M. Goeken, and R. A. Olmsted. 1994. Nucleotide sequence analysis of puma lentivirus (PLV-14): genomic organization and relationship to other lentiviruses. Virology 202:853-864. [DOI] [PubMed] [Google Scholar]
- 30.Leitner, T., and J. Albert. 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96:10752-10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logan, K. A., L. L. Sweanor, T. K. Ruth, and M. G. Hornocker. 1996. Cougars of the San Andres Mountains, New Mexico. New Mexico Department of Game and Fish, Albuquerque, N.Mex.
- 32.Lukashov, V. V., and J. Goudsmit. 2002. Recent evolutionary history of human immunodeficiency virus type 1 subtype B: reconstruction of epidemic onset based on sequence distances to the common ancestor. J. Mol. Evol. 54:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Maddison, W. P., and D. R. Maddison. 1992. MacClade: analysis of phylogeny and character evolution, version 3.0. Sinauer Associates, Sunderland, Mass.
- 33.Mindell, D. P. 1996. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proc. Natl. Acad. Sci. USA 93:3284-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 35.Moutouh, L., J. Corbeil, and D. D. Richman. 1996. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. USA 93:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller-Trutwin, M. C., S. Corbet, M. D. Tavares, V. M. Herve, E. Nerrienet, M. C. Georges-Courbot, W. Saurin, P. Sonigo, and F. Barre-Sinoussi. 1996. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology 223:89-102. [DOI] [PubMed] [Google Scholar]
- 37.Nerrienet, E., X. Amouretti, M. C. Muller-Trutwin, V. Poaty-Mavoungou, I. Bedjebaga, H. T. Nguyen, G. Dubreuil, S. Corbet, E. J. Wickings, F. Barre-Sinoussi, A. J. Georges, and M. C. Georges-Courbot. 1998. Phylogenetic analysis of SIV and STLV type I in mandrills (Mandrillus sphinx): indications that intracolony transmissions are predominantly the result of male-to-male aggressive contacts. AIDS Res. Hum. Retrovir. 14:785-796. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neil, L. L., M. J. Burkhard, L. J. Diehl, and E. A. Hoover. 1995. Vertical transmission of feline immunodeficiency virus. AIDS Res. Hum. Retrovir. 11:171-182. [DOI] [PubMed] [Google Scholar]
- 40.O'Neil, L. L., M. J. Burkhard, and E. A. Hoover. 1996. Frequent perinatal transmission of feline immunodeficiency virus by chronically infected cats. J. Virol. 70:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olmsted, R. A., R. Langley, M. E. Roelke, R. M. Goeken, D. Adger-Johnson, J. P. Goff, J. P. Albert, C. Packer, M. K. Laurenson, T. M. Caro, et al. 1992. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J. Virol. 66:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orenstein, J. M., C. Fox, and S. M. Wahl. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857-1861. [DOI] [PubMed] [Google Scholar]
- 43.Packer, C., S. Alitzer, M. Appel, E. Brown, J. Martenson, S. J. O'Brien, M. Roelke-Parker, R. Hofmann-Lehmann, and H. Lutz. 1999. Viruses of the Serengeti: patterns of infection and mortality in lions. J. Anim. Ecol. 68:1161-1178. [Google Scholar]
- 44.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 45.Paul-Murphy, J., T. Work, D. Hunter, E. McFie, and D. Fjelline. 1994. Serologic survey and serum biochemical reference ranges of the free-ranging mountain lion (Felis concolor) in California. J. Wildl. Dis. 30:205-215. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, N. C., E. W. Ho, M. L. Brown, and E. W. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 47.Pelletier, E., W. Saurin, R. Cheynier, N. L. Letvin, and S. Wain-Hobson. 1995. The tempo and mode of SIV quasispecies development in vivo calls for massive viral replication and clearance. Virology 208:644-652. [DOI] [PubMed] [Google Scholar]
- 48.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 49.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primatol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 51.Preston, B. D., and J. P. Dougherty. 1996. Mechanisms of retroviral mutation. Trends Microbiol. 4:16-21. [DOI] [PubMed] [Google Scholar]
- 52.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 53.Rigby, M. A., E. C. Holmes, M. Pistello, A. Mackay, A. J. Brown, and J. C. Neil. 1993. Evolution of structural proteins of feline immunodeficiency virus: molecular epidemiology and evidence of selection for change. J. Gen. Virol. 74:425-436. [DOI] [PubMed] [Google Scholar]
- 54.Robertson, D. L., P. M. Sharp, F. E. McCutchan, and B. H. Hahn. 1995. Recombination in HIV-1. Nature 374:124-126. [DOI] [PubMed] [Google Scholar]
- 55.Rodrigo, A. G., and J. Felsenstein. 1999. Coalescent approaches to HIV-1 population genetics, p. 233-272. In K. A. Crandall (ed.), The molecular evolution of HIV. Johns Hopkins University Press, Baltimore, Md.
- 56.Roelke, M. E., D. J. Forrester, E. R. Jacobson, G. V. Kollias, F. W. Scott, M. C. Barr, J. F. Evermann, and E. C. Pirtle. 1993. Seroprevalence of infectious disease agents in free-ranging Florida panthers (Felis concolor coryi). J. Wildl. Dis. 29:36-49. [DOI] [PubMed] [Google Scholar]
- 57.Sala, M., and S. Wain-Hobson. 2000. Are RNA viruses adapting or merely changing? J. Mol. Evol. 51:12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 59.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 60.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shpaer, E. G., and J. I. Mullins. 1993. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J. Mol. Evol. 37:57-65. [DOI] [PubMed] [Google Scholar]
- 62.Sodora, D. L., E. G. Shpaer, B. E. Kitchell, S. W. Dow, E. A. Hoover, and J. I. Mullins. 1994. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J. Virol. 68:2230-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62a.Swofford, D. L. 2002. PAUP* 4.0b10: phylogenetic analysis using parsimony (and other methods). Sinauer Associates, Sunderland, Mass.
- 63.Wahl, S. M., and J. M. Orenstein. 1997. Immune stimulation and HIV-1 viral replication. J. Leukoc. Biol. 62:67-71. [DOI] [PubMed] [Google Scholar]
- 64.Wolfe, N. D., A. A. Escalante, W. B. Karesh, A. Kilbourn, A. Spielman, and A. A. Lal. 1998. Wild primate populations in emerging infectious disease research: the missing link? Emerg. Infect. Dis. 4:149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103-110. [DOI] [PubMed] [Google Scholar]
- 66.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 67.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]