Abstract
RNA interference (RNAi) is a flexible gene silencing mechanism that responds to double-stranded RNA by suppressing homologous genes. Here, we report the characterization of RNAi effector complexes (RISCs) that contain small interfering RNAs and microRNAs (miRNAs). We identify two putative RNA-binding proteins, the Drosophila homolog of the fragile X mental retardation protein (FMRP), dFXR, and VIG (Vasa intronic gene), through their association with RISC. FMRP, the product of the human fragile X locus, regulates the expression of numerous mRNAs via an unknown mechanism. The possibility that dFXR, and potentially FMRP, use, at least in part, an RNAi-related mechanism for target recognition suggests a potentially important link between RNAi and human disease.
Keywords: dFXR; VIG; RNA interference; Ago-2, miRNA; Ago-1
RNA interference (RNAi) encompasses a suite of homology-dependent gene silencing mechanisms that are triggered by double-stranded RNA (dsRNA). RNAi is an evolutionarily conserved response, and mechanistically related processes exist in plants, animals, and fungi (Bernstein et al. 2001b). A combination of genetic approaches in Caenorhabditis elegans, Arabidopsis, Neurospora, and Chlamydomonas, along with biochemical analyses in Drosophila extracts has begun to reveal a mechanistic framework for RNAi (Hammond et al. 2001b). Silencing is initiated when dsRNA triggers are processed into small interfering RNAs (siRNAs), which are ∼21 nucleotides in length in Drosophila (Hamilton and Baulcombe 1999; Tuschl et al. 1999; Elbashir et al. 2001). This reaction is catalyzed by a group of related RNase III enzymes, now known as the Dicer family (Bernstein et al. 2001a). The siRNAs are incorporated into an effector complex, the RNA-induced silencing complex (RISC), which uses siRNAs as a guide to select complementary mRNA substrates (Hammond et al. 2000).
In general, RNAi acts at the posttranscriptional level and is associated with destabilization of mRNAs with complementarity to the silencing trigger (Fire et al. 1998). Recent studies suggest that some endogenously encoded small dsRNAs (known collectively as microRNAs, or miRNAs) can regulate the expression of endogenous protein-coding genes at the level of protein synthesis without message degradation (Olsen and Ambros 1999). The dependency of both siRNA- and miRNA-mediated silencing on Argonaute-family proteins and Dicer shows that the miRNA and siRNA pathways share conserved components and likely have related mechanisms. Recently, more than 100 miRNAs have been cloned from C. elegans (Lau et al. 2001; Lee and Ambros 2001), human (Lagos-Quintana et al. 2001; Mourelatos et al. 2002), mouse (Lagos-Quintana et al. 2002), Drosophila (Lagos-Quintana et al. 2001), and plants (Llave et al. 2002; Reinhart et al. 2002; A.A. Caudy, M. Myers, G.J. Hannon, and S.M. Hammond, unpubl.). Based on their prevalence, miRNAs are likely to play broad and important roles in gene regulation, possibly guiding complexes involved in various aspects of RNA metabolism to their cognate substrates. Both to address the underlying mechanism of siRNA-guided degradation and to investigate the degree to which the mechanisms of siRNA and miRNA-mediated silencing overlap, we have sought to determine the composition of RNAi-effector complexes.
Results and Discussion
Biochemical purification of the RNAi effector nuclease, RISC, revealed the Drosophila Argonaute-2 (Ago-2) protein as a core component of this complex (Hammond et al. 2001a). In an effort to identify additional RISC components, we performed large-scale biochemical purification, following both RISC activity and Ago-2 protein by Western blotting. After five purification steps, a number of additional proteins also consistently co-purified (Fig. 1A). Among those were two RNA-binding proteins, VIG and the Drosophila homolog of the human Fragile X Mental Retardation Protein [FMRP in human, dFXR in Drosophila (Wan et al. 2000); Fig. 1B,C]. Each protein was identified in multiple independent preparations of purified RISC.
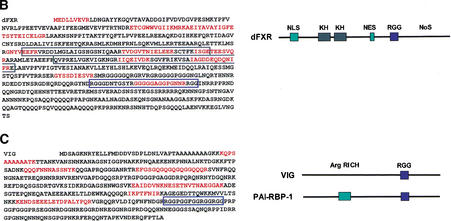
Figure 1.

Identification of dFXR [the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP)], and VIG as RISC components. (A) Coomassie-stained polyacrylamide gel showing purified RISC fractions from a Source Q column. (B) Peptides obtained from microsequencing are indicated in red on the dFXR protein sequence. The KH (hnRNP K homology) domains are boxed in black and the RGG box is indicated in blue (accession no. dFXR NP_611645). (C) Peptides obtained from microsequencing are indicated in red on the VIG protein sequence; the RGG box is indicated in blue. At right, a comparison of the domain structure of VIG to the human homolog PAI-RBP-1 (accession nos. VIG NP_523572 and PAI-RBP-1 NP_056455).
The VIG protein is encoded from within an intron of the Vasa gene. This protein has no recognizable protein domains, other than an RGG box, a motif that is known to bind RNA. VIG is an evolutionarily conserved protein, with homologs in C. elegans, Arabidopsis, mammals, and Schizosaccharomyces pombe. Little is known regarding the function of this protein family. However, a human homolog of VIG, PAI-RBP-1, was originally identified as a protein with affinity for an AU-rich element in the 3′-UTR of the plasminogen activator inhibitor (PAI) RNA, which regulates its stability (Heaton et al. 2001).
A human ortholog of dFXR, FMRP, is encoded from a locus on the X chromosome that is epigenetically silenced in Fragile X syndrome (Eberhart et al. 1996). FMRP is expressed not only in neuronal cells but also in numerous other tissues, and Fragile X patients display additional phenotypes, including macroorchidism (Rudelli et al. 1983). The human genome also encodes two additional FMRP homologs, FXR1 and FXR2 (Kirkpatrick et al. 2001). Fragile X family members each contain two copies of a KH domain (hnRNP K homology) and an RGG box. All three of these domains have been proposed to bind RNA.
To test the association of VIG and dFXR with RISCs, we expressed epitope-tagged proteins in Drosophila S2 cells. Notably, both Vig and dFXR co-fractionated with Ago-2 and RISC activity in an ∼500-kD complex, as predicted if all three were components of RISC (Fig. 2A). Both RISC and dFXR pellet in a high-speed centrifugation and can be extracted from the P100 pellet by treatment with 0.4 M KOAc (Fig. 2B). Essentially, all expressed dFXR co-fractionated with RISC through subsequent ion exchange columns (Fig. 2C; data not shown). In accord with this hypothesis, immunoprecipitation of dFXR resulted in co-precipitation of Ago-2 protein (Fig. 2D). To test whether the interaction was bridged by binding separately to a common mRNA, we treated immunoprecipitations with RNAse A. Interaction between Ago-2 and dFXR was resistant to treatment of the immunoprecipitates with RNAse A up to concentrations of 10 mg/mL (data not shown). Similarly, Ago-2 was also recovered in immunoprecipitations of epitope-tagged VIG (Fig. 2E), and VIG and dFXR were present in mutual immunoprecipitations (data not shown).
Figure 2.
dFXR [the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP)] and VIG associate with RISC. (A) S2 cells were transfected with a combination of luciferase double-stranded RNA (dsRNA), his-tagged VIG, and T7-tagged dFXR1. Ribosomal extracts were size-fractionated on Superose 6 and Western blotted. (B) Western blots of Argonaute-2 (Ago-2) and tagged dFXR in cytoplasmic and high-salt ribosomal extract from cells expressing the indicated tagged constructs. (C) In a separate experiment, Flag-tagged dFXR-1 was transfected, extracted, and size-fractionated as in A, and fractions containing dFXR were further fractionated on Source S. Fractions were Western blotted for Ago-2 and Flag-dFXR. (D,E) Drosophila S2 cells were transfected with dsRNA and either a control vector or vectors directing the expression of either T7-VIG (D) or Flag-dFXR (E). Proteins were recovered by immunoprecipitation and analyzed by Western blot using a polyclonal antibody raised against the Ago-2 protein. (F) Drosophila S2 cells were transfected with radiolabeled luciferase dsRNA and the indicated control or T7-tagged expression constructs. Proteins were immunoprecipitated with anti-T7 affinity resin, and associated RNA was separated on denaturing 15% polyacrylamide gel.
The presence of VIG and dFXR in RISCs would predict that these proteins would also interact with siRNAs, another established component of RISCs (Hammond et al. 2000). To test this possibility, we co-transfected Drosophila S2 cells with 32P-labeled dsRNA triggers of ∼500 nt in length and either tagged VIG or dFXR. In each case, immunoprecipitation selectively recovered ∼21 nt siRNAs, even though these are a minor component in analyses of total RNA extracted from cells transfected with labeled silencing triggers (Fig. 2E; data not shown).
To examine the requirement for dFXR and VIG in RNAi, we used RNAi to reduce the expression of each of these gene products in S2 cells. This approach has been used previously to test the dependency of RNAi on Dicer and Argonaute family members in Drosophila cells, mammalian cells, and C. elegans (Grishok et al. 2000; Bernstein et al. 2001a; Hammond et al. 2001a; Paddison et al. 2002; Tabara et al. 2002) and as a means to identify components of the RNAi machinery in worms (Dudley et al. 2002). Whereas cells pretreated with control dsRNAs (e.g. GFP) silence reporter genes efficiently, pretreatment of cells with dsRNAs covering the coding sequences of either dFXR or VIG partially impaired silencing. In the former case, expression returned to 50% of the unsuppressed control, whereas in the latter case, luciferase was de-repressed to ∼70% of the control (Fig. 3). The partial effect of suppressing dFXR can be interpreted in several ways. First, dFXR could be an accessory factor for RISCs, affecting efficiency of the complex but not being absolutely required for RNAi. Second, dFXR could be essential to only a subset of RISCs, whereas other, perhaps KH domain, proteins replace dFXR in alternative versions of RISCs. Third, dFXR might not be required for RNAi per se; however, loss of dFXR could affect the long-term stability of some RISC complexes. Another Argonaute family member expressed in S2 cells, Ago-1, appears to have little effect on the efficiency of RNAi in S2 cells. Ago-1 can be biochemically separated from active RISCs (data not shown). However, we by no means exclude a role for Ago-1 in RNAi (see below) as previous genetic studies (Williams and Rubin 2002) have indicated that Ago-1 is required for efficient RNAi during embryogenesis.
Figure 3.
dFXR [the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP)] and VIG are required for efficient RNA interference in Drosophila S2 cells. Drosophila S2 cells were transfected with double-stranded RNAs (dsRNAs) corresponding to the indicated cDNAs. Three days later, cells were transfected with dsRNAs directed against luciferase (or GFP for controls) and with expression vectors for both firefly and renilla luciferases. Values shown represent the expression of luciferase in luciferase dsRNA-transfected cells relative to luciferase expression from cells transfected with control dsRNA.
The forgoing biochemical and genetic studies are consistent with a model in which VIG and dFXR are associated with RISCs. We therefore tested the possibility that a sequence-specific nuclease activity might co-immunoprecipitate with either of these components. Cells were co-transfected with a dsRNA homologous to luciferase and with expression constructs for tagged versions of dFXR, VIG, or Ago-1. Control cells were transfected with a GFP expression construct. Immunoprecipitates were tested for RISC activity against substrates homologous to the dsRNA trigger and against nonhomologous substrates. Although immunoprecipitates from the GFP-transfected cells and immunoprecipitates of Ago-1 and Vig showed some specific activity against a synthetic luciferase mRNA, immunoprecipitates of dFXR substantially degraded the cognate but not the noncognate substrate. In the case of dFXR, >80% of the homologous substrate was degraded in 1 h of incubation (Fig. 4).
Figure 4.
dFXR [the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP)] immunoprecipitates contain RISC activity. Drosophila S2 cells were transfected with expression vectors for the indicated T7-tagged cDNAs and double-stranded RNAs corresponding to luciferase. Protein was immunoprecipitated with anti-T7 tag affinity resin, and this resin was incubated for the indicated times with radiolabeled single-stranded RNA (ssRNA) for target (luciferase), or control (GFP). RISC activity was indicated by loss of targeted RNA.
The majority of Fragle X mutations occur in the 5′-UTR of the gene and represent a trinucleotide repeat expansion that is thought to act by targeting FMR1 for epigenetic silencing. However, one point mutation in the FMRP protein itself has also been linked to the disease (I304N; De Boulle et al. 1993). This hypomorphic mutation in the human protein causes defects in ribosome association and RNA binding (Tamanini et al. 1996; Feng et al. 1997). We constructed an analogous mutation in the second KH domain of the Drosophila protein, I307N, and asked whether this mutation affected association with the RNAi machinery. Biochemical fractionation showed a significant alteration in the ability of the mutant dFXR to associate with RISC and Ago-2 (Fig. 2B), with ∼30% of the mutant protein shifting from the ribosome pellet (where Ago-2 localizes) to a soluble cytoplasmic form. This fraction is completely devoid of Ago-2, and Ago-2 cannot be found in association with cytoplasmic dFXR.
The specific biochemical role of the human protein FMRP is not known; however, numerous studies suggest that it regulates the expression of a large number of genes at the level of protein synthesis (Bardoni and Mandel 2002). Recent studies of Dicer function (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001) and of Argonaute family members (Grishok et al. 2001) in C. elegans have revealed that endogenously encoded small hairpin RNAs (generically, miRNAs) regulate the expression of protein coding genes via pathways related to RNAi. The presence of dFXR in RISCs suggests the hypothesis that FMRP or FMRP family members may regulate gene expression via a RISC-like complex.
Little is known of the effector complexes that contain miRNAs. A recent report has shown association of miRNAs in human cells with a 15S complex containing Gemin3, Gemin4, and eIF2c, a member of the Argonaute protein family (Mourelatos et al. 2002). Gemin3 is a DEAD-box family RNA helicase, and a helicase has previously been predicted as an essential activator of siRNA-containing complexes in Drosophila (Nykanen et al. 2001). Also, in C. elegans two DExH/D box helicases, one of which is Dicer, associate with Rde-4 and are required for RNAi (Tabara et al. 2002). Gemin4 lacks both recognizable protein motifs (Charroux et al. 2000) and an identifiable homolog in the Drosophila genome. Thus, the relationship between the previously characterized miRNP complex and RISC remains unclear, which led us to further characterize the association between miRNAs and RISCs.
A significant portion of two known miRNAs, mir2b and mir13a (Lagos-Quintana et al. 2001), in Drosophila S2 cells are found in association with polyribosomes, as would be predicted for small RNAs that regulate protein synthesis (data not shown). Furthermore, a functional connection between siRNAs and ribosome association has been made in trypanosomes (E. Ullu, pers. comm.). As was previously shown for RISCs (Hammond et al. 2000), miRNAs can be extracted from a high-speed pellet from S2 cell extracts (which contains polysomes) using buffers containing elevated salt concentrations (Fig. 5A).
Figure 5.
dFXR- [the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP)] and VIG-based RISC complexes contain microRNAs (miRNAs). (A) S2 cells were lysed hypotonically, and the nuclei were removed by centrifugation. The supernatant was pelleted at low salt, producing a S100 and P100. The P100 was extracted with high salt and centrifuged again to yield a high-salt P100 and a high-salt S100. RNA was extracted from equivalent portions of each and analyzed by Northern blotting with an oligonucleotide complementary to the Drosophila miRNA 13a. (B) S2 cells were soaked in luciferase double-stranded RNA, and extracts were prepared. Ribosomes were pelleted, salt extracted, and precipitated under low-salt conditions. The precipitates were dissolved again at high salt and size-fractionated on Superose 6. Column fractions were analyzed by Western blotting for Argonaute-1 (Ago-1) and Argonaute-2 (Ago-2) and by Northern blotting for miR2b, mir13a, and luciferase RNA. (C) S2 cells were transfected with T7-tagged FXR and his-tagged Vig expression constructs and size-fractionated. Fractions corresponding to the Ago-2 peak were immunoprecipitated using anti-T7 agarose beads and analyzed by Northern blotting for miR2b. Control immunoprecipitations were from similarly fractionated untransfected cells. (D) Drosophila S2 cells were transfected with the indicated tagged expression constructs or a GFP control. Proteins were collected by immunoprecipitation using anti-tag agarose beads. The beads were extracted with Trizol to prepare RNA and Northern blotted for miR2b.
Extracted material was fractionated by size-exclusion chromatography, and the behavior of miRNAs and siRNAs was compared with that of known RISC subunits. As we previously reported (Hammond et al. 2000), the majority of siRNAs co-fractionate with Ago-2 and with VIG and dFXR. In contrast, the majority of miRNAs fractionate in an ∼250-kD complex, although ∼20% of miRNAs are present in fractions containing RISC (Fig. 5B). However, we could not rule out the possibility that this represents a tail of the 250-kD peak, given the nonquantitative nature of Western blotting. Considering the established role of dFXR in the regulation of endogenous protein-coding genes, we examined the possibility that miRNAs might be present in dFXR-containing complexes. Immunoprecipitates of dFXR were prepared from column fractions using appropriate affinity resins. Notably, mir2b was detected in dFXR immunoprecipitates but not in control immunoprecipitates prepared using the same affinity resins from Ago-2- and miRNA-containing fractions of extracts from untransfected cells (Fig. 5C). Similarly, VIG could be used to immunoprecipitate mir2B directly from extracts (Fig. 5D).
The forgoing data suggest that miRNAs associate with RISCs that also contain dFXR and VIG. This is consistent with recent data suggesting that miRNAs can direct cleavage by RISC, given appropriate homologous substrates (Hutvagner and Zamore 2002). However, the observation of an ∼250-kD peak containing the majority of miRNAs raised questions regarding the composition of these smaller complexes. Another Argonaute family member, Ago-1, showed co-fractionation of this protein with the major miRNA peak both on sizing columns (Fig. 5B) and subsequently on ion exchange columns (data not shown). To test the association of Ago-1 with miRNAs, we performed immunoprecipitation experiments with an epitope-tagged Ago-1 protein. Both mir2b and mir13a could be detected in association with Ago-1 (Fig. 5D; data not shown). Furthermore, Ago-1 immunoprecipitates also contained siRNAs (Fig. 2F). Reciprocal immunoprecipitations and Western blots indicate that dFXR and Vig do not detectably associate with Ago-1 (data not shown).
The forgoing data raises the possibility that siRNAs and miRNAs are distinguished to some extent and preferentially, although not exclusively, assembled into separate, functional effector RNPs. Recently, it has been shown that Ago-1 is required for efficient interference with endogenous genes in Drosophila embryos (Williams and Rubin 2002). Notably, Ago-1 complexes in S2 cells are similar in size to the active effector complexes, RISC*, that have previously been detected in embryo lysates (Nykanen et al. 2001). Our failure to detect substantial RISC activity in Ago-1 complexes and our failure to detect a substantial dependence on Ago-1 for RNAi in S2 cells could reflect developmental differences in the utilization of Ago family members or experimental differences in the assays used to detect RISCs, namely measuring loss of substrate (in our case) versus endonucleolytic cleavage (as in Nyanken et al. 2001).
The identification of the Drosophila homolog of the fragile X protein as a RISC subunit suggests that disruptions in RNAi-related pathways may contribute to human disease. The fragile X protein has previously been implicated in the regulation of gene expression at the level of protein synthesis (Brown et al. 2001; Darnell et al. 2001). In Drosophila, a bona fide dFXR regulatory target has been identified as Futsch, a Map1B homolog (Zhang et al. 2001). Indeed, dFXR and Futsch are epistatic with respect to the neuronal defects of dFXR-mutant flies, although they are not epistatic with respect to circadian defects (Dockendorff et al. 2002). Many miRNAs in Drosophila show complementarity to common sequence motifs that negatively regulate gene expression at the posttranscriptional level (Lai 2002). These K boxes, Brd boxes, and GY boxes are present in numerous mRNAs. Notably, a K box and a GY box are found in the 3′-UTR of Futsch. However, we cannot yet definitively link a sequenced miRNA to suppression of Futch expression. In humans, FMRP has been isolated as a component of large cytoplasmic mRNA complexes and, through this association, has been implicated in the regulation of numerous mRNAs. None of the several proteins known to bind FMRP has been implicated in RNAi. However, there are many possible resolutions to this seeming contradiction. First, FMRP could function through several completely independent mechanisms. Second, FMRP could be an accessory factor for rather than an integral component of RISCs, using RISCs for identifying its cognate targets that may be regulated via mechanisms that involve its interaction with other proteins. This would be consistent with the partial effects of dFXR depletions on RNAi efficiency in S2 cells.
The precise mechanisms through which FMRP is delivered to its regulatory targets is unclear. Previously, selection of random RNA oligonucleotides implicated that FMRP binds G-quartet structures that lie adjacent to short RNA helices (Brown et al. 2001; Darnell et al. 2001). This interaction required only the RGG motif and did not involve the KH domain. Thus, it was proposed that FMRP recognized and regulated G-quartet-containing mRNAs. Here, we propose an alternative mechanism of regulation by fragile X protein. Our results are consistent with FMRP1 being targeted to its substrates as part of RNAi complexes, which are guided by miRNAs. Recognition of G-quartets might be an important determinant of further specificity or might be critical for translational regulation. We further propose that FMRP might be one of many distinct protein subunits that join RISCs, depending on the tissue, subcellular localization, and the developmental stage. In this way, RISCs could function as a flexible platform on which might be constructed a variety of regulatory machines that alter gene expression at the level of mRNA metabolism, at the level of protein synthesis, and at the level of genome structure.
Materials and methods
Cloning and expression
Ago-1, dFxr, and VIG coding sequences were amplified from full-length cDNAs by PCR, with N-terminal (Ago-1) or C-terminal (VIG, dFxr) epitope tags introduced. T7 (Novagen), Flag M2(Sigma), and hexahistidine tags were introduced, as indicated in the figure legends. PCR products were cloned into the pMT vector (Invitrogen), a CuSO4-inducible insect expression vector. The I307N dFxr mutant was made using the Stratagene QuikChange kit. Drosophila S2 cells were cultured and transfected as described. (Hammond et al. 2000)
Immunoprecipitation
Transfected cells were lysed using 0.5% NP-40, 150 mM NaCl, 20 mM Hepes (pH 7.0), 2 mM MgCl2, and 1 mM DTT. Proteins were immunoprecipitated using anti-T7 beads (Novagen) or anti-Flag M2 beads (Sigma). Immunoprecipitates were washed in the IP buffer containing 400 mM NaCl to extract ribosomes and were processed; Western blotting was as previously described (Hammond et al. 2001a). For Northern blotting, RNA from immunoprecipitates was extracted with Trizol (Invitrogen), separated on a 15% denaturing urea 1× TAE polyacrylamide gel, and electroblotted to Hybond N. Membranes were UV crosslinked and hybridized at 42°C in moderate stringency hybridization buffer (10 g/L BSA, 1 mM EDTA, 7% SDS, 15% formamide, and 0.4 M NaPO4 at pH 7). Washes were at 42°C in 1× SSC, 0.1% SDS. For siRNA immunoprecipitations, S2 cells were transfected with uniformly 32P-labeled dsRNA corresponding to the first 800 bp of the GL2 luciferase gene (Invitrogen) and the indicated expression construct. For RISC activity immunoprecipitations, transfected cells were immunoprecipitated as above, but the final wash was with buffer F (2 mM MgCl2, 20 mM Hepes at pH 7, 110 mM KOAc, and 1 mM DTT). The beads were left in a small volume of buffer F, and ATP was added to 1 mM, RNAsin was added to 20 U/mL, and 1000 cpm of 32P-labeled synthetic RNA transcript (as indicated) was added. Beads were removed at the indicated time points, and RNA was extracted with Trizol.
RISC purification
For protein identification, RISC activity was purified ∼10,000-fold from Drosophila S2 cells, as described in Hammond et al. (2001a). Column fractions were TCA precipitated and run on a 4%–20% SDS-PAGE gradient gel. Protein bands were excised and analyzed by Matrix Assisted Laser Desorption Ionization and Liquid Chromatograph Quadropole Mass Spectrometry.
RNAi of RNAi
Drosophila S2 cells were transfected with 4 μg of a mixture of 500 nt dsRNAs covering the entire indicated cDNA. Three days after transfection, cells were further transfected with 4 μg of a 10:1 mixture of pGL3 control firefly luciferase expression vector (Promega) and pRLSV40 Renilla luciferase expression vector (Promega) and 25 ng dsRNA corresponding to firefly luciferase or GFP. Three days later, luciferase activity was measured using the Dual Luciferase kit (Promega). Ratios of firefly activity to Renilla activity were calculated. For each targeted gene, the expression in the cells undergoing RNAi against luciferase was normalized against control cells undergoing RNAi against GFP.
Acknowledgments
We thank T. Uemura for the very kind gift of Ago-1 antisera and D. Conklin, E. Bernstein, and M. Carmell for critical reading of the manuscript. A.A.C. is a George A. and Marjorie H. Anderson Fellow of the Watson School of Biological Sciences and is a predoctoral fellow of the Howard Hughes Medical Institute. Supported in part by National Institutes of Health grant RO1-GM62534 (G.J.H.) and by a grant from Genetica, Inc. G.J.H. is a Rita Allen Foundation scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1025202.
References
- Bardoni B, Mandel JL. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr Opin Genet Dev. 2002;12:284–293. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001a;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001b;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4: A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc Natl Acad Sci. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart DE, Malter HE, Feng Y, Warren ST. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2: a link between genetic and biochemical analyses of RNAi. Science. 2001a;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001b;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the type-1 plasminogen activator inhibitor mRNA. J Biol Chem. 2001;276:3341–3347. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G. and Zamore, P.D. 2002. A MicroRNA in a multiple-turnover RNAi enzyme complex. Science (in press). [DOI] [PubMed]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick LL, McIlwain KA, Nelson DL. Comparative genomic sequence analysis of the FXR gene family: FMR1, FXR1, and FXR2. Genomics. 2001;78:169–177. doi: 10.1006/geno.2001.6667. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. The Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudelli RD, Jenkins EC, Wisniewski K, Moretz R, Byrne J, Brown WT. Testicular size in fetal fragile X syndrome. Lancet. 1983;1:1221–1222. doi: 10.1016/s0140-6736(83)92498-4. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tamanini F, Meijer N, Verheij C, Willems PJ, Galjaard H, Oostra BA, Hoogeveen AT. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]