Abstract
Oligonucleotides containing 5-(N-aminohexyl)carbamoyl-modified uracils have promising features for applications as antigene and antisense therapies. Relative to unmodified DNA, oligonucleotides containing 5-(N-aminohexyl)carbamoyl-2′-deoxyuridine (NU) or 5-(N-aminohexyl)carbamoyl-2′-O-methyluridine (NUm), respectively exhibit increased binding affinity for DNA and RNA, and enhanced nuclease resistance. To understand the structural implications of NU and NUm substitutions, we have determined the X-ray crystal structures of DNA:DNA duplexes containing either NU or NUm and of DNA:RNA hybrid duplexes containing NUm. The aminohexyl chains are fixed in the major groove through hydrogen bonds between the carbamoyl amino groups and the uracil O4 atoms. The terminal ammonium cations on these chains could interact with the phosphate oxygen anions of the residues in the target strands. These interactions partly account for the increased target binding affinity and nuclease resistance. In contrast to NU, NUm decreases DNA binding affinity. This could be explained by the drastic changes in sugar puckering and in the minor groove widths and hydration structures seen in the NUm containing DNA:DNA duplex structure. The conformation of NUm, however, is compatible with the preferred conformation in DNA:RNA hybrid duplexes. Furthermore, the ability of NUm to render the duplexes with altered minor grooves may increase nuclease resistance and elicit RNase H activity.
INTRODUCTION
Chemical modification of nucleic acids is being studied extensively as an approach for the development of nucleic acid-based therapies, such as antigene, antisense and small interfering RNA (siRNA) methods and for applications in biotechnology. In the case of the antigene method, oligonucleotides are aimed at DNA molecules in the single-stranded (1) or double-stranded state (2) to block gene transcription. On the other hand, the goal with antisense oligonucleotides is to form a hybrid duplex with the mRNA, thereby inhibiting gene expression at the level of translation (3). Regardless of the target nucleic acids and the levels at which they inhibit gene expression, it is desirable for these putative oligonucleotide therapies to possess high target binding affinity and specificity, resistance against nuclease degradation and cell permeability. The efficacy of antisense oligonucleotides can be enhanced by activating RNase H to cleave only the RNA strand of the RNA:DNA hybrid duplex.
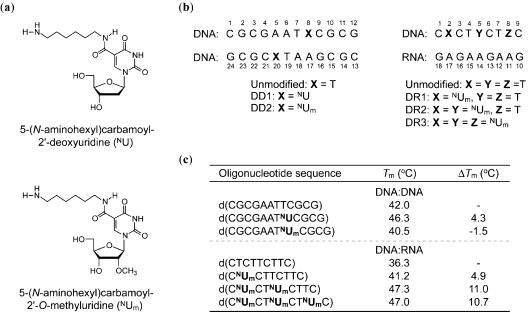
In view of these, a wide variety of oligonucleotides containing modifications at the nucleobase (4–11), the sugar ring or the phosphodiester backbone have been introduced (12). The most common phosphodiester-modified oligonucleotide is phosphorothioate DNA (PS DNA) in which one of the non-bridging phosphate oxygen atoms is replaced with sulfur. PS DNAs are easy to synthesize, and have increased nuclease resistance compared to unmodified DNA oligonucleotides (13). However, there are also some disadvantages of PS DNAs, such as low binding affinity for RNA targets (14) and non-sequence-specific activity (15,16). On the contrary, polyamines are known to have high binding affinity for DNA and to increase duplex (17) and triplex stabilities (18). Thus, we synthesized oligonucleotides carrying various polyamines at the nucleobases (19–23) or at the sugar rings (24–26). Some of them stabilized duplexes and triplexes and enhanced nuclease resistance. In particular, incorporation of 5-(N-aminohexyl)carbamoyl-2′-deoxyuridine (NU; Figure 1a) into DNA strands increased the thermal stability of the corresponding DNA:DNA duplexes by ∼4.3°C (20) (see Figure 1c). Since 2′-modifications are known to increase binding affinity for RNA and to improve nuclease resistance (27), we also synthesized oligonucleotides containing 5-(N-aminohexyl)carbamoyl-2′-O-methyluridine (23) (NUm in Figure 1a). Such oligonucleotides formed stable hybrid duplexes with complementary RNAs, with an increase of ∼3.9°C relative to their unmodified DNA:RNA counterparts. They also showed slightly higher binding affinities for RNA compared to the oligonucleotides containing NU. However, NUm residues decreased binding affinity for DNA (Figure 1c). Interestingly, the oligonucleotides containing NUm further exceed the improved resistance of those containing NU against degradation by both exo- and endo-nucleases. The hybrid duplexes formed between DNA strands containing NUm and RNA could act as substrates for RNase H, on the condition that the NUm residues are separated by at least five continuous unmodified 2′-deoxyribonucleotides.
Figure 1.
Structures of the modified nucleoside analogs (a), sequences and numbering schemes (b), and thermal denaturation of the DNA:DNA and DNA:RNA duplexes (c). The thermal denaturation of the duplexes was performed as described in (23).
In this paper, we report the X-ray crystal structures of B-form DNA:DNA duplexes containing either NU or NUm, and of an A-form DNA:RNA hybrid duplex containing NUm. Based on these duplex structures, we analyzed the correlations between the conformational changes brought about by the NU and NUm residues and the desirable therapeutic properties, such as target binding affinity and nuclease resistance. In addition, we determined the effects of the NU and NUm residues on the minor groove dimensions and hydration structures, since such parameters are expected to affect recognition by nucleases and RNase H.
MATERIALS AND METHODS
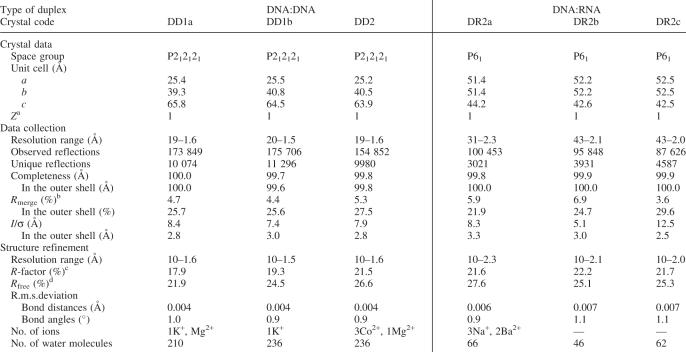
Synthesis, purification and crystallization
The oligonucleotides with sequences shown in Figure 1b were synthesized on a DNA/RNA synthesizer (Applied Biosystem Model 392), as described (23). The reverse-phase high-performance liquid chromatography (HPLC) purified oligonucleotides were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Initial screenings of the crystallization conditions were performed using the hanging drop vapor diffusion method, equilibrating 2 μl droplets against 1 ml of a reservoir solution. The optimized conditions for growing the two DD1 (DD1a and DD1b) and DD2 crystals were as follows. DD1a: 0.4 mM DNA, 20 mM sodium cacodylate (pH 6.0), 40 mM potassium chloride, 2.5 mM magnesium chloride, 1.5 mM spermine tetrahydrochloride and 5% (v/v) 2-methyl-2,4-pentanediol (MPD), equilibrated against 45% (v/v) MPD. DD1b: 0.75 mM DNA, 20 mM sodium cacodylate (pH 6.0), 40 mM potassium chloride, 6 mM spermine tetrahydrochloride and 5% (v/v) MPD, equilibrated against 35% (v/v) MPD. DD2: 0.75 mM DNA, 20 mM sodium cacodylate (pH 5.5), 40 mM sodium chloride, 5 mM magnesium chloride, 5 mM cobalt hexamine and 5% (v/v) MPD, equilibrated against 35% (v/v) MPD. Crystals for DR1 and DR3 were obtained, but they were not suitable for X-ray experiments. Only DR2 was successfully crystallized under three conditions, and the crystals will be referred to as DR2a, DR2b and DR2c, hereafter. The optimized crystallization conditions were as follows. DR2a: 0.5 mM DNA:RNA hybrid, 25 mM sodium cacodylate (pH 7.0), 50 mM sodium chloride, 50 mM barium chloride, 5 mM spermine tetrahydrochloride and 5% (v/v) MPD, equilibrated against 35% (v/v) MPD. DR2b: 0.5 mM DNA:RNA hybrid, 25 mM sodium cacodylate (pH 7.0), 25 mM lithium chloride, 5 mM spermine tetrahydrochloride and 5% (v/v) MPD, equilibrated against 35% (v/v) MPD. DR2c: 0.5 mM DNA:RNA hybrid, 25 mM sodium cacodylate (pH 7.0), 25 mM lithium chloride, 2.5 mM spermine tetrahydrochloride and 7.5% (v/v) MPD, equilibrated against 35% (v/v) MPD.
Data collection
All crystals were picked up from their droplets with a nylon loop (Hampton Research) and transferred into liquid nitrogen (100 K). X-ray data for all three DNA:DNA crystals and for the DR2a and DR2b crystals were taken at the BL18b beamline of Photon Factory in Tsukuba (λ = 1.00 Å). The CCD detector was positioned 100, 150 and 170 mm away from the DNA:DNA, DR2a and DR2b crystals, respectively. Each set of the patterns, using 1° oscillation and a total range of 180°, was processed with the program DPS/MOSFLM (28–31). For the DR2c crystal, the diffraction data were collected on the NW12 beamline of Photon Factory (λ = 1.00 Å). Data for DR2c were recorded on a CCD detector positioned 180 mm away from the crystal. A total of 180 frames were taken with 1° oscillation, and the patterns were also processed with DPS/MOSFLM. All intensity data were scaled and merged, and finally converted into independent structure factors using the programs SCALA and TRUNCATE in the CCP4 suite (32). The crystal data and data collection statistics are listed in Table 1.
Table 1.
Crystal data, statistics of data collection and statistics of structure refinement
aNumber of duplexes in the asymmetric unit.
bRmerge = 100 × Σhklj|Ihklj − <Ihklj>| / Σhklj<Ihklj>.
cR-factor = 100 × Σ‖Fo| − |Fc‖ / Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
dCalculated using a random set containing 10% of observations that were not included throughout refinement (51).
Structure determination and refinement
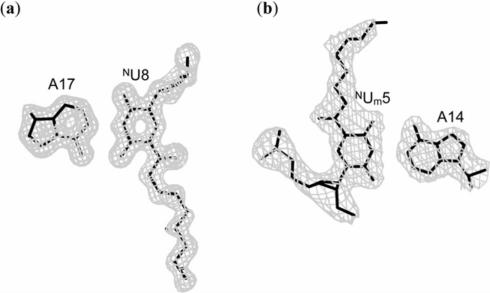
Initial phases were derived by molecular replacement with the program AMoRe (33) using the atomic coordinates of the corresponding unmodified DNA:DNA (34) and DNA:RNA (35) duplexes as probes. The molecular structures were constructed and modified on a graphic workstation with the program QUANTA (Accelrys Inc.). The atomic parameters were refined with the program CNS (36) through a combination of rigid-body, crystallographic conjugate gradient minimization refinement and B-factor refinements, followed by interpretation of an omit map at every nucleotide residue. Newly defined patches for the NU and NUm residues were used. The electron densities of some atoms in the aminohexyl chains of the NU and NUm residues were poor, but their positions derived with appropriate conformations were used in the subsequent refinements. The statistics of structure refinements are summarized in Table 1. An example of the quality of the final electron density map is depicted in Figure 2a. All global and local helical parameters, including torsion angles and pseudorotation phase angles of ribose rings, were calculated using the program 3DNA (37).
Figure 2.
Final 2|Fo| − |Fc| maps contoured at 1σ level for the base pairs: NU8:A17 in DD1b (a) and NUm5:A14 in DR2a (b).
RESULTS AND DISCUSSION
DNA:DNA duplexes
Overall features of the DNA:DNA duplexes and sugar conformations of the NU and NUm residues.
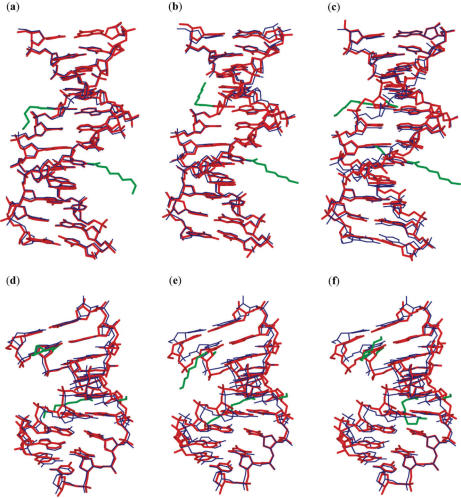
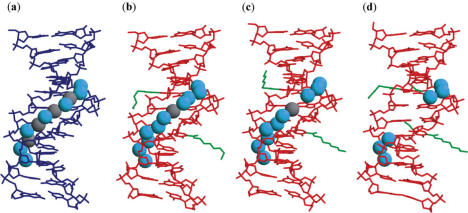
The average local helical parameters for the modified and unmodified DNA:DNA duplexes, as well as for the high-resolution A- and B-form DNA:DNA duplexes (38) are listed in Table 2. These parameters show that all three modified DNA:DNA duplexes adopt the B-form conformation. The modified DNA:DNA crystals are isomorphous to the unmodified crystal with similar unit cell dimensions (34). Least-squares superpositions of the DD1a, DD1b and DD2 structures onto the unmodified DNA:DNA duplex show the r.m.s. differences to be 0.31, 0.46 and 0.81 Å, respectively. Closer inspection of the superposed structures (Figure 3a–c) revealed significant geometric deviations between the backbones of the DD2 and unmodified duplexes. The largest deviations occur at the locations of the NUm residues, attested by the changes in the backbone torsion angles (data not shown).
Table 2.
Average local helical parameters of DNA:DNA and DNA:RNA duplex structures
| Structure | x-disp. (Å) | Inclination (°) | helical twist (°) | helical rise (Å) |
|---|---|---|---|---|
| DNA:DNA | ||||
| DD1a | −0.2 | 2 | 36 | 3.3 |
| DD1b | 0.0 | 1 | 36 | 3.3 |
| DD2 | −0.2 | 1 | 36 | 3.2 |
| Unmodified DNA:DNAa | −0.2 | 3 | 36 | 3.3 |
| DNA:RNA | ||||
| DR2a | −4.0 | 12 | 31 | 2.8 |
| DR2b | −4.0 | 12 | 31 | 2.9 |
| DR2c | −4.0 | 12 | 31 | 2.9 |
| Unmodified DNA:RNAb | −4.0 | 12 | 32 | 3.0 |
| B-DNAc | 0.05 | 2.1 | 36.5 | 3.29 |
| A-DNAc | −4.17 | 14.7 | 32.5 | 2.83 |
Figure 3.
Superimpositions of the DD1a (a), DD1b (b) and DD2 (c) duplexes with the unmodified DNA:DNA duplex structure, and of the DR2a (d), DR2b (e) and DR2c (f) duplexes with the unmodified DNA:RNA duplex structure. The present duplexes are shown in thick red lines while the unmodified duplex is shown in thin blue lines. The aminohexyl, carbamoyl and methoxyl groups are colored green.
In the unmodified DNA:DNA duplexes, the deoxyriboses of the T8 and T20 residues adopt the C1′-exo and the C2′-endo conformations, respectively (see Supplementary Figure S1). The sugar conformations of the NU8 and NU20 residues in DD1a and DD1b are O4′-endo and C1′-exo, respectively, and thus differ only slightly from their unmodified counterparts. On the contrary, the sugar groups of both NUm8 and NUm20 in DD2 adopt the C3′-endo conformation, which is typical in A-form duplexes. Although, some residues in the modified and unmodified DNA:DNA duplexes have the same C3′-endo conformation, the changes at these 8th and 20th positions in DD2 represent the biggest shifts in sugar conformation. Subsequently, such shifts in the sugar conformation affects the geometry of the backbone and could be the reason behind the destabilization of the DNA:DNA duplex as evidenced by the decreased Tm value of DD2 (Figure 1c).
Conformational features and interactions of the aminohexyl chains in the DNA:DNA duplexes
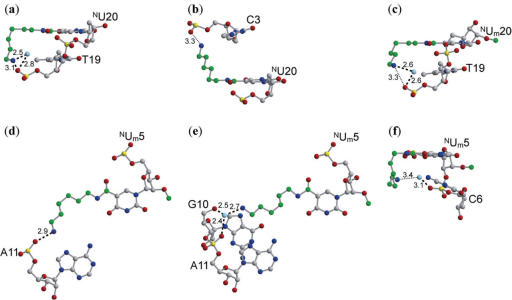
In all the modified residues, the amide group of the carbamoyl modification is associated in the syn conformation relative to the O4 carbonyl atom of the uracil base, possibly forming a planar six-membered ring through an N–H...O hydrogen bond, and allowing the long aminohexyl chain to project towards the major groove. The aminohexyl chain, nevertheless, exhibits conformational flexibility and as such, is capable of interacting with intrastrand and interstrand residues as well as with adjacent duplexes and solvent molecules (Figure 4). For instance, the NU20 and NUm20 residues of DD1a and DD2, respectively, extend towards the 5′-direction, and could interact with neighboring T19 residues on the same strand (Figure 4a and c). In contrast, the NUm20 residue of DD1b protrudes towards the 3′-direction, and could interact with the interstrand C3 residue (Figure 4b). In all these interactions involving the modified residues at the 20th position, the terminal ammonium groups on the aminohexyl chains are associated to the phosphate groups. Thus, the positive charge on the aminohexyl chain could neutralize the negative charge on the phosphate backbone. It is possible that these neutralizations contribute to the stability of DNA:DNA duplex formation (39). There are several reports on synthetic polyamines neutralizing phosphate backbones in DNA:DNA duplexes, but this is the first case in which a polyamine attached to the base group demonstrates such neutralization.
Figure 4.
Intraduplex interactions involving the 20th residues in the DD1a (a), DD1b (b) and DD2 (c) DNA:DNA duplexes, and the 5th residues in the DR2a (d), DR2b (e) and DR2c (f) DNA:RNA duplexes. The carbon atoms in the aminohexyl, carbamoyl and methoxyl modifications are colored green, and the water molecules are colored pale blue. Broken and dotted lines indicate possible hydrogen bonds and van der Waals interactions, respectively. The values indicated are in angstroms (Å).
Minor groove widths and hydration of the DNA:DNA duplexes.
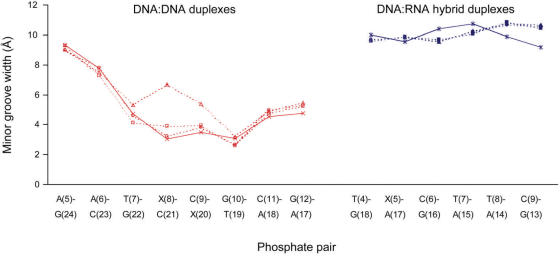
As is characteristically seen for AT tracts in DNA (34), narrowing of the minor groove of the DD1a and DD1b duplexes are observed (Figure 5). The minor grooves of these NU containing DNA:DNA structures are similar to that of the unmodified duplex. On the other hand, the DD2 structure has a minor groove that is wider than that of the unmodified structure, with the widths at the NUm8:C21 and C9: NUm20 phosphate pairs being greater than their counterparts in the unmodified structure by 3.6 and 1.9 Å, respectively.
Figure 5.
The minor groove widths in the DD1a (open square), DD1b (○), DD2 (open triangle) and unmodified (multi) DNA:DNA duplexes, and in the DR2a(filled square), DR2b (filled circle), DR2c (filled triangle) and unmodified (asterisk) DNA:RNA hybrid duplexes. The minor groove width is defined as the distance between the closest interstrand phosphates, diminished by 5.8 Å to account for the van der Waals radii of the phosphate groups (50). X is thymine in the unmodified duplexes and NU or NUm in the modified duplexes.
Hydration structures with cations in the minor grooves are said to stabilize B-form DNA:DNA duplexes. Those that are associated with the AT tracts of the minor grooves of unmodified DNA:DNA duplexes are composed of the primary and the secondary layers (Figure 6a). Similar hydration structures were also found in DD1a and DD1b (Figure 6b and c). In these NU containing duplexes, however, a few solvent molecules are slightly displaced or lacking. In contrast, only 6 of the 11 solvents in the primary and secondary layers were found in DD2 (Figure 6d). The hydration structure is peculiar to AT tracts in DNA because of their unusually narrow minor grooves. Thus, it is possible that the widened minor groove of DD2 hindered the formation of the regular hydration structure.
Figure 6.
Hydration structures in the minor grooves of the unmodified (a), DD1a (b), DD1b (c) and DD2 (d) DNA:DNA duplexes. The unmodified duplex is shown in blue lines while the present duplexes are shown in red lines. The aminohexyl, carbamoyl and methoxyl groups are colored green. In the unmodified duplex, the cyan spheres are water molecules and the gray spheres are solvent molecules partially occupied by sodium ions and water molecules. In DD1a and DD1b, the water molecules are in cyan, and the potassium ions are in gray.
Changes in the minor groove dimension and hydration are also believed to affect nuclease resistance. Cleavage of DNA:DNA duplexes by DNase I, an endonuclease, is presumably influenced by the minor groove width, with the duplexes having wider grooves being more DNase I resistant (40). The excellent nuclease resistance conferred by the NUm can thus be partly ascribed to the ability of the 2′-O-methyl modification to widen the minor groove. Other duplexes containing 2′-modifications were found to contain hydration networks that span the modifications, sugar rings and phosphate backbones (41–43), which may also enhance nuclease resistance. In DD2, a water molecule is hydrogen bonded to the O2′ atom of the methoxyl groups of NUm8 and NUm20 (see Supplementary Figure S2a and b). In the latter residue, the water is hydrogen bonded to another one, and together they link the O2′ methoxyl atoms and the phosphate groups to support the preferred C3′-endo pucker. However, the methyl group is surrounded by four water molecules, which are separated by long distances (3.3–4.0 Å). This finding suggests that the methyl group hinders the entrance and interaction of water molecules in the region between the uracil base and the sugar ring. Thus, the 2′-O-methyl substitution could induce steric hindrance and interfere with nuclease recognition of the phosphate backbone.
DNA:RNA hybrid duplex
Overall structural features of the DNA:RNA hybrid duplex and sugar conformations of the NUm residues
The NUm containing DNA:RNA hybrid duplex adopts the A-form conformation, with all of the average local helical parameters being similar to those for the unmodified and the high-resolution A-form duplex structures (Table 2). The present and the unmodified DNA:RNA (35) crystals belong to the same space group, but have slightly different cell dimensions. Superimpositions of the DR2a, DR2b and DR2c duplex structures onto that of the unmodified duplex structure yield r.m.s.d. of 0.82, 1.0 and 1.1 Å, respectively (Figure 3d–f). To compare the strand conformations based on the locations of the NUm residues, the DNA and RNA strands were separately superimposed onto the unmodified ones. The resulting average r.m.s. differences for individual strands are also small at 0.72 and 0.89 Å, respectively.
Most of the sugars in the DR2a, DR2b and DR2c, including those of the NUm residues, adopt the C3′-endo pucker similar to those of the unmodified hybrid duplexes (see Supplementary Figure S1). This mostly unified A-form character of the sugars in the DNA:RNA hybrid duplexes is similarly observed in the X-ray structures of other DNA:RNA hybrids (44,45). On the contrary, in solution structures of DNA:RNA hybrid duplexes, the sugar conformations of the deoxyribonucleotides have conformations that were intermediate between C2′- and the C3′-endo, while those of the ribonucleotides were C3′-endo (46–48).
Conformational features and interactions of the aminohexyl chains in the DNA:RNA hybrid duplex structures.
Similar to the case in the DNA:DNA duplexes, the carbamoyl modifications of the NUm residues are associated in the syn conformation to the uracil base. Furthermore, flexibility of the aminohexyl chains was observed. All the aminohexyl chains in the NUm5 residues may be involved in interactions with residues in the same DNA:RNA hybrid duplex, with or without water mediation (Figure 4). The terminal ammonium cations of the aminohexyl chains of NUm5 in DR2a and in DR2c could interact with the phosphate oxygen atoms of the A11 (Figure 4d) and the C6 (Figure 4f) residues, respectively. In DR2b, the aminohexyl chain could form interactions not only with the backbone oxygen atom of A11 but also with that of G10 (Figure 4e). Again, these neutralizations of the phosphate negative charges could lead to the stable DNA:RNA hybrid duplex formations, which are demonstrated by the increased Tm values (Figure 1c).
In addition to stabilizing DNA:RNA hybrid duplexes, the terminal ammonium cations on the aminohexyl chains could also render single-stranded DNAs with resistance to exonucleases. A crystallographic study on the complexes between DNA polymerase I Klenow fragment (a 3′ exonuclease) and oligonucleotides containing 2′-O-aminopropyl substituents revealed that the exonuclease resistance of such oligonucleotides is partly brought about by the interference of the positively charged aminopropyl substituent with the metal ion binding required for exonuclease activity (49). The aminohexyl chains carrying a positive charge at their termini can protect oligonucleotides against exonuclease degradation by occupying the sites for binding of the essential metal ions or by interacting with the 3′ phosphate groups, which is exemplified in the DR2c structure.
Minor groove widths and hydration in the DNA:RNA hybrid duplex structures.
Figure 5 shows that in the present DNA:RNA hybrid duplex structures the minor grooves are narrower at the regions where the 2′-O-methyl modifications protrude compared to those in the unmodified hybrids. It has been proposed that RNase H recognizes DNA:RNA hybrids that have minor groove widths that is between that of the A- and B-form duplexes (35,46). Although, the minor groove widths we observed are still not compatible with that required for RNase H recognition, further investigation on the dimensional changes in the minor grooves of NUm containing DNA:RNA hybrid duplexes is necessary. Our previous study revealed that a minimum of five consecutive gap residues between NUms is essential to constitute a substrate of Escherichia coli RNase H. Thus, X-ray structural analyses on DNA:RNA hybrid duplexes that satisfy this gap requirement may shed light into the mechanism of RNase H activation by NUm containing DNA:RNA hybrids.
As in the NUm containing DNA:DNA duplexes, water molecules were found to link the O2′ methoxyl atoms of the NUm5 residues and the phosphate oxygen atoms of the C9 residues in the neighboring duplex in DR2a, DR2b and DR2c (see Supplementary Figure S2). In addition, these waters are also bound to the O3′ atom of NUm. Again, these bound water molecules may help improve nuclease resistance by steric hindrance, but the protruding methyl group could interfere with RNase H activation.
CONCLUSION
In this study, we have determined and analyzed the three crystal structures of NU or NUm containing DNA:DNA duplexes and the three crystal structures of an NUm containing DNA:RNA duplex. These are the first X-ray structures of duplexes containing polyamines attached to nucleotide bases. Our analysis provides insights into the origins of improved target binding affinity, nuclease resistance and ability to elicit RNase H activity, as well as clues for the development of the next-generation antigene and antisense oligonucleotides. Interactions that occur between the aminohexyl chains and the phosphate backbones illustrate the neutralizations of the negative charges on the backbone, and explain the increased affinity of NU or NUm containing oligonucleotides for nucleic acid targets. The conformations of the sugar rings in the modified residues support the assumption that duplex stability is affected by structural homogeneity. The NU residues adopt conformations that are closer to those of unmodified B-form DNA residues, and could thus be incorporated into antigene DNAs without compromising DNA target binding affinity. On the other hand, NUm residues prefer the C3′-endo conformation, which is typical for A-type RNA residues. Therefore, NUms are suitable for incorporation into antisense DNAs. In addition, the widths and the hydration structures in the minor grooves of NUm containing duplexes are altered by the 2′-O-methyl modifications. These alterations on the minor grooves could subsequently lead to changes in the binding modes of nucleases and RNase H. Further studies on the structures of complexes of potential therapies, their targets and these minor groove-binding enzymes could deepen our understanding on the mechanisms of nuclease resistance and RNase H activity.
DATA BANK ACCESSION CODES
The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the ID codes 2DP7, 2DPB, 2DPC, 2DQO, 2DQP and 2DQQ for DD1a, DD1b, DD2, DR2a, DR2b and DR2c, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
The authors are grateful to M. Suzuki, N. Igarashi and A. Nakagawa for help with data collection. This work was supported in part by Grants-in-Aid for Scientific Research (No.12480177 and 14035217) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding to pay the Open Access publication charges for this article.
Conflict of interest statement. None declared.
REFERENCES
- 1.Armitage B.A. Antigene leaps forward through an open door. Nature Chem. Biol. 2005;1:185–186. doi: 10.1038/nchembio0905-185. [DOI] [PubMed] [Google Scholar]
- 2.Hélène C., Thuong N.T., Harel-Bellan A. Control of gene expression by triple helix-forming oligonucleotides. The antigene strategy. Ann. N. Y. Acad. Sci. 1992;660:27–36. doi: 10.1111/j.1749-6632.1992.tb21054.x. [DOI] [PubMed] [Google Scholar]
- 3.Crooke S.T. Antisense strategies. Curr. Mol. Med. 2004;4:465–487. doi: 10.2174/1566524043360375. [DOI] [PubMed] [Google Scholar]
- 4.Chatake T., Ono A., Ueno Y., Matsuda A., Takenaka A. Crystallographic studies on damaged DNAs. I. An N6-methoxyadenine residue forms a Watson–Crick pair with cytosine residue in a B-DNA duplex. J. Mol. Biol. 1999;294:1215–1222. doi: 10.1006/jmbi.1999.3303. [DOI] [PubMed] [Google Scholar]
- 5.Chatake T., Hikima T., Ono A., Ueno Y., Matsuda A., Takenaka A. Crystallographic studies on damaged DNAs. II. N6-methoxyadenine can present two alternate faces for Watson–Crick base-pairing, leading to pyrimidine transition mutagenesis. J. Mol. Biol. 1999;294:1223–1230. doi: 10.1006/jmbi.1999.3304. [DOI] [PubMed] [Google Scholar]
- 6.Chatake T., Sunami T., Ono A., Ueno Y., Matsuda A., Takénaka A. Crystallization and preliminary X-ray analysis of a DNA dodecamer of d(CGCGmo6AATCCGCG) containing 2′-deoxy-N6-methoxyadenosine: change in crystal packing with different humidity. Acta Crystallogr. 1999;D55:873–876. doi: 10.1107/s0907444998014048. [DOI] [PubMed] [Google Scholar]
- 7.Hossain M.T., Chatake T., Hikima T., Tsunoda M., Sunami T., Ueno Y., Matsuda A., Takenaka A. Crystallographic studies on damaged DNAs: III. N4-methoxycytosine can form both Watson–Crick type and wobbled base pairs in a B-form duplex. J. Biochem. 2001;130:9–12. doi: 10.1093/oxfordjournals.jbchem.a002967. [DOI] [PubMed] [Google Scholar]
- 8.Hossain M.T., Sunami T., Tsunoda M., Hikima T., Chatake T., Ueno Y., Matsuda A., Takenaka A. Crystallographic studies on damaged DNAs IV. N4-methoxycytosine shows a second face for Watson–Crick base-pairing, leading to purine transition mutagenesis. Nucleic Acids Res. 2001;29:3949–3954. doi: 10.1093/nar/29.19.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsunoda M., Karino N., Ueno Y., Matsuda A., Takénaka A. Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2′-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers? Acta Crystallogr. 2001;D57:345–348. doi: 10.1107/s0907444900017583. [DOI] [PubMed] [Google Scholar]
- 10.Hossain M.T., Kondo J., Ueno Y., Matsuda A., Takenaka A. X-Ray analysis of d(CGCGAATTXGCG)2 containing a 2′-deoxy-N4-methoxycytosine residue at X: A characteristic pattern of sugar puckers in the crystalline state of the Dickerson-Drew type DNA dodecamers. Biophys. Chem. 2002;95:69–77. doi: 10.1016/s0301-4622(01)00249-6. [DOI] [PubMed] [Google Scholar]
- 11.Tsunoda M., Kondo J., Karino N., Ueno Y., Matsuda A., Takenaka A. Water mediated Dickerson-Drew-type crystal of a DNA dodecamer containing 2′-deoxy-5-formyluridine. Biophys. Chem. 2002;95:227–233. doi: 10.1016/s0301-4622(01)00259-9. [DOI] [PubMed] [Google Scholar]
- 12.Freier S.M., Altmann K.-H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckstein F. Nucleoside phosphorothioates. Annu. Rev. Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 14.Kibler-Herzog L., Zon G., Uznanski B., Whittier G., Wilson W.D. Duplex stabilities of phosphorothioate, methylphosphonate and RNA analogs of two DNA 14mers. Nucleic Acids Res. 1991;19:2979–2986. doi: 10.1093/nar/19.11.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guvakova M.A., Yakubov L.A., Vlodavsky I., Tonkinson J.L., Stein C.A. Phosphorothioate oligonucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J. Biol. Chem. 1995;270:2620–2627. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- 16.Rockwell P., O′Connor W.J., King K., Goldstein N.I., Zhang L.M., Stein C.A. Cell-surface perturbations of the epidermal growth factor and vascular endothelial growth factor receptors by phosphorothioate oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 1997;94:6523–6528. doi: 10.1073/pnas.94.12.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabor H. Protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry. 1962;1:496–501. doi: 10.1021/bi00909a021. [DOI] [PubMed] [Google Scholar]
- 18.Hampel K.J., Crosson P., Lee J.S. Polyamines favor DNA triplex formation at neutral pH. Biochemistry. 1991;30:4455–4459. doi: 10.1021/bi00232a012. [DOI] [PubMed] [Google Scholar]
- 19.Ono A., Haginoya N., Kiyokawa M., Minakawa N., Matsuda A. A novel and convenient post-synthetic modification method for the synthesis of oligodeoxyribonucleotides carrying amino linkers at the 5-position of 2′-deoxyuridine. Bioorg. Med. Chem. Lett. 1994;4:361–366. [Google Scholar]
- 20.Haginoya N., Ono A., Nomura Y., Ueno Y., Matsuda A. Synthesis of oligodeoxyribonucleotides containing 5-(N-aminoalkyl)carbamoyl-2′-deoxyuridines by a new postsynthetic modification method and their thermal stability and nuclease-resistance properties. Bioconjug. Chem. 1997;8:271–280. doi: 10.1021/bc970021r. [DOI] [PubMed] [Google Scholar]
- 21.Ueno Y., Kumagai I., Haginoya N., Matsuda A. Effects of 5-(N-aminohexyl)carbamoyl-2′-deoxyuridine on endonuclease stability and an ability of oligodeoxynucleotide to activate RNase H. Nucleic Acids Res. 1997;25:3777–3782. doi: 10.1093/nar/25.19.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueno Y., Mikawa M., Matsuda A. Synthesis and properties of oligodeoxynucleotides containing 5-[N-[2-[N,N-Bis(2-aminoethyl) amino]ethyl]carbamoyl]-2′-deoxyuridine and 5-[N-[3-[N,N-Bis(3- aminopropyl)amino]propyl]carbamoyl]-2′-deoxyuridine. Bioconjug. Chem. 1998;9:33–39. doi: 10.1021/bc9701508. [DOI] [PubMed] [Google Scholar]
- 23.Ito T., Ueno Y., Komatsu Y., Matsuda A. Synthesis, thermal stability and resistance to enzymatic hydrolysis of the oligonucleotides containing 5-(N-aminohexyl)carbamoyl-2′O-methyluridines. Nucleic Acids Res. 2003;31:2514–2523. doi: 10.1093/nar/gkg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono A., Dan A., Matsuda A. Synthesis of oligonucleotides carrying linker groups at the 1′-position of sugar residues. Bioconjug. Chem. 1993;4:499–508. doi: 10.1021/bc00024a012. [DOI] [PubMed] [Google Scholar]
- 25.Kanazaki M., Ueno Y., Shuto S., Matsuda A. Highly nuclease-resistant phosphodiester-type oligodeoxynucleotides containing 4′α-C-aminoalkylthymidines form thermally stable duplexes with DNA and RNA. A candidate for potent antisense molecules. J. Am. Chem. Soc. 2000;122:2422–2432. [Google Scholar]
- 26.Ueno Y., Karino N., Matsuda A. Synthesis of oligodeoxynucleotides containing 6α-[N-(aminoalkyl)carbamoyloxy]-carbocyclic-thymidines and the thermal stability of the duplexes and their nuclease-resistance properties. Bioconjug. Chem. 2000;11:933–940. doi: 10.1021/bc0000684. [DOI] [PubMed] [Google Scholar]
- 27.Lesnik E.A., Guinosso C.J., Kawasaki A.M., Sasmor H., Zounes M., Cummins L.L., Ecker D.J., Cook P.D., Freier S.M. Oligodeoxynucleosides containing 2′-O-modified adenosine: synthesis and effects on stability of DNA:RNA duplexes. Biochemistry. 1993;32:7832–7838. doi: 10.1021/bi00081a031. [DOI] [PubMed] [Google Scholar]
- 28.Leslie A.G.W. Molecular data processing. In: Moras D., Podjarny A.D., Thierry J.C., editors. Crystallography Computing 5, From Chemistry to Biology. Oxford, UK: Oxford University Press; 1992. pp. 50–61. [Google Scholar]
- 29.Steller I., Bolotovsky R., Rossmann M.G. An algorithm for automatic indexing of oscillation images using Fourier analysis. J. Appl. Crystallogr. 1997;30:1036–1040. [Google Scholar]
- 30.Rossmann M.G., van Beek C.G. Data processing. Acta Crystallogr. D. 1999;55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 31.Powell H.R. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta Crystallogr. D. 1999;55:1690–1695. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- 32.Pflugrath J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 34.Shui X., McFail-Isom L., Hu G.G., Williams L.D. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 35.Xiong Y., Sundaralingam M. Crystal structure of a DNA·RNA hybrid duplex with a polypurine RNA r(gaagaagag) and a complementary polypyrimidine DNA d(CTCTTCTTC) Nucleic Acids Res. 2000;28:2171–2176. doi: 10.1093/nar/28.10.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Lu X.J., Olson W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey S.C., Heinemann U., Lu X.J., Neidle S., Shakked Z., et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda A., Ueno Y., Takenaka A. Thermal stability and nuclease-resistance properties of oligonucleotides having an aminoalkyl side chain at the nucleobase and sugar moieties. In: Schinazi R.F., Liotta D.C., editors. Frontiers in Nucleoside and Nucleic Acids. MA, USA: IHL Press; 2004. pp. 549–576. [Google Scholar]
- 40.Suck D. DNA recognition by DNase I. J. Mol. Recognit. 1994;7:65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- 41.Tereshko V., Portmann S., Tay E.C., Martin P., Natt F., Altmann K.-H., Egli M. Correlating structure and stability of DNA duplexes with incorporated 2′-O-modified RNA analogues. Biochemistry. 1998;37:10626–10634. doi: 10.1021/bi980392a. [DOI] [PubMed] [Google Scholar]
- 42.Teplova M., Minasov G., Tereshko V., Inamati G.B., Cook P.D., Manoharan M., Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nature Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 43.Prhavc M., Prakash T.P., Minasov G., Cook D.P., Egli M., Manoharan M. 2′-O-[2-[2-(N,N-Dimethylamino)ethoxy]ethyl] modified oligonucleotides: symbiosis of charge interaction factors and stereoelectronic effects. Org. Lett. 2003;5:2017–2020. doi: 10.1021/ol0340991. [DOI] [PubMed] [Google Scholar]
- 44.Horton N.C., Finzel B.C. The structure of an RNA/DNA hybrid: a substrate of the ribonuclease activity of HIV-1 reverse transcriptase. J. Mol. Biol. 1996;264:521–523. doi: 10.1006/jmbi.1996.0658. [DOI] [PubMed] [Google Scholar]
- 45.Kopka M.L., Lavelle L., Han G.W., Ng H.-L., Dickerson R.E. An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H. J. Mol. Biol. 2003;334:653–665. doi: 10.1016/j.jmb.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 46.Fedoroff O.Y., Salazar M., Reid B.R. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J. Mol. Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 47.Gyi J.I., Lane A.N., Conn G.L., Brown T. Solution structures of DNA·RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry. 1998;37:73–80. doi: 10.1021/bi9719713. [DOI] [PubMed] [Google Scholar]
- 48.Hantz E., Larue V., Ladam P., Le Moyec L., Gouyette C., Dinh T.H. Solution conformation of an RNA-DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand. Int. J. Biol. Macromol. 2001;28:273–284. doi: 10.1016/s0141-8130(01)00123-4. [DOI] [PubMed] [Google Scholar]
- 49.Teplova M., Wallace S.T., Tereshko V., Minasov G., Symons A.M., Cook P.D., Manoharan M., Egli M. Structural origins of the exonuclease resistance of a zwitterionic RNA. Proc. Natl Acad. Sci. USA. 1999;96:14240–14245. doi: 10.1073/pnas.96.25.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conner B.N., Takano T., Tanaka S., Itakura K., Dickerson R.E. The molecular structure of d(ICpCpGpG), a fragment of right-handed double helical A-DNA. Nature. 1982;295:294–299. doi: 10.1038/295294a0. [DOI] [PubMed] [Google Scholar]
- 51.Brünger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structure. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]