Abstract
How eukaryotes specify their replication origins is an important unanswered question. Here, we analyze the replicative organization of yeast rDNA, which consists of ∼150 identical repeats, each containing a potential origin. Using DNA combing and single-molecule imaging, we show that functional rDNA origins are clustered and interspersed with large domains where initiation is silenced. This repression is largely mediated by the Sir2p histone-deacetylase. Increased origin firing in sir2Δ mutants leads to the accumulation of circular rDNA species, a major determinant of yeast aging. We conclude that rDNA replication is regulated epigenetically and that Sir2p may promote genome stability and longevity by suppressing replication-dependent rDNA recombination.
Keywords: DNA replication, DNA combing, Sir2, recombination, aging
Eukaryotic chromosomes are replicated from multiple origins, which are activated sequentially throughout the S phase of the cell cycle. In the yeast Saccharomyces cerevisiae, as probably in other eukaryotes, most origins are not used every cell cycle, some of them being totally silent in normal growth conditions (Fangman and Brewer 1991; Pasero and Schwob 2000). A growing body of evidence indicates that chromosomal context is a major determinant of origin activity (Cimbora and Groudine 2001; Heun et al. 2001; Méchali 2001; Gerbi and Bielinsky 2002). However, how chromatin structure modulates origin activity is still poorly understood, and the biological significance of this regulation remains to be established.
The ribosomal DNA cluster of S. cerevisiae, which consists of 100–200 identical copies of a 9.1-kb unit, is an interesting model system to address these questions. Indeed, every unit contains a potential origin of DNA replication (Fig. 1A,B), but only ∼20% of these elements were shown to be active in a single cell cycle (Linskens and Huberman 1988; Fangman and Brewer 1991). Because sequences are identical, epigenetic mechanisms likely control origin activity in the rDNA. However, owing to its repeated nature, the organization of replicons in this locus has been difficult to address using conventional approaches and leads to divergent interpretations (Walmsley et al. 1984; Saffer and Miller 1986).
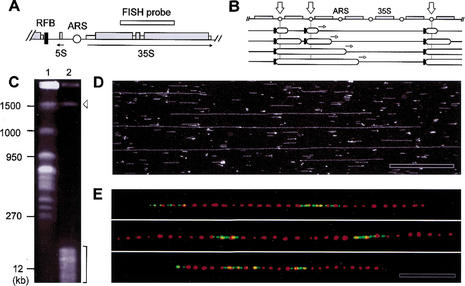
Figure 1.
Mapping of replication sites on individual rDNA fibers by dynamic molecular combing. (A) Map of the 9.1-kb rDNA unit. (ARS) Autonomously replicating sequence; (RFB) replication fork barrier. (B) Replication in the rDNA is monodirectional because of RFBs (black bars) located upstream of active origins. Stalled forks persist until they merge with rightward-moving forks (open arrows; Brewer and Fangman 1988; Linskens and Huberman 1988). (C) Purification of the rDNA cluster by PFGE after fragmentation of the yeast genome with endonucleases. (Lane 1) Intact chromosomes; (lane 2) digested with BamHI and XhoI; (arrowhead) rDNA cluster (1.5 Mb); (bracket) fragmented genomic DNA. (D) Ribosomal DNA fibers combed on silanized coverslips after restriction digestion. DNA was stained with YOYO-1 and visualized by fluorescence microscopy. (E) Representative rDNA fibers labeled in early S phase with BrdU. Individual rRNA genes were detected by FISH with a DIG-labeled 35S rDNA probe (red), and newly replicated regions (green) were visualized by indirect immunofluorescence. Bar, 50 kb. Background signals were removed for clarity (see Supplementary Fig. 1, see Supplementary Material at http://www.genesdev.org, for raw data). The size distribution of BrdU tracks and gaps is shown in Supplementary Figure 1 (see Supplementary Material at http://www.genesdev.org).
Here we use novel technologies based on BrdU incorporation in yeast and DNA combing to visualize replication origins on single rDNA molecules. Their distribution is nonrandom and dependent on the conserved histone deacetylase Sir2p, which is known to silence transcription and extend life span (Defossez et al. 2001; Gasser and Cockell 2001). Because yeast aging is linked to rDNA replication via the formation of extrachromosomal rDNA circles (Guarente 2000), our results suggest that Sir2p may promote genome stability and yeast longevity by modulating the number and distribution of rDNA replication origins.
Results and Discussion
We present here the first quantitative analysis of the distribution of replication origins in the repeated rDNA array of S. cerevisiae. A combination of bromodeoxyuridine (BrdU) incorporation (Lengronne et al. 2001) and the recently developed technique of dynamic molecular combing (DMC; Michalet et al. 1997; Herrick et al. 2000) was used to visualize replication origins on individual rDNA fibers. To this end, thymidine-kinase-expressing (TK+) yeast cells were arrested in G1 with α-factor, released synchronously into S phase in the presence of BrdU, and then chased with deoxythymidine to label only early-replicating sequences (Supplementary Fig. 1, see Supplementary Material at http://www.genesdev.org). Cells were then harvested in G2/M, and chromosomal DNA was purified and combed on silanized coverslips (Michalet et al. 1997). This procedure generated long, parallel DNA fibers, spread with a uniform extension of 2 kb/μm (Michalet et al. 1997). To facilitate the analysis of rDNA fibers, the yeast genome was fragmented prior to combing with restriction enzymes that do not cut within rDNA repeats, releasing intact, ∼1.5-Mb rDNA fragments (Fig. 1C,D). Individual repeats were revealed along combed fibers by hybridization with a 35S probe (Fig. 1E, red signals). The center-to-center distance between FISH probes was measured for 40 molecules encompassing a total of 12.8 Mb. All measured distances (Mean = 9.1 kb, SD = 0.8; Supplementary Fig. 1C, see Supplementary Material at http://www.genesdev.org) fall close to the exact size of rDNA repeats (9.1 kb), thereby confirming the uniform extension of the DNA molecules stretched by DMC.
To visualize newly replicated regions, rDNA fibers were subsequently processed for both FISH and anti-BrdU immunofluorescence. BrdU tracks of 3 rDNA units on average were detected, with large gaps lacking active origins (Fig. 1E; Supplementary Fig. 1D, see Supplementary Material at http://www.genesdev.org). To see whether these tracks reflect the simultaneous activation of contiguous origins, elongation of DNA synthesis was blocked by the addition of hydroxyurea (HU). Under these conditions, forks stall ∼9 kb away from origins in genomic DNA (Fig. 2A). Because replication is monodirectional in the rDNA (Fig. 1B), we anticipated that a single fork would not replicate more than one repeat (9.1 kb) before stalling. However, significantly longer signals (18 ± 14 kb; Fig. 2B) were detected, suggesting that active origins are clustered in the rDNA. Statistical analysis of a large number of molecules (>12 Mb in total) confirmed that initiation sites are not randomly distributed (P < 0.05) but, rather, form clusters of 2–3 adjacent units, a pattern that is reminiscent of the organization of transcriptionally active genes at this locus (Muller et al. 2000). This replication pattern contrasts strikingly with other regions of the genome, where replication initiation sites are more evenly spaced every 46 kb (±16) on average (Lengronne et al. 2001). This is best illustrated in Figure 2D, which shows the junction between rDNA repeats and unique sequences on Chromosome XII. We conclude that rDNA replication, perhaps because of its unidirectional or repeated nature, has requirements that imposed a specific replicative organization.
Figure 2.
Distribution of active origins in the rDNA cluster of HU-arrested cells. Wild-type cells (E1000) were released from α-factor into S phase and exposed for 90 min to 0.2 M HU and 0.4 mg/mL BrdU. BrdU-labeled rDNA fibers were analyzed by DMC, as described in Figure 1. (A) Size distribution of BrdU tracks (b) and gaps (a) in single-copy sequences of the yeast genome. A total of 12.3 Mb of DNA fibers devoid of rDNA signals were analyzed. To allow comparison with rDNA, where replication is monodirectional, the distance covered by only one fork is represented. (B) BrdU tracks (b) and gaps (a) in rDNA fibers. A set of representative fibers is shown in Supplementary Figure 2 (see Supplementary Material at http://www.genesdev.org). (C) Example of long rDNA fiber (830 kb) showing very large gaps (>100 kb). (D) Junction between the rDNA cluster and flanking sequences on Chromosome XII. Here, digestion with BamHI and XhoI was omitted. (Red) FISH probes; (green) BrdU. Bar, 50 kb.
The 35S probe used in the previous experiments only provides the relative position of active origins within the array. To see whether the same replication origins are used from fiber to fiber, a larger FISH probe located immediately upstream of the rDNA cluster on Chromosome XII was used to align rDNA fibers relative to the edge of the array. Ten representative fibers, encompassing ∼200 kb, are shown in Figure 3A. No recurrent pattern could be detected, suggesting that rDNA origin selection is a rather dynamic process that might be newly set up every cell cycle. However, it should be noticed that an epigenetic memory of origin usage persisting only for a few generations would not have been detected in this study, which used relatively large populations (≥106) of cells.
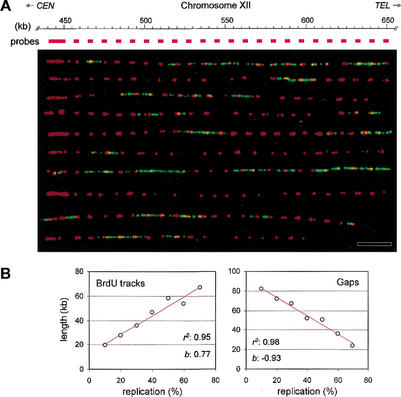
Figure 3.
Spatial and temporal distribution of initiation events in yeast rDNA. (A) Alignment of BrdU-labeled rDNA fibers relative to the edge of the array. Genomic DNA prepared from HU-arrested wild-type cells (E1000) was analyzed by DMC as described in Materials and Methods. A 10-kb PCR fragment, located immediately upstream of the array (pos. 441,034–450,816 bp) was used as a FISH probe. Representative fibers are shown. (Red) FISH probes; (green) BrdU. Bar, 50 kb. (B) Wild-type cells (E1000) were released synchronously into S phase from an α-factor arrest. Samples were collected at regular intervals throughout the S phase, and rDNA replication was analyzed by DMC as described in Figure 2. The mean length of BrdU tracks and gaps was scored for 48 rDNA molecules (average size, 470 kb; total, 22.4 Mb) and was plotted as a function of fiber substitution (10%–80%). The coefficient of regression (r2) and the slope (b) of the regression line (red) are indicated for each set of data.
The most striking feature of replicating rDNA fibers is the presence of large gaps lacking active origins (Fig. 2B,C). In HU-arrested cells, we observed that 28% of these gaps are larger than 60 kb and represent more than 70% (in size) of unreplicated rDNA (Fig. 2B). This pattern differs significantly from that expected if origins were randomly distributed (P < 0.05). It also reflects a unique property of rDNA, as under the same experimental conditions, large gaps represent only 3% of unreplicated DNA in other regions of the genome (Fig. 2A). To check whether large gaps might contain late-firing origins, we recorded how the rDNA replication pattern evolves during a synchronous S phase. To this aim, we measured the lengths of BrdU tracks and gaps and plotted them against the extent of rDNA fiber replication. We reasoned that if rDNA is only replicated from early origins, the mean length of BrdU tracks should increase regularly as S phase proceeds, whereas gap size should decrease proportionally. Indeed, this showed a linear relationship between the extent of replication of rDNA fibers and the length of tracks and gaps (Fig. 3B), indicating that most rDNA origins fire in early S phase. However, we also noticed that the mean gap size shrank slightly faster than the growth of BrdU tracks (slopes: −0.9 and 0.8, respectively), suggesting that a few late origins might get activated from within the large gaps. The fraction of rDNA origins (∼25%) that we found activated in HU-arrested cells also corresponds to the total number of rDNA origins estimated by other methods (Linskens and Huberman 1988; Fangman and Brewer 1991). We can therefore conclude that S. cerevisiae rDNA is mostly replicated from early origins, as recently shown in Schizosaccharomyces pombe (Kim and Huberman 2001).
Because initiation at most rDNA ARS elements appeared to be down-regulated, we next addressed the nature of the mechanism involved in this repression. The silencing protein complex Sir2/3/4p together with Rap1p represses both the transcription of adjacent genes and the initiation of adjacent origins at telomeres (Stevenson and Gottschling 1999; Gasser and Cockell 2001). One of these factors, the histone deacetylase Sir2p, is also found in the nucleolus, where it modulates chromatin structure and represses reporter gene expression (Fritze et al. 1997; Gotta et al. 1997). Because Sir2p is physically bound to the region containing the rDNA ARS (Gotta et al. 1997; Hoppe et al. 2002), we have tested whether it could affect initiation of DNA replication at this locus (Fig. 4A). To this aim, the distribution of active rDNA origins was analyzed in wild-type and sir2Δ cells. This analysis revealed that almost twice as many rDNA origins fired in HU-arrested sir2Δ cells relative to wild-type cells (+80%; Fig. 4A,B). Strikingly, BrdU tracks were not significantly longer in sir2Δ cells, but the proportion of large (>60 kb) unreplicated gaps dropped from 28% in wild-type cells (Fig. 2B) to only 4% in the mutant (Fig. 4B; Supplementary Table 1, see Supplementary Material at http://www.genesdev.org). We can therefore conclude that, in sir2Δ cells, additional origins are activated from within the large gaps normally repressed by Sir2p (Fig. 4C). To confirm the effect of Sir2p on rDNA replication by an independent method, we monitored initiation at rDNA origins in wild-type and sir2Δ cells by 2D gel electrophoresis (Brewer and Fangman 1987). The quantitation of the bubble to Y-arc ratio in seven independent experiments revealed a 62% (±21%) increase of origin activity in the sir2Δ mutant relative to wild-type cells (Fig. 5A,B,E). Moreover, the ectopic expression of SIR2 in sir2Δ cells restored the repression at rDNA origins (Fig. 5B). Taken together, these data indicate that Sir2p affects both the number and the distribution of active origins in the rDNA by maintaining large domains of rDNA in a state refractory for the initiation of DNA replication.
Figure 4.
Sir2p affects both the firing efficiency and distribution of replication origins in the rDNA. Wild-type and sir2Δ hmlΔ cells (E1385) were synchronized in G1 with α-factor and released into S phase in the presence of HU. Ribosomal DNA fibers were analyzed by DMC as described previously. (A) Representative rDNA fibers. (Red) FISH; (green) BrdU. Bar, 50 kb. (B) Size distribution of BrdU tracks (b) and gaps (a) in sir2Δ rDNA fibers. (C) Model of the distribution of active origins in the rDNA of wild-type and sir2Δ cells (see text).
Figure 5.
2D gel analysis of rDNA ARS activity in wild-type and sir2Δ cells. Genomic DNA was prepared from congenic wild-type (E1000) and sir2Δ (E1244) cells, and replication intermediates were examined by 2D gel electrophoresis. (A) Schematic presentation of replication intermediates expected for a StuI–XbaI fragment (3.5 kb) containing the ARS element. (Bubble arc) active origins; (Y-arc) passive replication. (B) The proportion of active origins (arrowhead) increases by 60% in sir2Δ cells, but wild-type levels are restored when SIR2 is ectopically expressed in sir2Δ cells from a galactose-inducible promoter. Recombination intermediates are 20 times less abundant than replication bubbles in wild-type cells (Zou and Rothstein 1997; Ivessa et al. 2000) and are not detectable here. (C) Replication intermediates expected for an XbaI fragment (5.5 kb) centered around the RFB. X-shaped molecules correspond to converging replication forks. (D) Stalled and converged forks (arrowheads) are 43% more abundant in sir2Δ cells than in wild-type cells. (E) Quantitation by phosphor imaging of the relative amount of bubbles (lane 1) and stalled forks (lane 2) in wild-type (light gray) and sir2Δ (dark gray) cells. (F) Extrachromosomal rDNA circles (ERCs) accumulate in sir2Δ cells. Undigested genomic DNA from wild-type and sir2Δ cells was separated on a 0.7% agarose gel, transferred onto a membrane, and probed with rDNA. (Arrowheads) ERCs; (arrow) rDNA array. (G) The orc2-1 mutation reduces the accumulation of ERCs in sir2Δ cells. Genomic DNA from wild-type (E1000), sir2Δ (E1244), fob1Δ (E1410), orc2-1 (E1314), and sir2Δ orc2-1 (E1612) cells was analyzed as described in Supplementary Figure 3 (see Supplementary Material at http://www.genesdev.org), and the fraction of rDNA in circles was quantitated by phosphor imaging.
Sir2p is also known as a key regulator of life span in different species, but it is not clear exactly how Sir2p promotes longevity at the molecular level (Defossez et al. 2001). In yeast, the short life span of sir2Δ mutants is attributable mainly to the accumulation of extrachromosomal rDNA circles (ERCs; Kaeberlein et al. 1999). Because deletion of the FOB1 gene, which is required for fork arrest at RFBs (Kobayashi and Horiuchi 1996; Defossez et al. 1999; Johzuka and Horiuchi 2002), suppresses both the accumulation of ERCs and the premature aging of sir2 mutants, it has been proposed that Sir2p somehow stabilizes stalled forks to prevent DNA breaks and recombination (Kaeberlein et al. 1999). Our finding that Sir2p controls the frequency of origin firing within the rDNA sheds new light on this process. Indeed, given that RFBs are located immediately leftward of every active origin (Fig. 1A), more initiation will inevitably translate into more arrested forks, which are highly prone to recombination (Defossez et al. 1999; Ivessa et al. 2000; Rothstein et al. 2000). Although the causal link between RFB activity and ERCs formation remains to be established (Ward et al. 2000; Johzuka and Horiuchi 2002), it is therefore tempting to speculate that Sir2p acts indirectly to limit the formation of ERCs by reducing the number of active origins, and consequently of stalled forks. To test this model, we monitored the amount of stalled forks and rDNA circles in wild-type and sir2Δ cells. As shown in Figure 5D, the number of stalled and converged forks increased significantly in sir2Δ cells (43% ± 7%). This correlated with a sharp elevation of ERCs in sir2Δ cells (Fig. 5F), as shown earlier (Kaeberlein et al. 1999). To substantiate an eventual causal link between increased origin firing and accumulation of rDNA circles, we then tested whether an unrelated mutation that lowers replication initiation frequency (such as in the origin recognition complex, ORC) might suppress the accumulation of ERCs in sir2Δ cells. Hence, sir2Δ orc2-1 cells were grown at permissive temperature, conditions that reduce origin activity by ∼50% (Liang et al. 1995). Interestingly, this led to a threefold reduction in the amount of ERCs (Fig. 5G), suggesting that the formation of circular rDNA species is coupled to the frequency of origin firing. Therefore, we propose that Sir2p suppresses rDNA recombination and extends yeast life span at least partly by lowering the number of active origins within the rDNA. Furthermore, because its deacetylase activity is regulated by NAD+ levels (Imai et al. 2000; Defossez et al. 2001), an attractive possibility could be that Sir2p modulates rDNA replication and recombination (generating ERCs as a by-product) to expand/contract the size of the rDNA array according to the metabolic activity of cells (Kobayashi et al. 1998).
In most eukaryotes, the rDNA is composed of long tandem repeats that are replicated pseudo-monodirectionally because of the presence of polar RFBs next to the 35S transcription termination site. It is generally believed that RFBs evolved to avoid or limit collision between RNA and DNA polymerases, however at the cost of increased recombination at arrested forks (Hyrien 2000). One way to dampen the latter would be to organize the replication of this locus so as to limit in space or time the number of arrested forks. We report here that replication origins are not distributed evenly in the rDNA, but form clusters of 2–3 consecutive units that are separated by large regions where origins are silenced. This organization differs from the rest of the yeast genome and depends on the histone deacetylase Sir2p. The increased number of active origins and shorter distance between arrested forks in sir2Δ mutants may largely account for some of their phenotypes. Indeed, the link between rDNA replication, recombination, and genomic instability is corroborated by other mutations, such as in the Rrm3p DNA helicase, which also induce replication pausing and rDNA recombination (Ivessa et al. 2000). Furthermore, our results indicate that even in yeast, the choice of replication origins can be dictated by epigenetic mechanisms, which are susceptible to rapid and metastable variations. In this context, it is interesting to note that phenotypic switching in Candida albicans is controlled by an Sir2p ortholog and via events that include rDNA recombination and chromosome rearrangements (Perez-Martin et al. 1999). More practically, we believe that the single-molecule approach described here will be especially valuable to map replication origins in higher eukaryotes, where cell-to-cell variations of origin usage might be common and epigenetic selection the rule rather than the exception.
Materials and methods
Strains and synchronization procedures
Congenic E1000 (MATa, ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 GAL, ura3::URA3/GPD-TK7x), E1244 (MATa, sir2::HIS3, TK+), E1385 (MATa, sir2::HIS3, hml::LEU2, TK+), E1410 (MATa, fob1::URA3, TK+), E1314 (MATa, orc2-1, TK+), and E1612 (MATa, sir2::HIS3, hml:: LEU2, orc2-1, TK+) cells were grown in complete synthetic medium at 25°C (Rose et al. 1990). Cells were synchronized in G1 with 2 μg/mL α-factor (E1000 and E1385) or by centrifugal elutriation (E1244) and were released into S phase in the presence of 0.4 mg/mL BrdU by the addition of 50 μg/mL pronase (Calbiochem). Incubation in 0.2 M hydroxyurea for 90 min after release from the G1 block was used to block elongation (Lengronne et al. 2001). To induce SIR2 in sir2Δ cells, the E1244 strain was transformed with a plasmid bearing SIR2 under the control of a galactose-inducible promoter [gift of M. Cockell (Institut Suisse de Recherche Expérimentale sur le Cancer, Lausanne, Switzerland)] and grown overnight in synthetic medium complemented with 1% raffinose and 0.3% galactose.
DNA combing
Genomic DNA was purified in agarose plugs, stained with YOYO-1 (Molecular Probes), and resuspended in 50 mM MES (pH 5.5) at a concentration of 200 ng/mL in a Teflon reservoir. DNA combing was performed as described previously (Michalet et al. 1997; Lengronne et al. 2001). Silanized coverslips (22 × 22 mm) were dipped into the DNA solution for 5 min and pulled out at a constant speed (0.3 mm/sec). Coverslips were baked at 60°C overnight, mounted on microscope slides, and stored at −20°C until use. We verified that replication intermediates in HU-exposed samples did not interfere with DNA combing by comparing the distribution of rDNA probes along linear and branched rDNA fibers.
FISH and BrdU detection
Coverslips were incubated for 30 min in 1 N NaOH to denature the DNA and neutralized in PBS (pH 7.5). Hybridization was performed at 37°C overnight in 2× SSC, 50% formamide, 10% dextran sulfate, 50 mM sodium phosphate (pH 7.0) with 1 ng/mL of a DIG-labeled 35S probe (3 kb). Coverslips were washed with 2× SSC, 50% formamide, and 2× SSC, and were processed for immunofluorescence. DIG and BrdU epitopes were detected, respectively, with a mouse monoclonal antibody (Roche) and a rat monoclonal antibody (Sera Lab) as described (Lengronne et al. 2001).
Statistical and computer analyses
For each experiment, a total of >12 Mb of rDNA fibers (∼250 kb on average) were analyzed. Images were recorded with a Leica DMRA microscope coupled to a Hamamatsu CCD camera, and signals were measured with MetaMorph tools (Universal Imaging Corp.). Physical distances were converted into base pairs (1 pixel = 340 bp = 170 nm) using λ DNA molecules as size standard. To test whether the distribution of active rDNA origins differed from a random distribution, a set of 2000 virtual rDNA arrays (150 repeats each) was generated with a Visual Basic program. The active/inactive state of replication origins was sequentially drawn for the 150 repeats, using initiation rates derived from experimental observations (Supplementary Table 1, see Supplementary Material at http://www.genesdev.org). For each theoretical array, the occurrence of single origins and origin clusters (e.g., 2, 3, 4 contiguous active origins) was recorded. Observed frequencies were considered as significantly different from random (P < 0.05) when they fell outside of the full range of frequencies (95%) drawn for the 2000 random molecules. For example, in wild-type cells exposed to HU, solitary origins appeared to be significantly less abundant than expected from a random distribution, whereas clusters of 2 to 3 origins were statistically enriched.
Pulsed-field gel electrophoresis, 2D gels, and ERC analysis
PFGE was performed as described (Lengronne et al. 2001). 2D gel analysis was performed as indicated in Brewer and Fangman (1987). To monitor the amount of ERC species, genomic DNA was isolated following a glass-beads procedure (Rose et al. 1990), and samples were separated on a 0.7% agarose gel run at 1 V/cm for 22 h in 1× TAE. Alternatively, genomic DNA was prepared in agarose plugs to limit shearing of chromosomes (Lengronne et al. 2001). Quantification of autoradiograms was performed by storage phosphor imaging (Amersham).
Acknowledgments
We thank S. Gasser, D. Gilbert, and P.A. Defossez for comments and stimulating discussions. Critical reading of the manuscript by R. Hipskind, R. Feil, and members of the Schwob lab is gratefully acknowledged. We are grateful to M. Cockell for plasmids and to P. Ravel for help with statistical analysis. Thanks also go to J. Herrick, C. Conti, A. Lengronne, and M. Marilley for their contribution to this work. P.P. thanks ARC (Association pour la Recherche contre le Cancer). This work was supported by grants from CNRS (Physique et Chimie du Vivant) and ARC to E.S. and from NIH (R21CA81721) to A.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.232902.
References
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- ————— A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Cimbora DM, Groudine M. The control of mammalian DNA replication: A brief history of space and timing. Cell. 2001;104:643–646. [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Defossez PA, Lin SJ, McNabb DS. Sound silencing: The Sir2 protein and cellular senescence. Bioessays. 2001;23:327–332. doi: 10.1002/bies.1047. [DOI] [PubMed] [Google Scholar]
- Fangman WL, Brewer BJ. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Bielinsky AK. DNA replication and chromatin. Curr Opin Genet Dev. 2002;12:243–248. doi: 10.1016/s0959-437x(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Herrick J, Stanislawski P, Hyrien O, Bensimon A. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J Mol Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-M, Huberman JA. Regulation of replication timing in fission yeast. EMBO J. 2001;20:6115–6126. doi: 10.1093/emboj/20.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Linskens MH, Huberman JA. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchali M. DNA replication origins: From sequence specificity to epigenetics. Nat Rev Genet. 2001;2:640–645. doi: 10.1038/35084598. [DOI] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: Stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Muller M, Lucchini R, Sogo JM. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol Cell. 2000;5:767–777. doi: 10.1016/s1097-2765(00)80317-2. [DOI] [PubMed] [Google Scholar]
- Pasero P, Schwob E. Think global, act local—How to regulate S phase from individual replication origins. Curr Opin Genet Dev. 2000;10:178–186. doi: 10.1016/s0959-437x(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J, Uria JA, Johnson AD. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes & Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- Saffer LD, Miller OL., Jr Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol Cell Biol. 1986;6:1148–1157. doi: 10.1128/mcb.6.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley RM, Johnston LH, Williamson DH, Oliver SG. Replicon size of yeast ribosomal DNA. Mol Gen Genet. 1984;195:260–266. doi: 10.1007/BF00332757. [DOI] [PubMed] [Google Scholar]
- Ward TR, Hoang ML, Prusty R, Lau CK, Keil RL, Fangman WL, Brewer BJ. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: Shared sequences but independent activities. Mol Cell Biol. 2000;20:4948–4957. doi: 10.1128/mcb.20.13.4948-4957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]