Abstract
Fragile X syndrome is a common form of inherited mental retardation caused by the loss of FMR1 expression. The FMR1 gene encodes an RNA-binding protein that associates with translating ribosomes and acts as a negative translational regulator. In Drosophila, the fly homolog of the FMR1 protein (dFMR1) binds to and represses the translation of an mRNA encoding of the microtuble-associated protein Futsch. We have isolated a dFMR1-associated complex that includes two ribosomal proteins, L5 and L11, along with 5S RNA. The dFMR1 complex also contains Argonaute2 (AGO2) and a Drosophila homolog of p68 RNA helicase (Dmp68). AGO2 is an essential component for the RNA-induced silencing complex (RISC), a sequence-specific nuclease complex that mediates RNA interference (RNAi) in Drosophila. We show that Dmp68 is also required for efficient RNAi. We further show that dFMR1 is associated with Dicer, another essential component of the RNAi pathway, and microRNAs (miRNAs) in vivo, suggesting that dFMR1 is part of the RNAi-related apparatus. Our findings suggest a model in which the RNAi and dFMR1-mediated translational control pathways intersect in Drosophila. Our findings also raise the possibility that defects in an RNAi-related machinery may cause human disease.
Keywords: fragile X syndrome, FMR1, RNAi, RNA helicase, miRNA
Fragile X syndrome is the single most common form of inherited disease causing mental retardation with a prevalence estimated at ∼1 in 4000 male births and 1 in 8000 female births (for review, see Hagerman 2002). Cognitive deficits reported in fragile X children range from mild to severe, and behavioral disturbances include social and attention deficits, autistic-like behaviors, unusual responses to sensory stimuli, hyperactivity, and abnormal sleep (Gould et al. 2000; Hagerman 2002). In most cases, the syndrome is caused by a trinucleotide repeat expansion in the 5′ untranslated region of the fragile X mental retardation 1 gene (FMR1; for review, see Imbert et al. 1998; O'Donnell and Warren 2002). An expansion of the CGG repeat is associated with abnormal DNA methylation of both a nearby CpG island and the repeat itself. As a result, the FMR1 locus becomes silent at the transcriptional level and thus no translation occurs (Siomi et al. 1993; Verheij et al. 1993). It is therefore clear that the pathophysiological mechanisms leading to the symptoms in fragile X syndrome can be elucidated by studying the function of the FMR1 gene.
Pathology in the brains of fragile X patients and Fmr1 knockout mice show the presence of abnormal dendritic spines reminiscent of a maturation delay (Greenough et al. 2001; Irwin et al. 2001). Because the FMR1 protein (FMRP) is an RNA-binding protein that associates with polyribosomes, it is thought to be related to posttranscriptional regulation of gene expression in a manner critical for the correct development of neurons (Inoue et al. 2000; O'Donnell and Warren 2002). Indeed, biochemical studies suggest that FMRP acts as a negative regulator of translation (Laggerbauer et al. 2000; Li et al. 2000; Schaeffer et al. 2001). These observations suggest that by modulating mRNA translation and consequently protein synthesis, FMRP is important for the formation and function of synapses. However, to what extent and how it might effect translation in vivo is unknown. The fruit fly Drosophila melanogaster has proven to be a powerful tool for the genetic dissection of biochemical pathways. Because many genes are conserved between flies and humans, including genes that regulate complex behaviors such as learning and memory, as well as entire pathways of development and oncogenesis (Patel 1994; Miklos and Rubin 1996; Rubin et al. 2000), Drosophila could be used, therefore, to define the molecular pathways leading to human neurological diseases and to identify the genes involved (Fortini and Bonini 2000). In the Drosophila genome, there is a single gene, dFMR1, that is homologous to FMR1 (Wan et al. 2000). Drosophila and vertebrate FMR1 proteins share a number of topographical landmarks (Wan et al. 2000), including two types of RNA-binding motifs, namely, two KH domains and an RGG box (Siomi and Dreyfuss 1997). Moreover, they show similar biochemical properties such as RNAbinding and ribosome-association (Wan et al. 2000).Importantly, genetic studies demonstrate that dFMR1 has a role in the regulation of synapse growth and functions (Zhang et al. 2001; Dockendorff et al. 2002; Morales et al. 2002; Inoue et al. 2002), probably by acting as a translational repressor of an mRNA encoding Futsch, which is the fly homolog of the microtubule-associated protein MAP1B (Zhang et al. 2001). Therefore, examining the role of FMRP in the fruit fly is a promising approach providing significant insights into the function of FMRP.
RNA interference (RNAi) is the process of sequence-specific posttranscriptional gene silencing (PTGS) in a variety of organisms, initiated by the introduction of double-stranded (ds) RNA that is homologous in sequence to the silenced gene (Fire et al. 1998; for review, see Cogoni and Macino 2000; Vance and Vaucheret 2001; Waterhouse et al. 2001; Hannon 2002; Hutvagner and Zamore 2002). During RNAi, introduced dsRNAs are processed into small RNAs of ∼21–22 nucleotides (nt) that have been termed small interfering RNAs (siRNAs) because they direct the cleavage of complementary mRNA targets (Zamore et al. 2000; Elbashir et al. 2001). RNAi pathways share features with a developmental gene regulatory pathway that involves natural dsRNA-encoding genes, recently named microRNA (miRNA) genes (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Natural miRNA genes encode ∼70-nt hairpin precursor RNAs that are processed into mature 22-nt miRNAs whose expression is often developmentally regulated. miRNAs are thought to regulate mRNA translation because the founding members of the miRNA gene family, lin-4 and let-7, encode miRNA products that repress translation during Caenorhabditis elegans development by base pairing with complementary sequences located in the 3′ UTRs of their target mRNAs (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000). Although siRNA and miRNA pathways are thought to be distinct, studies in animals have revealed that the multidomain RNase III-related enzyme Dicer is required for processing both siRNAs and miRNAs, therefore producing RNA species of similar sizes (Bernstein et al. 2001; Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). Furthermore, both the siRNA pathway and miRNA pathway require members of the Argonaute family [also referred to as the PAZ-and-Piwi domain (PPD) family, or the RDE-1 family; Tabara et al. 1999; Catalanotto et al. 2000; Fagard et al. 2000; Hammond et al. 2001]. In C. elegans, RNAi requires the Argonaute family member rde-1 (Tabara et al. 1999), whereas two homologs of rde-1, named alg-1 and alg-2, are required for the processing and function of the lin-4 and let-7 miRNAs (Grishok et al. 2001). Argonaute family members have also been implicated in PTGS and development in fungi, plants, C. elegans, Arabidopsis, and Drosophila (Bohmert et al. 1998; Tabara et al. 1999; Catalanotto et al. 2000; Fagard et al. 2000; Hammond et al. 2001; Harris and Macdonald 2001; Kennerdell et al. 2002; Pal-Bhadra et al. 2002; Williams and Rubin 2002). Very recently, human eIL2C2, a human Argonaute protein, has been found in an RNA–protein complex, the miRNP, that contains miRNAs (Mourelatos et al. 2002). Therefore, these observations suggest the speculation that the distinct Argonaute family protein associated with siRNAs or miRNAs regulates the maturation and function of these small RNAs. In Drosophila, RNAi is mediated by the RNA-induced silencing complex (RISC), a sequence-specific, multicomponent ribonucleoprotein complex that acts as an siRNA-directed endonuclease, recognizing and cleaving the complementary mRNA (Hammond et al. 2001). Argonaute2 (AGO2), an Argonaute family protein, was found to be part of the RISC nuclease and to copurify with siRNAs and Dicer (Hammond et al. 2001). However, other cellular components that associate with AGO2 have not been identified.
Here we report the identification of a novel ribonucleoprotein (RNP) complex that contains dFMR1, AGO2, a Drosophila homolog of p68 RNA helicase (Dmp68), and two ribosomal proteins, L5 and L11, along with 5S ribosomal RNA (rRNA). dFMR1 interacts directly with L5 and L11, and forms a ternary complex with the L5/5S rRNA. We show that Dmp68 is required for efficient RNAi. dFMR1 also physically interacts with Dicer and miRNAs; therefore, dFMR1 is in an RNAi-related apparatus. Our findings suggest a model in which the RNAi and dFMR1-mediated translational control pathways intersect in Drosophila.
Results
Isolation of a dFMR1-associated complex
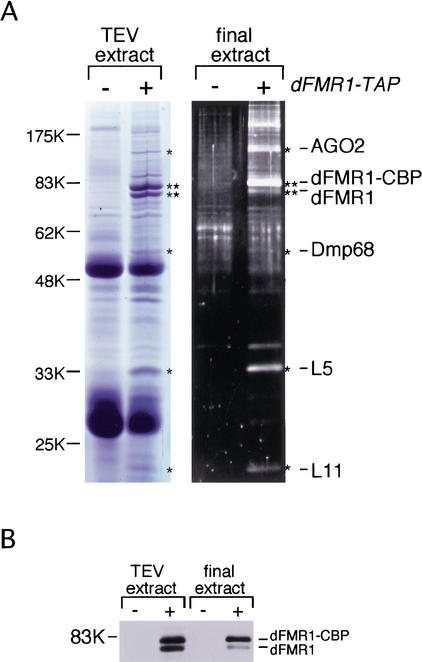
To better understand the biological function of dFMR1, we used a tandem affinity purification (TAP) method (Rigaut et al. 1999) to identify molecules that interact with dFMR1 in vivo. dFMR1-TAP and associated molecules were recovered from a cytoplasmic lysate of S2 cells expressing dFMR1-TAP. Protein contents in the tobacco etch virus (TEV) protease and the final extracts were visualized on SDS-PAGE gels by staining with Coomassie Blue and SYPRO Ruby, respectively (Fig. 1A). Several protein bands were observed specifically in the dFMR1-TAP plus lanes. Four proteins were found by mass spectrometric analysis to be two ribosomal proteins, L5 and L11, AGO2 (Hammond et al. 2001) and a Drosophila homolog of p68 RNA helicase (Ford et al. 1988; Flybase annotation no. CG10279; we refer to the protein as Dmp68). The presence of endogenous dFMR1 in both extracts supports our previous findings that dFMR1 forms homodimers in vitro and in vivo (H. Siomi and M.C. Siomi, in prep.; Fig. 1B).
Figure 1.
Protein components of TAP-purified dFMR1 complex from S2 cells. (A) The protein components in the TEV and the final extracts obtained from S2 cells expressing dFMR1-TAP and the parental cells (dFMR1-TAP minus) were resolved on SDS-PAGE and visualized by Coomassie Blue and SYPRO Ruby staining, respectively. Four of the distinct bands observed only in the dFMR1-TAP plus lanes (indicated with an asterisk) were analyzed by mass spectrometry and were found to be AGO2, Dmp68, and ribosomal proteins L5 and L11 as indicated at right. The protein bands indicated with two asterisks correspond to dFMR1-CBP (a converted form of dFMR1-TAP after TEV cleavage) and endogenous dFMR1 (see panel B). (B) Western blot analysis on the TEV and the final extracts using anti-dFMR1 antibody. The bands corresponding to dFMR1-CBP and endogenous dFMR1 are indicated at right.
dFMR1 interacts with AGO2, Dmp68, and ribosomal proteins in vivo and in vitro
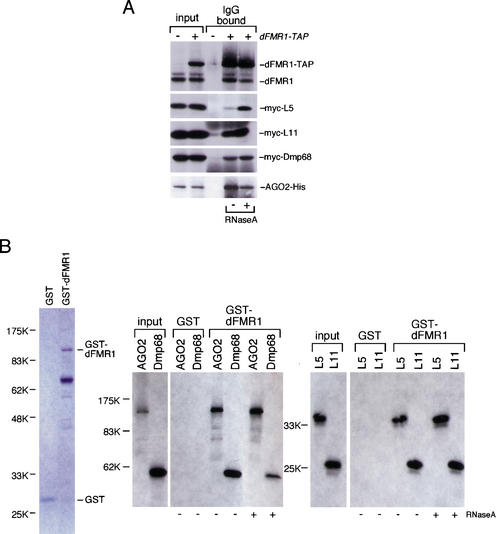
To confirm the in vivo association with dFMR1, either myc-L5, myc-L11, myc-Dmp68, or AGO2-His were coexpressed with dFMR1-TAP in S2 cells. Cytoplasmic lysates of the transfected cells were prepared and subjected to IgG-bead binding. Western blots with anti-myc or anti-His antibody revealed that each protein tested specifically copurified with dFMR1 (Fig. 2A). The interactions were not abolished by ribonuclease A (RNase A) treatment, suggesting that the associations of dFMR1 with L5, L11, AGO2, and Dmp68 are mediated by protein–protein interactions (Fig. 2A). We further investigated whether these proteins can bind to dFMR1 in vitro. L5, L11, AGO2, and Dmp68 were produced by an in vitro translation system and used in binding assays with recombinant dFMR1 fused to glutathione S-transferase (GST; Fig. 2B). GST-dFMR1 interacted with all the proteins tested, whereas GST itself showed no detectable bindings. In addition, RNase A treatment did not affect the in vitro interactions (Fig. 2B). Although the assay we used could not distinguish between direct and indirect interactions among dFMR1, L5, L11, AGO2, and Dmp68 (but also see below for L5 and L11), nevertheless, these results demonstrate that the association of dFMR1 with L5, L11, AGO2, and Dmp68 occurs both in vivo and in vitro and that RNA molecules do not mediate the association.
Figure 2.
Confirmation of dFMR1 interactions with AGO2, Dmp68, and ribosomal proteins L5 and L11. (A) Cytoplasmic lysate prepared from S2 cells expressing dFMR1-TAP with either AGO2-His, myc-Dmp68, myc-L5, or myc-L11 was incubated with IgG beads and a Western blot performed on the IgG bound fractions using anti-myc and anti-His antibodies. All proteins found as dFMR1-interacting proteins (Fig. 1A) were detected in the bound fractions. RNase A treatment showed no effect on the binding, indicating that the associations occur through protein–protein interactions. The presence of endogenous dFMR1 in the IgG bound fraction was determined by the Western blot using anti-dFMR1 antibody. (B) 35S-labeled AGO2, Dmp68, and ribosomal proteins L5 and L11 were produced by an in vitro transcription and translation system in the presence of [35S]methionine and incubated with either GST-dFMR1 or GST itself immobilized on glutathione-Sepharose resin. After extensive washing, the bound fractions were resolved on a polyacrylamide gel and the proteins labeled with 35S visualized by autoradiography. All proteins tested were detected only in the bound fractions of GST-dFMR1, demonstrating that they interact with dFMR1 in vitro. RNase A treatment showed no effect on the bindings. Coomassie Blue stainings of GST and GST-dFMR1 used in this experiment are shown at left.
dFMR1 associates with ribosomes through interaction with L5 and L11
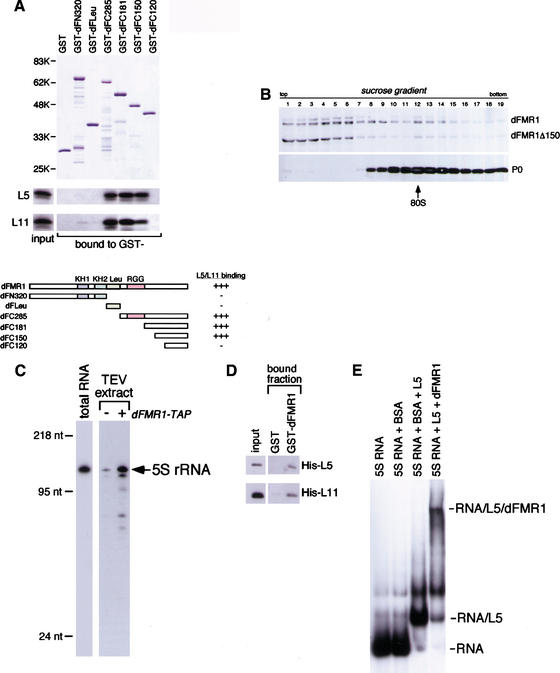
That the large ribosomal subunit proteins, L5 and L11, associate with dFMR1 is consistent with our previous finding that FMRP is associated with the 60S ribosomal subunit (Siomi et al. 1996). To determine the regions in dFMR1 that mediate its interactions with L5 and L11, we made several dFMR1 deletion mutants and performed binding assays. Unexpectedly, GST-dFleu, which contains the region of dFMR1 equivalent to the ribosomal-binding domain in human FMRP, bound neither L5 nor L11 (Fig. 3A). In contrast, dFC285, containing the C-terminal 285 amino acids of dFMR1, did bind both of the proteins (Fig. 3A). A further delineation revealed that dFC150 was capable of interacting with both L5 and L11, but dFC120 was not, demonstrating that dFC150 contains the critical region for the interactions (Fig. 3A). To address the question of whether L5 and/or L11 might act as bridging molecules between dFMR1 and ribosomes, dFMR1 lacking the dFC150 region (dFMR1Δ150) was expressed in S2 cells. Fractionating the cytoplasmic lysate by sedimentation on a linear sucrose gradient revealed that dFMR1Δ150 was less abundant in the monosome/polysome fractions, but accumulated near the top of the gradient, whereas endogenous dFMR1 were detected in the monosome/polysome fractions as expected (Fig. 3B). Although it is conceivable that other proteins within the dFMR1 complex interact with the ribosomal L5 and L11, which interact with the N-terminal part of dFMR1, these results indicate that the dFC150 region in dFMR1, the binding domain to L5 and L11, plays an important role in the association of dFMR1 with ribosomes in vivo.
Figure 3.
Analysis of dFMR1 interaction with the ribosomal proteins L5 and L11. (A) Delineation of dFMR1 to determine the binding domains with L5 and L11. A fragment, dFLeu, containing the region equivalent to the ribosome-binding domain in hFMR1 interacted with neither L5 nor L11. A delineated fragment, dFC150, interacted with 35S-labeled L5 and L11 as in the case of dFC181, whereas dFC120 did not, demonstrating that dFC150 is the L5 and L11 binding domain in dFMR1. (B) The binding domain to L5 and L11 confers to dFMR1 the activity of interacting with ribosomes. A truncated mutant of dFMR1 lacking the binding domains with L5 and L11 (dFMR1Δ150) was expressed in S2 cells and the cytoplasmic lysate was subjected to sedimentation on a linear density sucrose gradient. Western blots were performed on the fractions using anti-dFMR1 antibody. (C) Northern blot on TEV extracts obtained from cytoplasmic lysates with and without dFMR1-TAP. After protease K treatment of the TEV extracts, RNA molecules were recovered and resolved on a 10% denaturing gel containing 6 M urea. A Northern blot was then performed using a riboprobe specific for 5S rRNA. The total RNA lane contains the total RNA isolated from the parental S2 cells. Mass markers are indicated at left. (D) dFMR1 is able to interact directly with L5 and L11 in vitro. GST pull-down assays were carried out using bacterially expressed His-L5 and His-L11, and Western blots with anti-His antibody were performed. (E) The ternary complex formation of 5S rRNA/L5/dFMR1 in vitro. 5S rRNA was labeled with [32P]UTP. When dFMR1 was incubated with preformed L5/5S rRNA, the RNA band was super-shifted. The migration of free 5S rRNA, 5S rRNA plus BSA, or 5S rRNA plus L5 are shown.
dFMR1 interacts directly with L5 bound to 5S RNA
L5 is a 5S rRNA-binding protein. Therefore, we tested if the dFMR1 complex contains 5S rRNA. Northern blot analysis on RNA molecules isolated from the TEV extract revealed that 5S rRNA is a component of the complex (Fig. 3C). We then used gel mobility shift assays to find whether dFMR1 interacts with the L5/5S rRNA complex. The ability of bacterially produced L5 and L11 to interact directly with bacterially expressed recombinant dFMR1 had been ascertained by binding assays (Fig. 3D). In gel-shift assays, addition of dFMR1 to the preformed L5/5S rRNA yielded a new complex containing both L5 and dFMR1, demonstrating that dFMR1 forms a ternary complex with the L5/5S rRNA (Fig. 3E). L5 and L11 are eukaryotic counterparts of bacterial ribosomal proteins L18 and L5, respectively, both of which are shown to locate on the surface of the 50S large ribosomal subunit along with 5S rRNA (Nissen et al. 2000). Therefore, it can be envisioned that the dFMR1 interaction with the ribosome takes place on its surface where L11 and L5/5S rRNA are located.
A conserved DEAD-box helicase required for RNAi
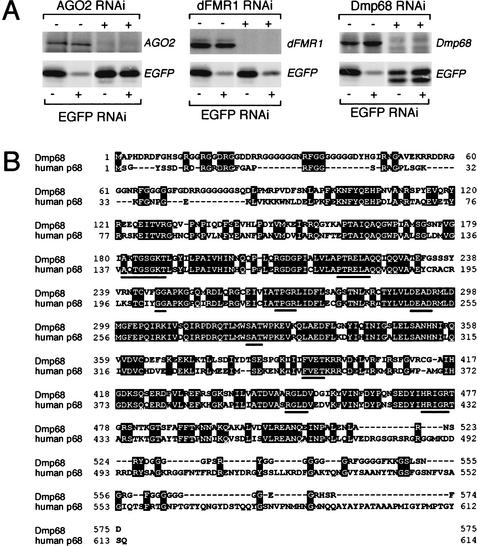
The identification of AGO2 as a dFMR1-interacting protein is particularly striking. AGO2 is a member of the Argonaute gene family and is an essential component for the RNA-induced silencing complex (RISC), a sequence-specific nuclease complex that mediates RNAi in Drosophila (Hammond et al. 2001). Therefore, the finding that dFMR1 forms a complex in vivo with AGO2 suggests that dFMR1 may function in RNAi. To test this, we used RNAi to suppress the endogenous proteins, much as had been done previously to establish a role for AGO2 in RNAi (Hammond et al. 2001). Suppression of ribosomal proteins L5 and L11 with specific dsRNAs made S2 cells so sick that we could not assess their roles in RNAi. However, treatment of S2 cells with dsRNAs homologous to AGO2, dFMR1, or Dmp68 markedly reduced the levels of these proteins (Fig. 4A). We then assessed the ability of these cells to carry out RNAi by transfection with enhanced green fluorescent protein (EGFP) expression plasmid in combination with an EGFP dsRNA. Suppression of AGO2 expression correlated with a pronounced reduction in the ability of cells to silence the reporter EGFP by RNAi as reported previously (Hammond et al. 2001; Fig. 4A). Interestingly, RNAi targeting Dmp68 resulted in inhibition of RNAi in S2 cells. These results suggest that the DEAD-box helicase Dmp68 not only interacts with dFMR1 but is also required for efficient RNAi in S2 cells. Dmp68 is a Drosophila ortholog of human p68 (Fig. 4B), which has recently been demonstrated to unwind short but not long dsRNAs in an ATP-dependent manner (Huang and Liu 2002). We conclude that at least two of the dFMR1-interacting proteins, AGO2 and Dmp68, are required for RNAi in cultured Drosophila S2 cells. In contrast, depletion of dFMR1 did not appear to affect the EGFP silencing (Fig. 4A). Therefore, although dFMR1 interacts strongly with AGO2 and Dmp68 in vivo, dFMR1 does not appear to be essential for efficient RNAi.
Figure 4.
AGO2 and Dmp68, but not dFMR1 alone, are essential for the RNAi pathway. (A) When AGO2 was suppressed by introducing specific dsRNA in S2 cells expressing EGFP, the ability of the cells to silence EGFP by RNAi was profoundly reduced as reported previously (Hammond et al. 2001). In contrast, when dFMR1 expression was suppressed by dFMR1 dsRNA, the EGFP silencing effect was unaffected, indicating that dFMR1 is not essential for the RNAi pathway. Interestingly, when Dmp68 expression was repressed, EGFP silencing by RNAi was completely abolished, indicating that Dmp68 is a novel protein playing an essential role in the RNAi pathway. (B) The amino acid sequence alignment of Dmp68 with human p68. Amino acids identical in Dmp68 and human p68 are indicated with black boxes and residues conserved within DEAD box RNA helicases are underlined.
dFMR1 is associated with an RNAi-related apparatus
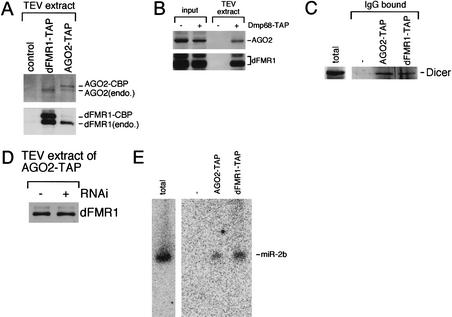
Recent work in numerous organisms has shown that RNAi shares features with a developmental gene regulatory mechanism that involves miRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). These small RNAs (siRNAs and miRNAs) are thought to be incorporated into silencing complexes that mediate mRNA destruction during RNAi and translational control during development, respectively. Therefore, it is suggested that a common processing machinery generates guide RNAs that mediate both RNAi and endogenous gene regulation (Hutvagner and Zamore 2002). AGO2 and Dmp68 are essential for RNAi in Drosophila (Fig. 4A). On the other hand, dFMR1 appears to be a translation repressor (Zhang et al. 2001). Because dFMR1 interacts with AGO2 and Dmp68 in vivo, we wanted to examine whether dFMR1 is also present in an AGO2- and/or Dmp68-associated complex. To do this, we expressed TAP-tagged AGO2 (AGO2-TAP) or Dmp68 (Dmp68-TAP) in S2 cells. Cytoplasmic lysate of the cells expressing AGO2-TAP or Dmp68-TAP was prepared and subjected to TAP purifications. In reciprocal assays, endogenous dFMR1 and AGO2 were found to associate with each other (Fig. 5A). In addition, endogenous AGO2 was copurified with AGO2-TAP. Endogenous dFMR1 and AGO2 were also found to be present in the Dmp68-associated complex (Fig. 5B). Because AGO2 can be coimmunoprecipitated with Dicer (Hammond et al. 2001), which initiates RNAi by processing dsRNA silencing triggers into siRNAs, and also processes miRNA precursors into mature miRNAs (Bernstein et al. 2001; Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001); therefore, we considered the possibility that dFMR1 might also interact physically with Dicer. Indeed, endogenous Dicer can be copurified not only with AGO2-TAP but also with dFMR1-TAP (Fig. 5C). Next, we tested whether the interaction between AGO2 and dFMR1 is changed upon RNAi induction. Extracts prepared from S2 cells expressing AGO2-TAP, either not exposed to dsRNA or exposed to dsRNA targeting the casein kinase II (CKII) β subunit (Saxena et al. 1987), were subjected to TAP purifications. Cells introduced with the CKIIβ dsRNA by soaking showed diminished levels of endogenous CKIIβ protein (data not shown). As shown in Figure 5D, dFMR1 remains associated with AGO2 after RNAi induction. It is well established that siRNAs associate with AGO2 during RNAi in S2 cells (Hammond et al. 2001). Therefore, these results indicate that dFMR1 may be a part of RISC. Finally, analogous to the human AGO2 ortholog (eIF2C2)-associated complex that contains a DEAD-box type RNA helicase and miRNAs (Mourelatos et al. 2002), we wanted to test whether miRNAs are also found in AGO2- and/or dFMR1-associated complexes. RNA molecules copurified with AGO2-TAP or dFMR1-TAP were recovered, dissolved on a 15% polyacrylamide denaturing gel, and subjected to Northern blot analysis. A known miRNA, miR-2b (Lagos-Quintana et al. 2001), in Drosophila S2 cells can be detected both in the AGO2- and dFMR1-associated complexes (Fig. 5E). Together, our data show that dFMR1 is present in a complex with components of RNAi and miRNAs in cultured Drosophila S2 cells.
Figure 5.
dFMR1 is associated with RNAi-related complexes. (A) An AGO2 complex formed in S2 cells contains dFMR1. The TEV extracts prepared from S2 cells expressing AGO2-TAP or dFMR1-TAP, and extracts from the parental cells were subjected to Western blots using anti-AGO2 and anti-dFMR1 antibodies. As expected, endogenous dFMR1 was found in both fractions of AGO2-TAP and dFMR1-TAP, confirming that AGO2 interacts with dFMR1 in vivo. Endogenous AGO2 was also copurified with AGO2-TAP. (B) A Dmp68 complex formed in S2 cells contains both dFMR1 and AGO2. The TEV extracts prepared from S2 cells expressing Dmp68-TAP, and extracts from the parental cells were subjected to Western blots using anti-dFMR1 or AGO2 antibodies. Endogenous dFMR1 and AGO2 were found in the Dmp68-associated complex, confirming that Dmp68 interacts with dFMR1 and AGO2 in vivo. (C) Dicer physically interacts with dFMR1. The TEV extracts prepared from S2 cells expressing AGO2-TAP or dFMR1-TAP, and extracts from the parental cells were subjected to Western blot using anti-Dicer antibodies (a kind gift of S. Hammond and G. Hannon; Bernstein et al. 2001). Endogenous Dicer was found in both fractions of AGO2-TAP and dFMR1-TAP, confirming that Dicer interacts with dFMR1 in vivo. The S2 control lane (indicated as total) contains cytoplasmic lysates of parental S2 cells. (D) dFMR1 remains associated with AGO2 during RNAi. AGO2-TAP TEV extracts prepared from S2 cells treated with and without dCKII β dsRNA were subjected to Western blot using anti-dFMR1 antibodies. The amount of dFMR1 bound to AGO2 was not affected by RNAi treatment, suggesting the possibility that dFMR1 is part of RISC. (E) An miRNA, miR-2b, is associated with dFMR1 and AGO2 in vivo. TAP-purifications were performed using cytoplasmic lysates of S2 cells expressing dFMR1-TAP or AGO2-TAP. RNAs were isolated from the purified complexes and analyzed by Northern blot as described (Lagos-Quintana et al. 2001; Lee and Ambros 2001). The blot was probed for the miR-2b (Lagos-Quintana et al. 2001).
Discussion
dFMR1 is thought to have important roles in the translation of some specific mRNAs such as futsch mRNA (Zhang et al. 2001). Although it is unclear how dFMR1 regulates translation of such mRNAs, our findings may hold some clues. Because dFMR1 interacts with ribosomal L5/5S rRNA and L11, all of which are located at the top of the 60S ribosomal subunit (Nissen et al. 2000), it is likely that this interaction brings dFMR1 to the 60S ribosomal subunit. Therefore, the association of dFMR1 with the 60S ribosomal subunit through direct interactions with ribosomal L5/5S rRNA and L11 may inhibit translation by preventing the assembly of initiation complexes or by giving rise to structural change of the ribosomes, which, in turn, influences a step(s) after translation initiation. Alternatively, because 5S rRNA is the only known RNA species that binds ribosomal proteins, including L5, and forms a 5S RNP before it is incorporated into the ribosomes (Steitz et al. 1988; Szymanski et al. 2000), dFMR1 may interact with the cytoplasmic nonribosome-associated 5S RNP (Steitz et al. 1988), which, in turn, influences the formation of the mature 60S ribosomal subunit. It is interesting to note in this context that only about half of the 5S rRNA molecules in mammalian cells are associated with the 60S ribosomal subunit (Knight and Darnell 1967) and that although 5S rRNA enhances ribosomal activity, it is not absolutely essential for it (Moore 1996; Nissen et al. 2000).
dFMR1 is present in a complex isolated from S2 cells, which also contains AGO2 and Dicer. AGO2 and Dicer are essential components of RNAi (Hammond et al. 2001). The interaction between dFMR1 and AGO2 remains constant before and after RNAi induction (Fig. 5D), suggesting that dFMR1 is part of RISC during RNAi. However, there is no evidence to support the notion that RISC formation is induced by treatment of S2 cells with dsRNA. As one of the functions of the RNAi apparatus is to silence transposons and repetitive sequences residing naturally in the Drosophila genome (Hannon 2002; Hutvagner and Zamore 2002), these cells are therefore likely to be full of pre-formed RISC complexes, irrespective of dsRNA treatment. Therefore, it is possible that dFMR1 is part of the pre-formed RISC complexes and remains to be part of the active RISC after ATP-dependent siRNA unwinding (Nykanen et al. 2001; Hutvagner and Zamore 2002).
The involvement of another dFMR1-interacting protein, Dmp68, in RNAi further suggests the close association of dFMR1 with RNAi. The p68 RNA helicase was first identified by cross-reaction with a monoclonal antibody that was originally raised against SV40 large T antigen two decades ago (Lane and Hoeffler 1980). The helicase plays important roles in cell proliferation and organ maturation (Iggo et al. 1991; Stevenson et al. 1998) and belongs to a large family of highly evolutionarily conserved proteins, the so-called DEAD-box family of putative ATPases and helicases (for review, see de la Cruz et al. 1999). Recent studies have revealed several RNA helicases including mut6 (Wu-Scharf et al. 2000), SDE3 (Dalmay et al. 2001), mut14 (Tijsterman et al. 2002), drh-1 (Tabara et al. 2002), and spindle-E (Kennerdell et al. 2002) required for RNAi and related posttranscriptional gene silencing (PTGS) pathways. Dmp68 is similar to, but clearly not an ortholog of these proteins. Therefore, Dmp68 is a novel component of RNAi. Because ATP-dependent unwinding of the siRNA duplex remodels the RISC to generate an active RISC in the RNAi pathway (Nykanen et al. 2001), Dmp68 may mediate the unwinding process. It is also conceivable that Dmp68 may be involved in downstream events such as target RNA recognition, as an RNA chaperone (Lorsch 2002) or an RNPase (Linder et al. 2001).
The connection that we have established between dFMR1, components of RNAi, miRNAs, and the general translation machinery is of considerable significance because they provide intriguing clues and possible connections to the function of dFMR1 and the pathways with which it may intersect. Recent work in numerous organisms has shown that RNAi shares features with a developmental gene regulatory mechanism that involves miRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). For example, both the foreign dsRNAs that trigger RNAi and the endogenous miRNA precursors that function in development are processed into small RNAs by Dicer (Bernstein et al. 2001; Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). Members of the Argonaute gene family are also involved in both the siRNA and miRNA pathways (Grishok et al. 2001). In C. elegans, Tabara et al. (2002) have shown that Dicer, the dsRNA-binding protein RDE-4, and a conserved DExH-box RNA helicase (DRH-1) are in a complex with RDE-1, an AGO2 ortholog. Furthermore, the human AGO2 ortholog, eIF2C2, is in a complex, the miRNP, that contains the DEAD-box RNA helicase Gem3 (Mourelatos et al. 2002). Therefore, Argonaute proteins appear to be in a complex that contains an RNA helicase(s), Dicer and small guide RNAs, and function in a variety of homology-dependent mechanisms that involve base-pairing between small guide RNAs and target mRNAs. Our findings that dFMR1 interacts with AGO2, Dmp68, Dicer, miRNAs, and the general translation machinery, provide a means to link RNAi enzymes to translational control pathways, and are also consistent with the fact that the RISC nuclease fractionates with ribosomes (Hammond et al. 2000, 2001).
It appears that dFMR1 is important for translational control, possibly mediated by RNAi-related components and miRNAs. Although recent studies have identified a list of mRNAs that are potential FMRP targets (Brown et al. 2001; Darnell et al. 2001), our results further suggest a model in which FMRP may not directly bind its mRNA targets but rather it is targeted to its mRNA substrates as part of RNAi-related apparatus, which are guided by miRNAs. How then might FMRP regulate translation of its mRNA targets? In the case of lin-4, a prototype of miRNAs, its mRNA targets (lin-14 and lin-28) are translationally repressed yet remain associated with polyribosomes (Olsen and Ambros 1999; Seggerson et al. 2002), suggesting a block at a step after translation initiation. FMRP may form an miRNP complex on its mRNA targets and the association of this complex with ribosomal L5/5S rRNA and L11 may inhibit translation at one or more postinitiation steps, including elongation, termination, or the release of functional protein as discussed above. Finally, we propose that fragile X syndrome may be the result of protein synthesis abnormality caused by a defect(s) in an RNAi-related apparatus.
Materials and methods
TAP purification from S2 cells
An expression vector containing genes encoding dFMR1, AGO2, or Dmp68 and a TAP tag (Rigaut et al. 1999) under the control of a metallothionein promoter (pRmHa vector) was transfected into S2 cells, and the expression of dFMR1-TAP, AGO2-TAP, or Dmp68-TAP fusion protein was induced by adding copper ion to the medium (Lafont et al. 1998). After incubation overnight, a cytoplasmic lysate of the cells was prepared in a buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Triton X-100, 2 μg/mL leupeptin, 2 μg/mL pepstatin, and 0.5% aprotinin. dFMR1-TAP, AGO2-TAP, or Dmp68-TAP and the associated materials within the lysate were purified with IgG beads (Amersham Bioscience). After extensive washing, the bound fraction was eluted by treating with TEV protease (Invitrogen). The TEV extract was then incubated with calmodulin-coated beads (Stratagene) and the final extract was obtained by adding EGTA to the beads.
Protein identification by peptide mass fingerprinting
Specific bands shown in Figure 1A were excised from the gel and digested in gel with trypsin. The masses of the tryptic peptides were measured with matrix-assisted laser desorption ionization-reflectron time-of-flight (MALDI-TOF) mass spectrometry (Shimazu Biotech, Japan).
Protein–protein interaction assays
To obtain cDNAs encoding ribosomal proteins L5 and L11, Dmp68 and AGO2, polyA(+) RNAs were purified from S2 cells and RT–PCR carried out using primers specifically designed for each clone. For the expression in S2 cells by transfection, the cDNAs were inserted into the pRmHa3 vector (Lafont et al. 1998). After coexpression of either myc-L5, myc-L11, myc-Dmp68, or AGO2-His with dFMR-TAP in S2 cells, the cytoplasmic lysate was prepared and incubated with IgG beads. After extensive washing, the bound materials were subjected to Western blots using anti-myc, anti-His (Santa Cruz), and anti-dFMR1 antibodies. RNase A treatment of the lysate was carried out for 30 min prior to the IgG bindings. To produce [35S]methionine-labeled proteins by a TnT in vitro transcription and translation kit (Promega), the cDNAs of L5 and L11, Dmp68 and AGO2 were inserted into an expression vector, pET28 (Novagen). The GST pull-down assays were carried out using GST-dFMR1 and GST itself that were bound to glutathione-Sepharose 4B resins (Amersham Bioscience) in a binding buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.5% Triton X-100. After incubation with the TnT products and extensive washing, the bound fractions were separated on a polyacrylamide gel and proteins labeled with 35S were visualized by autoradiography after treatment with Amplify Fluorographic Reagent (Amersham Bioscience). RNase A treatment was carried out by adding the enzyme to the binding mixture. To produce GST fusion proteins, either full-length or truncated dFMR1 cDNA was subcloned into a pGEX-5X expression vector (Amersham Bioscience). The fusion proteins, as well as GST itself, were induced and purified as described by the manufacturer. cDNAs of all the deletion mutants were produced by performing PCR using primers specifically designed for the truncation.
Sucrose gradient centrifugation
Cytoplasmic lysate of the cells expressing dFMR1Δ150 was resolved on a linear sucrose density gradient (5%–30%). The gradient was centrifuged at 4°C in a Beckman MLS-50 rotor at 40,000 rpm for 90 min. Following centrifugation, fractions were collected and subjected to Western blots using anti-dFMR1 and anti-P0 ribosomal protein antibodies (Uchiumi and Kominami 1997).
Northern blot analysis and gel mobility shift assay
TEV extracts plus/minus dFMR1-TAP were treated with proteinase K (0.5 mg/mL; Roche) at 37°C for 20 min, followed by a phenol/chloroform extraction and isopropanol precipitation. Resultant RNA molecules were resolved on a 10% acrylamide/6 M urea denaturing gel and subjected to a Northern blot using a specific riboprobe for 5S rRNA. Gel retardation assay was carried out using 5S rRNA labeled with [32P]UTP and His-tagged L5 and dFMR1 recombinant proteins produced in bacteria. Formation of L5/5S rRNA complex was carried out on ice for 30 min in the presence of 0.5 μg of BSA and 0.5 μg of unlabeled yeast RNA. His-dFMR1 was subsequently added to the L5/5S rRNA complex and further incubation carried out on ice for 30 min. The samples were then separated on a 6% nondenaturing gel and autoradiography was performed to visualize the complexes.
For miRNA Northern blots (Lagos-Quintana et al. 2001; Lee and Ambros 2001), RNA samples were run on 15% acrylamide denaturing urea gels and then transferred to Zeta-Probe GT membranes by electrophoresis. After transfer, they were UV-crosslinked and baked at 80°C for 1 h. Oligonucleotide probes were labeled with T4 polynucleotide kinase and [γ-32P]ATP and hybridized to the membranes at 42°C in 7% SDS, 0.2 M Na2PO4 (pH 7.0) overnight. Membranes were washed at 42°C twice with 2xSSPE, 0.1% SDS. The blots were exposed on BAS-MS 2040 imaging plate and signals were quantified using BAS-2500 (FUJIFILM).
Detection of Dicer in an AGO2 complex or a dFMR1 complex
dFMR1-TAP or GO2-TAP and the associated materials were purified with IgG beads (Amersham Bioscience). After extensive washing, the bound fractions were subjected to Western blots using anti-Dicer antibodies (a kind gift of S. Hammond and G. Hannon; Bernstein et al. 2001).
RNAi
Full-length EGFP cDNA and portions of cDNAs of AGO2, Dmp68, and dFMR1, each about 700 bp in size, were subcloned into pBluescript, and dsRNA were produced by in vitro T7 and T3 transcriptions, followed by annealing in water. dsRNAs of AGO2, Dmp68, or dFMR1 were introduced to S2 cells by soaking (Clemens et al. 2000). After 3 d, the cells were harvested and EGFP dsRNAs were then introduced into half of the cells. Both cell pools treated with and without EGFP dsRNA were cultured for another 4 d and subjected to Western blots using anti-AGO2, anti-dFMR1, MAD1 [anti-human p68 cross-reacting to Dmp68; a gift of F. Fuller-Pace (Department of Molecular and Cellular Pathology, University of Dundee, Ninewells Medical School, Dundee, U.K.)] and anti-EGFP antibodies. Anti-dFMR1 antibody was a gift of G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, PA). We also produced polyclonal anti-dFMR1 antibodies by immunizing mice with bacterially produced dFMR1 fragments. Anti-AGO2 polyclonal antibodies were raised in mice by immunizing with bacterially produced AGO2 fragments. dsRNA of CKII β subunit was produced by in vitro transcription, followed by annealing. CKII β dsRNA was then introduced by soaking into S2 cells that express AGO2-TAP fusion.
Acknowledgments
We thank G. Dreyfuss and L. Wan for dfmr1 cDNA and anti-dFMR1 antibodies; F. Fuller-Pace for anti-human p68 antibody MAD1; S. Hammond and G. Hannon for anti-Dicer antibodies; T. Uchiumi for anti-ribosomal protein P0 antibodies; B. Seraphin for the TAP plasmids; and F. Lafont for pRmHa plasmid. We thank Y. Nakahori and members of the Siomi laboratory for discussions and A. Azuma and Y. Kawamura for technical assistance. This work was supported by grants from the FRAXA Research Foundation; the Japan Society for the Promotion of Science (JSPS); the Ministry of Health, Labor and Welfare of Japan; and the Ministry of Education, Science, Sports, and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL siomi@genome.tokushima-u.ac.jp; FAX 81-88-633-9451.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1022002.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Ford MJ, Anton IA, Lane DP. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988;332:736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Bonini NM. Modeling human neurodegenerative diseases in Drosophila: On a wing and a prayer. Trends Genet. 2000;16:161–167. doi: 10.1016/s0168-9525(99)01939-3. [DOI] [PubMed] [Google Scholar]
- Gould EL, Loesch DZ, Martin MJ, Hagerman RJ, Armstrong SM, Huggins RM. Melatonin profiles and sleep characteristics in boys with fragile X syndrome: A preliminary study. Am J Med Genet. 2000;95:307–315. [PubMed] [Google Scholar]
- Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hagerman RJ. The physical and behavioral phenotype. In: Hagerman RJ, Hagerma PJ, editors. Fragile X syndrome, diagnosis, treatment, and research. Baltimore, MD: John Hopkins University Press; 2002. pp. 3–109. [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. RNAi: Nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Iggo RD, Jamieson DJ, MacNeill SA, Southgate J, McPheat J, Lane DP. p68 RNA helicase: Identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol. 1991;11:1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert G, Feng Y, Nelson DL, Warren ST, Mandel J-L. FMR1 and mutations in fragile X syndrome: Molecular biology, biochemistry, and genetics. In: Wells RD, Warren ST, editors. Genetic instabilities and hereditary neurological diseases. San Diego, CA: Academic Press; 1998. pp. 27–53. [Google Scholar]
- Inoue SB, Siomi MC, Siomi H. Molecular mechanisms of fragile X syndrome. J Med Invest. 2000;47:101–107. [PubMed] [Google Scholar]
- Inoue SB, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, Kobayashi S, Ishida N, Siomi H. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: A quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight E, Jr, Darnell JE. Distribution of 5S RNA in HeLa cells. J Mol Biol. 1967;28:491–502. doi: 10.1016/s0022-2836(67)80099-8. [DOI] [PubMed] [Google Scholar]
- Lafont F, Lecat S, Verkade P, Simons K. Annexin XIIIb associates with lipid microdomains to function in apical delivery. J Cell Biol. 1998;142:1413–1427. doi: 10.1083/jcb.142.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2000;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lane DP, Hoeffler WK. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature. 1980;288:167–170. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2000;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Tanner NK, Banroques J. From RNA helicases to RNPases. Trends Biochem Sci. 2001;26:339–341. doi: 10.1016/s0968-0004(01)01870-9. [DOI] [PubMed] [Google Scholar]
- Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- Miklos GL, Rubin GM. The role of the genome project in determining gene function: Insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- Moore PB. The structure and function of 5S ribosomal RNA. In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA and structure, evolution, processing and function in protein biosynthesis. Boca Raton, FL: CRC Press; 1996. pp. 199–236. [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Patel NH. Developmental evolution: Insights from studies of insect segmentation. Science. 1994;266:581–590. doi: 10.1126/science.7939712. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena A, Padmanabha R, Glover CV. Isolation and sequencing of cDNA clones encoding alpha and beta subunits of Drosophila melanogaster casein kinase II. Mol Cell Biol. 1987;7:3409–3417. doi: 10.1128/mcb.7.10.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel J-L, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interaction among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RJ, Hamilton SJ, MacCallum DE, Hall PA, Fuller-Pace FV. Expression of the “dead box” RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol. 1998;184:351–359. doi: 10.1002/(SICI)1096-9896(199804)184:4<351::AID-PATH1235>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Barciszewski J, Erdmann VA. 5S ribosomal RNA database Y2K. Nucleic Acids Res. 2000;28:166–167. doi: 10.1093/nar/28.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. rde-4 encodes a double-stranded RNA binding protein that interacts in vivo with RDE-1, DCR-1 and a conserved DExH-box helicase to direct RNA interference in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RH. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Kominami R. Binding of mammalian ribosomal protein complex P0.P1.P2 and protein L12 to the GTPase-associated domain of 28 S ribosomal RNA and effect on the accessibility to anti-28 S RNA autoantibody. J Biol Chem. 1997;272:3302–3308. doi: 10.1074/jbc.272.6.3302. [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants: Defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Verheij C, Bakker CE, de Graaff E, Keulemans J, Willemsen R, Verkerk AJ, Galjaard H, Reuser AJ, Hoogeveen AT, Oostra BA. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290:1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]