Abstract
This article describes the construction of a set of versatile expression vectors based on the In-Fusion™ cloning enzyme and their use for high-throughput cloning and expression screening. Modifications to commonly used vectors rendering them compatible with In-Fusion™ has produced a ligation-independent cloning system that is (1) insert sequence independent (2) capable of cloning large PCR fragments (3) efficient over a wide (20-fold) insert concentration range and (4) applicable to expression in multiple hosts. The system enables the precise engineering of (His6-) tagged constructs with no undesirable vector or restriction-site-derived amino acids added to the expressed protein. The use of a multiple host-enabled vector allows rapid screening in both E. coli and eukaryotic hosts (HEK293T cells and insect cell hosts, e.g. Sf9 cells). These high-throughput screening activities have prompted the development and validation of automated protocols for transfection of mammalian cells and Ni-NTA protein purification.
INTRODUCTION
The Oxford Protein Production Facility (OPPF) is a structural proteomics facility funded to produce high quality structural data for proteins from a diverse range of host organisms, including viruses, bacterial human pathogens and human proteins associated with the aetiology of human diseases such as cancers. As such, the OPPF target list contains many proteins that may be viewed as problematic to express and crystallize. In order to investigate these proteins either in large numbers or to investigate, in parallel, many multiple domains of smaller numbers of these proteins, a highly efficient cloning and expression screening strategy was required.
The prime characteristics of this process must be:
The ability to clone genes encoding proteins, or domains thereof, in a rapid and reliable parallel or high-throughput (HTP) fashion.
The ability to accurately determine the final constructs without the addition of extraneous/vector or restriction-site-derived amino acids to the expressed protein.
The process must be versatile in terms of insert sequence independence.
The process would preferably be single step, to give rapid and cost-effective vector construction.
The constructs should be suitable for expressing proteins from multiple hosts, i.e. a single vector capable of expression in E. coli, mammalian cell lines (e.g. HEK293T cells) and insect cell lines (e.g. Sf9 cells).
The expressed proteins must be capable of purification in HTP mode, i.e. they must all be fused to a common affinity purification ‘tag’ which may be removed, if desired, by enzymatic digest prior to crystallization.
The process should be amenable to automation.
Two options are available for constructing the expression vectors required for protein production, namely ligation-dependent and ligation-independent cloning. The former uses restriction enzyme digestion in combination with DNA ligation to produce the vectors, whereas the latter utilizes either some form of recombination or the production and annealing of single-stranded overhangs, to avoid the need to restriction digest the input DNA insert. Typically, in both cases the starting DNA is a PCR product corresponding to a complete open reading frame (ORF), or domains thereof, produced from either a genomic or cDNA template. The PCR primers incorporate either restriction enzyme recognition sites or the sequences required for ligation-independent cloning (LIC) reactivity. By using rare cutting restriction enzyme sites, ligation-based cloning has been used effectively for semi-automated high-throughput cloning (1). However, most projects have adopted ligation-independent cloning for the obvious reason that it is independent of the input sequence (2). A number of ligation-independent cloning methods are commercially available but these require either multiple rounds of cloning or substantial preparation of inserts and vector prior to cloning. In addition, these ligation-independent methods, without exception, introduce extra codons into the sequence. These are either predefined, for example the att recombination sites in the 2-step Gateway™ system (InVitrogen, Paisley, UK), or may be composed from only three of the four bases, where the fourth base acts as a ‘lock’ during single-strand production by the 3′ to 5′ processing activity of T4 polymerase, for example ligation-independent cloning [LIC—see (3,4) and also commercially available as the Radiance™ system Novagen, Nottingham, UK].
The heterologous expression of large, multi-domain mammalian or viral proteins in E. coli can be problematic, often producing either very low yields of the target protein or miss-folded protein, targeted to inclusion bodies. These problems highlight the necessity for expression screening in more than one host type (e.g. mammalian or insect cells in addition to E. coli). The current HTP cloning and expression platforms only accommodate this by the use of multiple host-specific vectors.
The limitations of existing systems led us to the development of a ligation-independent cloning method that satisfies our pipeline requirements and cloning specifications by utilizing the unique properties of the commercially available In-Fusion™ enzyme (Clontech–Takara Bio Europe, St. Germain en Laye, France). We have produced a versatile suite of vectors for the expression of proteins or protein domains without, or with minimal, extraneous recombination site, or vector-derived amino acids added to the expressed product. The vectors described here utilize multiple promoter systems such that a single construct may be screened for expression in E. coli, mammalian or insect hosts, thereby avoiding the need to make multiple, host-specific, vectors for each target. In combination with automated liquid handling, we show that the method enables rapid one-step cloning and rapid expression screening of recombinant proteins in E. coli, mammalian cells and insect cells (from baculovirus).
MATERIALS AND METHODS
Vector construction and preparation
The three-promoter vector pTriEx2 (Novagen) was used as the basis for construction of the pOPIN series of expression vectors (Table 1). The In-Fusion™-ready vectors described here include fusion tags for N-His6 plus a 3C cleavage site (5), N-His6-Glutathione-S-Transferase (GST) plus a 3C cleavage site, N-His6-Maltose Binding Protein (MBP) plus a 3C cleavage site, a C-terminal Lys-His6 or a secretion leader sequence in combination with C-terminal LysHis6. All of the N-terminal fusion tags are removable with the use of 3C protease and the histidine residues of the C-terminal tags are removable by Carboxypeptidase A to leave only the C-terminal lysine (6).
Table 1.
Summary of In-Fusion™ site sequences and characteristics of the pOPIN vectors presented in this article
| Vector | Fusion tag | Parent vector/ antibiotic resistance | Promoters/baculoviral recombination sites | Forward primer extension | Reverse primer extension |
|---|---|---|---|---|---|
| pOPINE | C-terminal … KHHHHHH | pTriEx2/ampicillin | T7lacO, CMV enhancer and β-actin promoter, p10 promoter/ lef-2 and 1629 baculo elements. | AGGAGATATACCATG† | GTGATGGTGATGTTT† |
| pOPINF* | N-terminal MAHHHHHHSSGLEVL FQ GP … GP … |
pTriEx2/ampicillin | T7lacO, CMV enhancer and ββ-actin promoter, p10 promoter/ lef-2 and 1629 baculo elements. | AAGTTCTGTTTCAGGGCCCG‡ | ATGGTCTAGAAAGCTTTA‡ |
| pOPING | N-terminal MGILPSPGMPALLSLV SLLSVLLMGCVA ET G … cleavable secretion leader and C-terminal … KHHHHHH ET G … cleavable secretion leader and C-terminal … KHHHHHH |
pTriEx2/ampicillin | (T7lacO-not used), CMV enhancer and β-actin promoter, p10 promoter/lef-2 and 1629 baculo elements. | GCGTAGCTGAAACCGGC | GTGATGGTGATGTTT |
| pOPINJ* | N-terminal MAHHHHHHSSG-GST- LEVLFQ ÞGP … ÞGP … |
pTriEx2/ampicillin | T7lacO, CMV enhancer and β-actin promoter, p10 promoter/lef-2 and 1629 baculo elements. | AAGTTCTGTTTCAGGGCCCG‡ | ATGGTCTAGAAAGCTTTA‡ |
| pOPINM* | N-terminal MAHHHHHHSSG-MBP- LEVLFQ GP … GP … |
pTriEx2 ampicillin | T7lacO, CMV enhancer and β-actin promoter, p10 promoter/lef-2 and 1629 baculo elements. | AAGTTCTGTTTCAGGGCCCG‡ | ATGGTCTAGAAAGCTTTA‡ |
 -represents the point of cleavage by 3C protease or signal peptidase (as appropriate). Vectors marked ‡ use the same primer extensions, enabling the same PCR product to be cloned into all marked vectors. Underlined sequences represent methionine initiation or stop codons (as appropriate) and may be excluded from the gene-specific primers.
-represents the point of cleavage by 3C protease or signal peptidase (as appropriate). Vectors marked ‡ use the same primer extensions, enabling the same PCR product to be cloned into all marked vectors. Underlined sequences represent methionine initiation or stop codons (as appropriate) and may be excluded from the gene-specific primers.
To enable blue/white screening of recombinant clones (blue colonies indicate the presence of non-linearized/non-recombinant parental vector) the lacZ insert from intact pDNR-Dual (Clontech–Takara Bio Europe) was amplified using KOD Hi-Fi polymerase according to the manufacturer's instructions (Novagen) and the following primer pairs: Efwd: 5′-GAGATATACCATGGCACACCATCACCACCATCACAGCAGCGGTACCGTCGACCCGACTG GAAAGCG-3′ versus Erev: 5′-ACTTAGTGATGGTGATGGTGATGTTTAAACTGGTCTAGAAAGCTTGGCGCC-3′ Ffwd: 5′-GAGATATACCATGGCACACCATCACCACCATCACAGCAGCGGTCTGGAAGTTCTGTTTCA GGGTACCGTCGACCCGACTGGAAAGCG-3′ versus Frev: 5′-ACTTAGTGATGGTGATGGTGATGTTTAAACTGGTCTAGAAAGCTTGGCGCC-3′.
PCR products were purified by agarose gel electrophoresis and gel extraction (Geneclean–Bio101, Morgan Irvine, CA, US). Products E and F were extended 3′ by amplification versus MscIrev primer: 5′-ccacaccagccaccaccttctga-3′ with pTriEx2 as template.
The extended products E and F were purified and digested with NcoI before ligation into NcoI/MscI cut pTriEx2. Ligation products were transformed into TAM1 cells (Activ Motif, Rixensart, Belgium) and screened for β-galactosidase activity on LB Agar plates supplemented with 50 µg/ml carbenicillin/0.2% w/v X-Gal and 1 mM IPTG. Colonies expressing β-galactosidase activity were picked, grown overnight in 1.5 ml LB supplemented with the appropriate antibiotic and the resulting plasmids extracted by standard methods.
pOPINE was created by ligation of the NcoI-digested extended product E into NcoI/MscI-cut pTriEx2. pOPINF* was created by ligation of the NcoI-digested extended product F into NcoI/MscI-cut pTriEx2. pOPINF was then created by the deletion of the sequence encoding the C-terminal Lys-His6 tag from pOPINF* by the ligation of a phosphorylated primer duplex into PmeI/MscI-cut pOPINF*: TriEx-CH6fwd: 5′-GTGATTAACCTCAGGTGCAGGCTGCCTATCAGAAGGTGGTGGCTGGTGTGG-3′ TriEx-CH6rev: 5′-CCACACCAGCCACCACCTTCTGATAGGCAGCCTGCACCTGAGGTTAATCAC-3′.
pOPINF was derived in order to increase the efficiency of cloning of N-His6-3C pOPINF* constructs by deletion of the sequence encoding the C-terminal Lys-His6 tag as, in a small number of In-Fusion™ reactions with pOPINF* a ‘fusion’ of the sequences encoding both N- and C-terminal His6 tags was observed (data not shown).
pOPING was generated by amplification of the µ-phosphatase secretion leader sequence from pHLsec (7) using sigpepfwd: 5′-CAAGCTTGCCACCATGGGGATC-3′ and sigpeprev: 5′-CGGGGTACCGGTTTCAGCTACGCAAC-3′ primers. The resulting 113 bp PCR product was gel purified, digested with NcoI and KpnI enzymes and ligated into NcoI and KpnI-cut, and purified, pOPINE (this digest removes the N-His6 site insert from pOPINE). This vector encodes the MGILPSPGMPALLSLVSLLSVLLMGCVA ETG secretion leader sequence (where
ETG secretion leader sequence (where  indicates the cleavage site for the eukaryotic signal peptidase enzyme).
indicates the cleavage site for the eukaryotic signal peptidase enzyme).
pOPINJ was generated by amplification of the N-His GST sequence from pDESTH6N15 (derived from the Gateway™ GST vector, pDEST15, Berrow unpublished data) using the following primers: pOPIN-GST-fwd: 5′ GAATTCCATGGCACATCACCATCACCATCACATGTCCCCT 3′ pOPIN-GST-rev: 5′ CGACGGTACCCTGAAACAGAACTTCCAGACCGCTGCTCAGATCCGATTTTGGAGGATG 3′ The resulting ∼700 bp PCR product was gel purified, digested with NcoI and KpnI, re-purified and ligated into NcoI and KpnI-cut pOPINE to produce pOPINJ. This vector encodes an N-terminal His6-GST-3C cleavable tag: MAHHHHHHSSG-GST-SSGLEVLFQ GP … (where
GP … (where  indicates the cleavage sites for 3C protease).
indicates the cleavage sites for 3C protease).
Similarly pOPINM was generated by amplification of the MBP sequence from pMAL2c (NEB, Hitchin, Hertfordshire, UK) using the following primer pairs MBPinffwd: 5′ ACCATCACAGCAGCGGCATGAAAATCGAAGAAGGTAAACTGG 3′ and MBP SSG 3C rev: 5′ GTCGACGGTACCCTGAAACAGAACTTCCAGACCGCTGCTAGTCTGCGCGTCTTTCAGGGC 3′ The resulting ∼1200 bp PCR product was gel purified and then extended 3′ by PCR using pOPINE as template and LACZ + 3′INF REV: 5′ CTGGTCTAGAAAGCTTGGCGCCATTCGCCATTCAG 3′ as the reverse primer.
The resulting ∼1500 bp PCR product was then gel purified and In-Fused into NcoI and HindIII-cut pOPINE (normal NcoI-KpnI cloning of this fragment was not thought possible due to the presence of an internal NcoI site, although recent checking of the pOPINM sequencing data reveals that the NcoI site has been previously removed c.f. the sequence available at NEB).
This vector encodes an N-terminal His6-MBP-3C cleavable tag: MAHHHHHHSSG-MBP-SSGLEVLFQ GP … (where
GP … (where  indicates the cleavage sites for 3C protease).
indicates the cleavage sites for 3C protease).
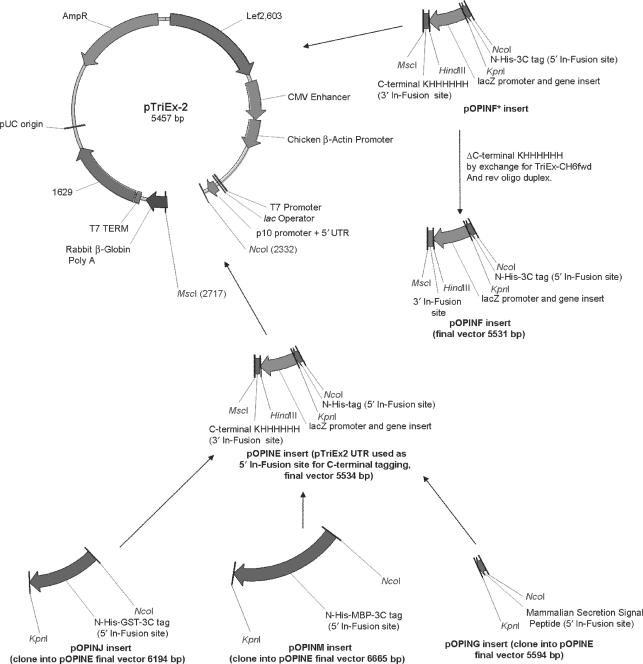
For full details of the fusion tags contributed by these vectors see Table 1, for a summary of the construction of these vectors see Figure 1.
Figure 1.
Vector derivations and maps. Derivation of the pOPIN vectors from pTriEx2. PCR fragments were prepared as described in the Materials and methods section and either ligated into the pTriEx2 vector or inserted by In-Fusion™. In cases where the pOPIN vector is not directly derivatized from pTriEx2, the intermediate vector is also shown. Features of the pTriEx2 vector retained in the pOPIN vector suite are: T7/lacO promoter/operator and terminator for inducible expression in E. coli harbouring the λ (DE3) prophage, CMV Enhancer/Chicken β-actin promoter and rabbit β-globin polyA site for efficient expression in mammalian hosts, p10 baculoviral promoter and 5′ UTR/ORF603 and ORF 1629 for efficient expression from/recombination into baculovirus respectively. The high-copy pUC origin of replication and β-lactamase (Ampicillin resistance marker) gene allow high-copy production of the vector in E. coli.
The pTriEx2 vector contains the hybrid CMV and Chicken β-actin promoter/enhancer combination (CAG) promoter that has been reported to give higher expression levels when compared to those vectors (e.g. pTriEx4) using CMV-derived promoter and enhancer (8). In addition this vector contains a Kozak consensus sequence (9) for efficient initiation of translation in eukaryotic hosts. The presence of the p10 baculoviral promoter and the flanking lef2 (ORF 603) and ORF1629 baculoviral recombination sites allow the construction of recombinant baculoviruses and, finally, a T7 polymerase promoter with lacO operator offers high level inducible expression in E.coli harbouring the λ (DE3) prophage (10).
The integrity of all the vectors was verified by sequencing (MWG Biotech, London, UK) before large-scale plasmid preparations were performed. Prior to their use in In-Fusion™ reactions, pOPINF, pOPINJ and pOPINM vectors were prepared by digestion with KpnI and HindIII, pOPINE by digestion with NcoI and PmeI and pOPING by digestion with KpnI and PmeI. All restriction digests were followed by agarose gel electrophoresis, gel extraction and purification before elution in 10 mM Tris pH 8.0 buffer. Linearized vectors were stored at −20°C in 10 µg aliquots (equivalent to one 96-well plate of In-Fusion™ reactions).
For full details of the fusion tags contributed by the pOPIN vectors see Table 1, for a summary of the construction of these vectors see Figure 1. and the Genbank Accession Numbers for these vectors are as follows … EF372394 (pOPING), EF372395, (pOPINJ), EF372396 (pOPINM), EF372397 (pOPINE), EF372398 (pOPINF).
In-Fusion™ cloning of PCR products
The appropriate primer extensions were used to enable In-Fusion™ cloning into the prepared pOPIN vectors to derive the desired His6-tagged proteins/constructs (see Table 1 for primer extension sequence details). PCR was performed in 50 µl reaction mixes using KOD Hi-Fi polymerase according to the manufacturer's instructions (Novagen) with 30 pmol of each forward and reverse primers, appended with the In-Fusion™ extensions as appropriate, and either 1 µl of the appropriate genomic DNA (50 ng/µl) or 1 µl of plasmid DNA (100 ng/µl) as template per reaction. The resulting PCR products for the Neisseria spp. test set were separated by electrophoresis on a 1.25% w/v agarose Tris/Borate/EDTA gel, visualized with SybrSafe™ (InVitrogen) and purified from the gel using the QIAquick™ 96 kit modified for gel extraction (Qiagen, Crawley, West Sussex, UK). Purified PCR products were eluted from the QIAquick™ 96 plates in 50 µl of Buffer EB, 10 mM Tris pH 8.0 buffer and yields assessed by gel electrophoresis.
All other PCR products were purified using AMPure magnetic beads (Beckman-Coulter, High Wycombe, Buckinghamshire, UK) according to the manufacturer's instructions, eluted in 50 µl of EB buffer (10 mM Tris pH 8.0) and yields assessed by gel electrophoresis.
About 5 µl of purified PCR product (range 10–200 ng in total) and 100 ng of the appropriately linearized pOPIN vector were mixed in the wells of an In-Fusion™ Dry-Down 96-well plate and incubated at 42°C for 30 min. All reactions were diluted 1:5 with T.E. Buffer (10 mM Tris pH 8.0, 1 mM EDTA) and 5 µl used to transform OmniMaxII T1-phage resistant cells (Invitrogen) in 96-tube format. Transformants were selected by plating on 24-well culture plates containing 1 ml of LB Agar/well, supplemented with the appropriate antibiotic/0.02% w/v X-Gal and 1 mM IPTG, and incubation overnight at 37°C. Generally, if cloning is successful, 90–100% of the colonies will be white, although this is entirely dependent on the quality of the initial batch linearization of the vectors as blues represent the undigested parental vector. White colonies (four per vector/insert combination) were used to inoculate 1.5 ml LB supplemented with the appropriate antibiotic in deepwell, 96-well plates. The cultures were grown overnight at 37°C, with shaking at 200 r.p.m., before harvesting by centrifugation at 5000g for 10 min at 4°C, pelleted cells were then used for plasmid preparation. Plasmids specifically for use in E. coli expression trials were prepared from the E. coli cultures in 96-well format using a QIAgen BioRobot 8000 and QIAgen Turboprep kits, according to the manufacturer's instructions (Qiagen). Plasmids used for transfection into either HEK293T or Sf9 cells were prepared under sterile conditions in a Class 2 flow cabinet using Wizard SV96 kits according to the manufacturer's instructions (Promega, Southampton, UK) and a QiaVAC96 manifold (Qiagen). Typically the plasmids prepared with the Wizard SV96 kits had a concentration of 125 ng/µl of DNA and an A260/280 ratio of ≥2.0. The resulting plasmids were screened using the PCR protocol described previously except that the forward primer in each case was replaced by a standard T7 forward primer (5′-TAATACGACTCACTATAGGG-3′), PCR products were analysed by electrophoresis on a 1.25% w/v Agarose Tris/Borate/EDTA gel.
Expression in E. coli
PCR verified expression constructs were transformed into either B834 (DE3) or Rosetta(DE3)LysS E. coli (Novagen) in 96-tube format as described for OmniMaxII transformations with 1% w/v Glucose replacing the X-Gal and IPTG reagents in the LB agar. All plates and subsequent media used for the culture of the Rosetta(DE3) LysS cells are identical to those described for B834(DE3) cells but also supplemented with 35 µg/ml chloramphenicol to maintain the pRareLysS plasmid. Plates were incubated for 18 h at 37°C before individual colonies were used to inoculate, 500 µl GS96 (Bio101, QBioGene, Cambridge, UK) supplemented with 0.05% v/v glycerol, 1% w/v glucose and 50 µg/ml carbenicillin in 96-well deep-well plates. The plates were sealed with gas-permeable adhesive seals and shaken at 225 r.p.m. at 37°C for 18 h. For IPTG induction of expression, 50 µl of each overnight culture was then used to inoculate (in four 24-well deep-well plates) 2.5 ml of GS96 supplemented with 50 µg/ml carbenicillin. The diluted cultures were grown at 37°C with shaking at 225 r.p.m., for 3 h before reducing the temperature to 20°C, addition of ITPG to a final concentration of 0.5 mM and shaking for a further 18 h at 20°C. For auto-induction of expression, 50 µl of each overnight culture was used to inoculate (in four 24-well deep-well plates) 2.5 ml of Overnight Express™ Instant TB media (Novagen) supplemented with 50 µg/ml carbenicillin. The diluted cultures were grown at 37°C with shaking at 225 r.p.m., for 3 h before reducing the temperature to 25°C and shaking for a further 24 h at 25°C.
A 1.5 ml aliquot of culture from each well was then transferred to a 2 ml 96-well deep-well plate using a Theonyx robot (Aviso Gmbh, Gera, Germany) and harvested by centrifugation at 6000 g for 10 min at 4°C before decanting of the waste media. Pelleted cells were frozen at −80°C for at least 30 min prior to screening for soluble His6-tag protein expression using either the Theonyx or BR8000 robotic platforms with standard Qiagen Ni-NTA magnetic bead protocols (as per manufacturer's instructions). Proteins purified by elution from the Ni-NTA beads were analysed on SDS-PAGE gels (Criterion™ 10–20% gradient gels—Biorad, Hemel Hempstead, UK or InVitrogen NuPAGE™ Novex 10% Bis-Tris Midi gels with MES buffer system) and visualized with SafeStain™ (InVitrogen). Scale-up and purification of proteins from E. coli were carried out as described earlier (11).
Expression in HEK 293T cells
HEK 293T cells were maintained in DMEM (Sigma) supplemented with 10% foetal calf serum (Sigma, Poole, UK), 1× non-essential amino acids and 1 mM glutamine (InVitrogen). For expression screening, cells were seeded in 24-well plates, at a density to give 75–80% confluent cells at the time of transfection, and transiently transfected using GeneJuice® (Novagen). The transfection protocol was implemented on a Tecan EVO75 two-probe liquid handling robot controlled by scripts written in-house using the Tecan EVOWare® software. The non-centric pipetting capability of this robot was used throughout in order to minimize disturbance of the cell monolayers during reagent addition and aspiration. The liquid-handling robot was contained in a Class 100 flow cabinet to maintain an aseptic environment for all of the robotic manipulations that were performed at room temperature. In brief, ∼1 µg of individual mini-prepped plasmid DNAs and 2 µl of GeneJuice™ (1.33 mg/ml) were pre-incubated for 30 min in V-bottom well polypropylene micro-titre plates in 60 µl of DMEM (Sigma) supplemented with 1× non-essential amino acids and 1 mM glutamine (InVitrogen). The DNA/GeneJuice® cocktail was then made up to 200 µl with DMEM supplemented with 2% foetal calf serum, 1× non-essential amino acids and 1 mM glutamine before addition to each well of the freshly (robotically) aspirated cells. After 30 min at room temperature, the contents of each well are made up to 1 ml by the addition of 800 µl of DMEM supplemented with 2% foetal calf serum, 1× non-essential amino acids and 1 mM glutamine before the plated cells are (manually) returned to a 37°C/5% CO2/95% air environment. Secretion of protein into the media was analysed by harvesting the culture media 96 h post-transfection, and running samples (10 μl) on SDS-PAGE. Following transfer to PVDF membranes, His-tagged proteins were detected by western blotting using an anti-His6 monoclonal antibody (clone BMG-His-1, Roche Diagnostics) in combination with an anti-Mouse-Horse-Radish Peroxidase secondary antibody (Pierce, Tattenhall, UK) and ECL (GE Healthcare, Chalfont St.Giles, UK). The expression of non-secreted/intracellular proteins was analysed by washing of the adherent cells in PBS (1 ml/well) followed by addition of 200 µl/well of NPI-10 (see standard Qiagen Ni-NTA purification protocol) supplemented with 1% v/v Tween 20 and 1000 KUnits/ml DNAse I (Sigma, Poole Dorset, UK). The plates were shaken (on the Qiagen BioRobot8000) for 15 min at 800 r.p.m., subjected to a single freeze–thaw cycle (to −80°C), followed by shaking for a further 15 min at 800 r.p.m. shake, dilution in SDS-PAGE sample buffer and analysis by western blotting as described for secreted proteins.
Scale-up and purification of proteins from HEK cells was carried out as follows. Cells were grown in roller bottles and transfected with plasmid DNA using polyethylenimine (PEI), as previously described (7,12). Secreted proteins were purified from the culture medium using an automated protocol, written in-house, implemented on an ÄKTA Xpress instrument (GE Healthcare, Uppsala, Sweden), Briefly, batches of media (100 ml) were passed through a 5 ml HisTrap column (GE Healthcare, Uppsala, Sweden) each followed by a 20 ml wash of 50 mM Tris-HCl, 500 mM NaCl, 30 mM imidazole, pH 7.5. This ‘batch loading’ methodology was adopted to prevent components of the eukaryotic cell media displacing the His-tagged protein of interest during the processing of large sample volumes (J.N., unpublisheddata). This sequence was automatically repeated until all the media had been loaded (completion of the loading was detected by the integral air sensor), the column was washed with a further 20 ml of loading buffer, before elution of bound proteins in 50 mM Tris-HCl, 500 mM NaCl, 500 mM imidazole, pH 7.5.The eluted protein peak was automatically collected in a sample loop and re-injected onto a pre-equilibrated HiLoad 16/60 Superdex 200 column (GE Healthcare, Uppsala, Sweden). Protein was subjected to size exclusion chromatography in 20 mM Tris-HCl, 200 mM NaCl, pH 7.5 and fractions collected using a peak detection algorithm. Fractions containing the protein of interest at >95% pure by SDS-PAGE were pooled, quantified (by A280 nm), analysed by mass spectrometry and stored at 4°C prior to use.
Construction of baculoviruses and expression in insect cell lines
Sf9 cells were grown in Sf900 II-SFM media and plated in 24-well plates at a density of 2 × 105 cells per 400 µl per well, and incubated for a minimum of 1 h prior to transfection at 27°C to allow attachment to the plate/substrate. Adherent cells were then co-transfected on a Tecan EVO75 two-probe liquid handling robot controlled by scripts written in-house using the Tecan EVOWare® software. In Brief, 4 µl (300–700 ng) of each pOPIN plasmid preparation (see Cloning section), 4 µl linearized Autographa californica bacmid (12), 4 µl of Insect GeneJuice™ (Novagen) and 13 µl of Sf900 II-SFM media were mixed and then pre-incubated for 30 min in V-well polypropylene micro-titre plates before addition to the adherent cell cultures. Primary viral stocks were manually harvested from the supernatant at 120 h post-transfection and the remaining adherent cells were lysed and analysed by SDS-PAGE and western blotting (as intracellular targets in HEK cells—see previous section).
RESULTS
Vector design
A suite of versatile expression vectors has been constructed to enable the rapid cloning of PCR products using the In-Fusion™ enzyme. The mechanism of this reaction has not been fully reported but relies on the presence of homology between extensions on the PCR product and the ends of a linearized vector; the optimal length for these homologous sequences is around 15 bp. Once these homologous extensions have been incorporated into the PCR product, no further processing of the insert is required prior to the In-Fusion™ reaction (a similarity with the Gateway™ system, but in contrast to PCR-LIC methods). By selection of the appropriate primer tag and linearized vector, a variety of constructs can be made incorporating either N- or C-terminal His6-purification tags for expression in E. coli, mammalian and insect cells from a single cloning operation (see Table 1). One feature of the In-Fusion™ reaction is the removal of any 3′ overhangs generated by linearization of the vectors, for example, the GTAC 3′ overhang created by KpnI-cleavage of the vectors is removed from the vector during the In-Fusion reaction. This allows an efficient restriction enzyme to be used for linearization and the simple re-constitution of the full 3C protease cleavage site (LEVLFQ GP-cleavage site denoted by arrow) by the addition of the last two bases of the glycine codon and the whole proline codon into the appropriate primer extension. This property of the In-Fusion™ enzyme is also exploited to re-constitute the signal peptidase cleavage site in the production of constructs for secreted products (pOPING—see Table 1). To exemplify the utility of the method described in this article, we report the results from three case studies in which five pOPIN vectors have been used for the parallel cloning and expression screening of multiple genes or gene fragments. Automated liquid handling steps have been implemented to produce a scaleable and sustainable process amenable for high-throughput operation. Typically, vector construction, and PCR verification, can be accomplished in three days and subsequent expression screening takes between three and five days depending upon whether E. coli, mammalian or insect cells are used to express the proteins. Thus the whole procedure can be carried out within two working weeks. The target genes used in these case studies are referred to by their construct number in the OPPF target database and full details are given in Supplementary Table 1.
GP-cleavage site denoted by arrow) by the addition of the last two bases of the glycine codon and the whole proline codon into the appropriate primer extension. This property of the In-Fusion™ enzyme is also exploited to re-constitute the signal peptidase cleavage site in the production of constructs for secreted products (pOPING—see Table 1). To exemplify the utility of the method described in this article, we report the results from three case studies in which five pOPIN vectors have been used for the parallel cloning and expression screening of multiple genes or gene fragments. Automated liquid handling steps have been implemented to produce a scaleable and sustainable process amenable for high-throughput operation. Typically, vector construction, and PCR verification, can be accomplished in three days and subsequent expression screening takes between three and five days depending upon whether E. coli, mammalian or insect cells are used to express the proteins. Thus the whole procedure can be carried out within two working weeks. The target genes used in these case studies are referred to by their construct number in the OPPF target database and full details are given in Supplementary Table 1.
Case study I: Expression of Neisseria transcription factors in E. coli
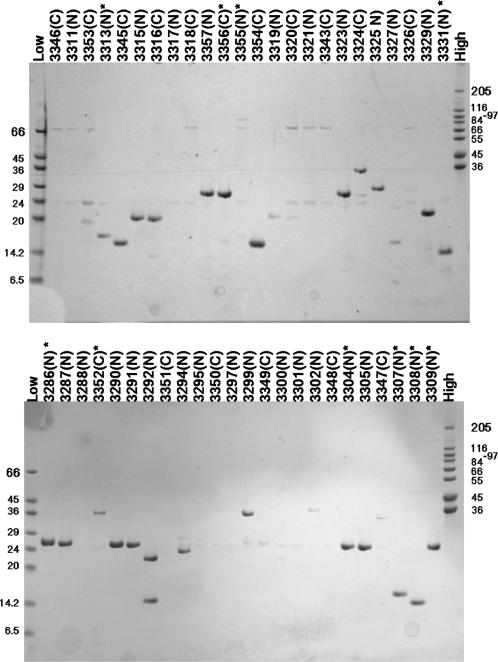
As part of a structural proteomics study of pathogenic Neisseria, twenty transcriptional regulators were selected from the annotated genomes of Neisseria meningitidis MC 58 (13) and Neisseria gonorrhoeae (unpublished data) for cloning and expression screening in E. coli in order to identify constructs suitable for protein production and subsequent crystallization experiments. Primer tags were selected to produce full-length constructs with either N-terminal or C-terminal His6 tags. In addition sub-domain versions of seven of the targets were designed with N-His6 or C- His6 tags to give a total of forty-eight constructs, thirty-one with N-His6 and seventeen with C-His6 (see Supplementary Table 1 for details). Forty-seven of the forty-eight Neisseria spp. gene sequences were successfully amplified from genomic DNA templates and the PCR products purified in a parallelized/HTP format without any significant losses. Purified PCR products were cloned by In-Fusion™ into either pOPINE or pOPINF to add C-His6 or N-His6-3C site tags to the gene sequences respectively. PCR screening of two clones for each construct gave forty-two expression vectors that increased to forty-four when a further two clones were analysed. Therefore, the overall cloning efficiency (in terms of construct coverage) after picking and testing four clones was 92% with no difference observed in the cloning efficiencies of pOPINE compared to pOPINF. The reason that four of the amplified sequences did not clone in this experiment is not clear, though the yield of two of the PCR products was relatively low. Expression screening of PCR verified clones in E. coli was carried out using an automated procedure, with soluble proteins purified at small-scale using Ni-NTA magnetic beads and analysed by SDS-PAGE. The results showed that twenty-seven of the forty-three expression vectors that had been constructed gave soluble protein, though three of these proteins ran at a lower molecular weight than expected on SDS-PAGE (OPPF 3292, 3354, 3323, Figure 2). Of the remaining vectors, no expression was detected in either soluble or insoluble fractions of the induced cultures (data not shown). All the vectors were sequence verified to ensure that the lack of expression was not due to errors in the sequences. Expression of full-length lysine response regulator (LysR) family genes appeared to be problematical, though interestingly, for the cysB gene, expression of a C-His protein was observed whereas no expression of the equivalent N-His construct was detected (Figure 2, OPPF 3352 versus OPPF 3288). This illustrates the value of producing both versions in parallel.
Figure 2.
Expression of Neisseria sp. proteins from either pOPINE (C-His6) or pOPINF (N-His6) in B834(DE3) E. coli. Plasmids were transformed into B834(DE3) cells, expression was induced by the addition of IPTG to the media and the resulting expression levels assayed by a Ni-NTA robotic screen followed by SDS-PAGE. The lane labels refer to unique OPPF identifiers (OPPF construct numbers; see Supplementary Table 1) followed by N or C to indicate the position of the His6 tag in the construct. The lane labelled ‘Low’ contains Low Range Sigma Markers (M3913) and the lane labelled ‘High’ contains High Range Sigma Markers (M3788), molecular marker masses are indicated in kDa. Construct numbers labelled * were selected for scale-up and purification (Table 2).
To assess the yield of protein following purification at largescale, ten constructs were grown to one-litre culture and purified using the semi-automated purification protocol described in the Materials and methods section. In all cases, the yields of purified protein were more than sufficient for both functional and structural studies with high yields obtained for both N-His-tagged and C-His-tagged proteins in the range of 10–90 purified protein mg/l culture (Table 2).
Table 2.
Yield of Neisseria proteins expressed in E. coli using pOPINE and pOPINF vectors
| OPPF No | Description | Construct | N or C terminal His tag | Molecular weight of protein including His tag (D) | Yield (mg/l) |
|---|---|---|---|---|---|
| 3286 | >NMB0381_1 cysB | 89–317aa | N | 27390 | 63 |
| 3352 | >NMB0381_1 cysB | Full length | C | 36224 | 10 |
| 3304 | >NMB1049_1 transcriptional regulator | 86–305aa | N | 26163 | 90 |
| 3307 | >NMB0595_1 phoP | 1–124aa | N | 15718 | 37 |
| 3308 | >NMB0595_1 phoP | 127–225aa | N | 13118 | 26 |
| 3309 | >NMB0810_1 TetR family transcriptional regulator | Full length | N | 26972 | 29 |
| 3313 | >NMC1118_1 transcriptional regulator | Full length | N | 17329 | 60 |
| 3356 | >NMA1884_1 transcriptional regulator | Full length | C | 27557 | 68 |
| 3355 | >NMB0075 | Full length | N | 85301 | 17 |
| 3331 | >NMB1891 | Full length | N | 13374 | 16 |
Case study II: Expression of human gene extra-cellular constructs as secreted proteins in HEK 293T cells
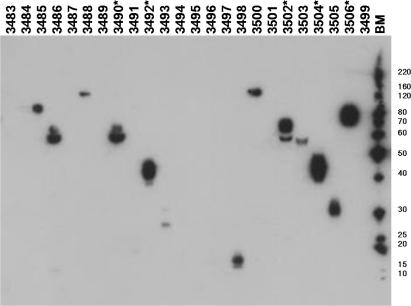
Multiple constructs of the human and mouse gene, KIAA0319, a cell surface glycoprotein recently associated with developmental dyslexia (14) were designed on the basis of bioinformatics analyses using the OPAL resource of bioinformatics tools developed in-house (15). The sequences were annotated with information on potential regions of high disorder (16), signal peptides and transmembrane domains/helices and twelve constructs were designed to cover the majority of the gene sequences, including domains and combinations thereof (see supplementary Table 1 for details). These were then amplified from both the human and mouse templates to give a total of twenty-four PCR products, which were subsequently cloned by In-Fusion™ into the pOPING vector which contains a resident secretion signal sequence. PCR screening of two clones for each construct gave twenty-one expression vectors, which again was improved when a further two clones were analysed, to give twenty-three constructs cloned (96%). In this experiment, the In-Fusion™ reaction was capable of cloning PCR products over a wide size range (450–2800 bp products). One PCR product that failed to clone was obtained in relatively low yield that may explain the lack of success with this construct. The pOPING plasmid mini-preparations were used directly in the automated HEK293T transfection protocol prior to detection by western blotting. Expression and secretion into the medium of thirteen of the twenty-four constructs was observed by western blotting though the levels of expression appeared to vary between constructs (Figure 3). To assess how immuno-reactivity in the western blot screen related to the yield of protein on scale-up, transient expression of a subset of these constructs was carried out at one-litre culture scale. These constructs were selected on the basis that they gave the strongest signals in the western blot screen (Figure 3). Following purification, the yield of protein from these constructs was measured by absorbance at 280 nm using extinction coefficients calculated for each sequence. Four of the five constructs gave yields of purified protein ≥0.5 mg/l culture, whereas one (OPPF 3502) yielded 0.2 mg/l. (Table 3). With the exception of OPPF 3502, these yields would be adequate for functional studies and with the use of sub-microlitre crystallization trials suitable for initial screening (17). Therefore, screening at small-scale has successfully identified constructs that would be suitable for further investigation.
Figure 3.
Expression of KIAA0319 proteins in HEK 293T cells using the pOPING vector. Domain and multi-domain constructs were screened for expression in HEK293T cells and subsequent secretion into the cell media analysed using SDS-PAGE and western blotting as described in the Materials and methods section. The lane labels refer to unique OPPF identifiers (OPPF contruct numbers: see Supplementary Table 1). The lane labelled BM contains the BenchMark™ ladder (InVitrogen 10747-012), molecular marker masses are indicated in kDa. Construct numbers labelled * were selected for scale-up and purification (Table 3).
Table 3.
Yield of KIAA0319 proteins expressed in HEK293T cells using the pOPING vector
| OPPF No | Description | Construct | N or C terminal His tag | Molecular weight of protein including His tag (D) | Yield (mg/l) |
|---|---|---|---|---|---|
| 3490 | Human KIAA0319 | 382–813aa | C | 47566 | 0.5 |
| 3492 | Human KIAA0319 | 382–623aa | C | 27197 | 1.5 |
| 3502 | Mouse KIAA0319 | 391–822aa | C | 47227 | 0.2 |
| 3504 | Mouse KIAA0319 | 391–632aa | C | 27065 | 1.0 |
| 3506 | Mouse KIAA0319 | 21–438aa | C | 46402 | 0.7 |
Case study III: Comparative expression of viral and human gene domains as fusion proteins in E. coli, HEK 293T and Sf9 cells
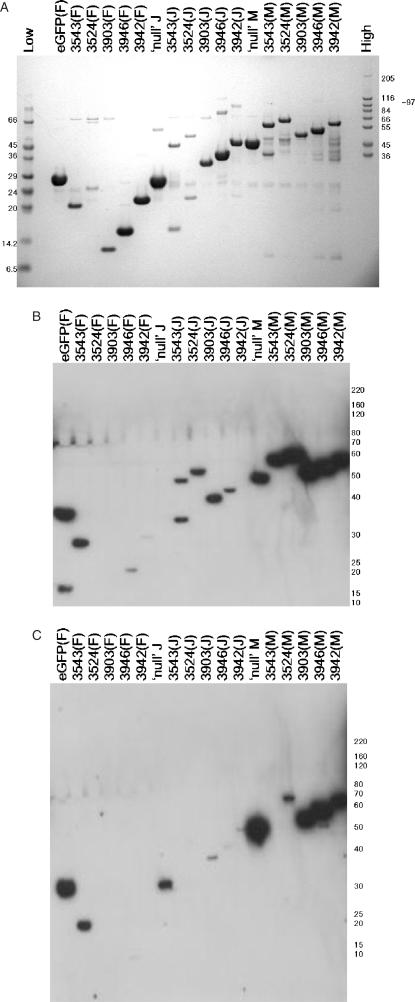
Five proteins (or domains thereof—see Supplementary Table 1 for details) were selected for expression trials in all three of the expression systems available and also as three different fusions, namely N-His-3C, N-His-GST (Glutathione-S-Transferase)-3C and N-His-MBP (Maltose Binding Protein)-3C) from pOPINF, pOPINJ and pOPINM, respectively. These fusion partners have been used extensively by structural genomics groups in attempts to render proteins ‘soluble’ in E. coli (see review (2)). A single batch of PCR product for each target was produced with the appropriate In-Fusion® extensions for cloning into pOPINF. The products were purified and cloned into pOPINF, pOPINJ and pOPINM (as described in the ‘Cloning’ section) and the resulting, PCR and sequence-verified, clones for each construct were then expressed in the three available hosts. For reference, ‘null’pOPINJ, ‘null’pOPINM vectors (i.e. vector with no target genes inserted) and eGFP in pOPINF were included in these screens. Protein expression in E. coli was evaluated using the Ni-NTA protocol, expression in HEK293T and Sf9 cells was evaluated in total protein samples by western blotting. The results demonstrate that, overall, E. coli gave the best coverage for expression of the five constructs in all three fusion formats (Figure 4 summarized in Table 4). The MBP fusions gave the strongest signals in the western blot screen for expression of the targets in both HEK293 and Sf9 cell, whereas the N-His versions performed the poorest. Further studies would be required to assess the solubility and characteristics of these proteins after cleavage from these large ‘solubilizing’ fusion partners.
Figure 4.
(A) Expression of viral and human protein/protein domains in Rosetta(DE3)LysS E. coli. Plasmids were transformed into Rosetta(DE3)pLysS cells, expression was auto-induced in TB Overnight Express™ and the resulting expression levels assayed by a Ni-NTA robotic screen, followed by SDS-PAGE. The lane labels refer to unique OPPF identifiers (OPPF construct numbers; see Supplementary Table 2) followed by the pOPIN vector name (pOPINF contributes an N-His-3C site fusion to the protein of interest, pOPINJ contributes an N-His-GST-3C site fusion to the protein of interest and pOPINM contributes an N-His-MBP-3C site fusion to the protein of interest). eGFP refers to enhanced GFP and ‘null’ refers to vector alone (i.e. vector with no target genes inserted), pOPINJ and pOPINM will however express N-His-GST (29.5 kDa) and N-His-MBP (44.3 kDa), respectively. The lane labelled ‘Low’ contains Low Range Sigma Markers (M3913) and the lane labelled ‘High’ contains High Range Sigma Markers (M3788), molecular marker masses are indicated in kDa. (B) Expression of viral and human protein/protein domains in HEK293T cells. Plasmids were robotically transfected into HEK293T cells and the resulting expression levels assayed by a SDS-PAGE and western blotting as described in the Materials and methods section. The lane labels are identical to panel A except that the standard molecular weight markers are replaced with marked positions of the BenchMark™ ladder (InVitrogen 10747-012), His-tagged molecular mass markers (masses in kDa) on the right. (C) Expression of viral and human protein/protein domains in Sf9 cells. Plasmids were robotically co-transfected into HEK293T cells with linearized A. californica bacmid and the resulting expression levels assayed by a SDS-PAGE and western blotting as described in the Materials and methods section. The lane labels are identical to panel A except that the standard molecular weight markers are replaced with marked positions of the BenchMark™ ladder (InVitrogen 10747-012), His-tagged molecular mass markers (masses in kDa) on the right.
Table 4.
Expression of proteins in multiple hosts from pOPINF, pOPINJ and pOPINM vectors
| OPPF number | Description of target gene | Expression as N-His6-3C fusion (from pOPINF) | Expression as N-His6-GST-3C fusion (pOPINJ) | Expression as N-His6-MBP-3C fusion (pOPINM) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expression host | E.coli | HEK293 | Sf9 | E.coli | HEK293 | Sf9 | E.coli | HEK293 | Sf9 | |
| 3524 | Lagos bat virus species ‘M’ protein (aa 1–202) | ✓ | ✓ | ✓ | ✓ | ✓ | X | ✓ | ✓ | X |
| 3543 | Lagos bat virus species ‘M’ protein (aa 111–202) | ✓ (weak) | ✓ | X | ✓ (weak) | ✓ | X | ✓ | ✓ | ✓ |
| 3903 | Human Fyn-SH3 Domain (aa 82–149) | ✓ | ✓ | X | ✓ | ✓ | ✓(weak) | ✓ | ✓ | ✓ |
| 3946 | Mouse Fyn-SH3-SH2 Domain (aa 82–248) | ✓ | ✓(weak) | X | ✓ | ✓ | X | ✓ | ✓ | ✓ |
| 3942 | Human Fyn-SH2 Domain (aa 148–248) | ✓ | ✓(weak) | X | ✓ | X | ✓(weak) | ✓ | ✓ | ✓ |
Right mark represents a clearly visible band on SDS-PAGE gels or western blots (weak signals are labelled), Cross mark represents no clearly visible bands for this construct/host pair.
Although based on a limited test set of proteins, these data demonstrate parallel, multiple host screening using the pOPIN vectors and that, combining this with the facility to generate different fusion tag formats, provides the strategy for subsequent scale-up experiments.
DISCUSSION
The In-Fusion™ cloning method was originally developed to produce a common entry vector for a multi-vector cloning system based on the cre-lox recombination (Clontech). As shown here and by others (18) (and Andrew Farmer: Clontech, personal communication), the enzyme can be used in combination with any 15 bp homology region to enable ligation-independent cloning of PCR products. We have exploited this property, in combination with single and multiple promoter vectors, to produce a simple and versatile system for expression of recombinant proteins. We have incorporated the method into a semi-automated pipeline for both vector construction and expression screening using standard laboratory liquid handling instruments. The pilot experiments in 96-well format showed cloning efficiencies of 89% after two clones per target tested, with 94% achievable with four clones tested, which we would recommend to maximize cloning efficiency. The reason(s) for some PCR products not cloning is not clear although the quality and quantity of the input DNA appears to be important. Over the last 12 months in the OPPF, we have used In-Fusion™ to construct a total of 661 vectors from 703 PCR products, an overall cloning efficiency of 94%. These results compare favourably with data reported by other HTP structural genomics consortia using either the Gateway™ recombinatorial system (e.g. 79% PCR product to expression clone efficiency (19,20)) or ‘classical’ restriction enzyme and ligation-dependent methods (e.g. 87% (1)).
Multi-promoter vectors have been used to express recombinant proteins in more than one host in parallel, typically baculovirus-infected insect cells and E. coli e.g. (21,22). Similarly, the pOPINE, pOPINF, pOPINJ or pOPINM vectors based on pTriEx2 (Novagen) described in this study can be used to survey expression in E. coli, insect cells, and mammalian cells from a single vector, which can be easily constructed in HTP mode with the associated savings in time and resources. Transient transfection of mammalian cells, notably COS7 and HEK293, is widely used to investigate the expression of eukaryotic proteins and can be readily scaled up for production purposes (7,12,23,24). We describe the derivation and use of a secretion vector based on the pOPINF/pTriEx2 vector backbone for the HTP expression of extra-cellular proteins and domains. In addition, the expression from single vector constructs in all three hosts (E. coli, HEK293T and Sf9 cells) has been demonstrated for five target proteins. This ability to screen in multiple hosts would enable rapid scale-up in the most appropriate host (determined by small-scale screening) without the necessity for additional rounds of sub-cloning. The transient transfection protocol (HEK293 cells) and co-transfection protocols (Sf9 cells), which make use of a commercial transfection reagents, have both been automated to enable consistent HTP operation for functional/structural proteomic applications. As far as we are aware, this is the first report of such implementations.
In summary, we have addressed the primary characteristics we set for an improved strategy for HTP cloning as follows:
The ability to clone genes-encoding proteins, or domains thereof, in a rapid and reliable parallel or high-throughput (HTP) fashion. Cloning efficiencies of >90% have been achieved in a parallel 96-well plate format using a generic vector in combination with the In-Fusion™ enzyme.
The ability to accurately determine the final constructs without the addition of extraneous/vector or restriction-site-derived amino acids to the expressed protein. A suite of vectors has been described which make use of In-Fusion™ cloning to precisely engineer the expressed sequence, for example, introducing an N-terminal His6 tag and 3C protease cleavage site to enable removal of the tag during purification. The utility of this format has been exemplified (Case study I).
The process must be versatile in terms of insert sequence independence. Three case studies have been described in which eighty-two expression vectors have been produced from a variety of target genes using the vector system (Case studies I, II, III).
The process would preferably be single-step, to give rapid and cost-effective vector construction. Vector construction involves a single-step reaction that enables the simple and rapid construction of vectors in parallel using a 96-well plate format. Further, certain PCR products are compatible with multiple fusion vector formats (e.g. N-His, N-His-GST and N-His-MBP—see examples in Case study III) thereby enabling cost-effective use of PCR primers.
The constructs should be capable of expressing proteins from multiple hosts, i.e. a single vector capable of expression in E. coli, mammalian cell lines (e.g. HEK293T cells) and insect cell lines (e.g. Sf9 cells). By adapting a multi-promoter vector a single cloning step has been used to produce a single vector suitable for expression in E. coli, mammalian and insect cells. The utility of this has been exemplified (Case study III).
The expressed proteins must be capable of purification in HTP mode, i.e. they must all be fused to a common affinity purification ‘tag’ which may be removed, if desired, by enzymatic digest prior to crystallization. All vectors, regardless of additional fusion protein expressed, add either an N-terminal or C-terminal His6 tag onto the expressed target protein to facilitate detection and purification. The purification of proteins expressed both intracellularly in E. coli and secreted from mammalian cells using a common purification strategy has been exemplified (Case studies I and II).
The process should be amenable to automation. Laboratory automation to carry out liquid handling tasks involved in the various stages of the expression screening experiments in all three hosts has been described.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
We would like to thank Andrew Farmer of Clontech for his advice on vector design and comments on the preparation of this manuscript and Ian Jones (Reading University) for advice on baculovirus expression and for the provision of the modified A. californica baculovirus bacmid. We also thank our collaborators, Nigel Saunders (Sir William Dunn School of Pathology, Oxford University) for providing Neisseria genomic DNA, Antonio Velayos for the KIAA0319 human and mouse cDNA clones, Louise Bird for the Fyn vectors, Radu Aricescu for the pHLSec vector template (Wellcome Trust Centre for Human Genetics, Oxford University), Hervé Bourhy (Pasteur Insitute, Paris) for the Lagos bat M protein cDNA. The Oxford Protein Production Facility is funded by the Medical Research Council, UK and is part of the Structural Proteomics IN Europe (SPINE) consortium (European Commission Grant No. QLG2-CT-2002-00988) and Vizier (European Commission FP6 contract: LSHG-CT-2004-511960) Funding to pay the Open Access publication charge was provided by the Medical Research Council, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klock HE, White A, Koesema E, Lesley SA. Methods and results for semi-automated cloning using integrated robotics. J. Struct. Funct. Genomics. 2005;6:89–94. doi: 10.1007/s10969-005-3084-1. [DOI] [PubMed] [Google Scholar]
- 2.Alzari P, Berglund H, Berrow NS, Blagova E, Busso D, Cambillau C, Campanacci V, Christodoulo E, Eiler S, et al. Implementation of semi-automated cloning and prokaryotic expression screening: the impact of SPINE. Acta Crystallogr. D Biol. Crystallogr. 2005;D62:1103–1113. doi: 10.1107/S0907444906029775. [DOI] [PubMed] [Google Scholar]
- 3.Aslandis Cd PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haun RS, Serventi IM, Moss J. Rapid, reliable ligation-independent cloning of PCR products using modified plasmid vectors. Biotechniques. 1992;13:515–518. [PubMed] [Google Scholar]
- 5.Cordingley MG, Register RB, Callahan PL, Garsky VM, Colonno RJ. Cleavage of small peptides in vitro by human rhinovirus 14 3C protease expressed in Escherichia coli. J. Virol. 1989;63:5037–5045. doi: 10.1128/jvi.63.12.5037-5045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmeyer H, Gawehn K, Grassl M. Methods of Enzymatic Analysis. Vol. 1. New York: Academic Press Inc.; 1974. [Google Scholar]
- 7.Aricescu AR, Weixian L, Jones EY. A time and cost efficient system for high level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 9.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 10.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 11.Ren J, Sainsbury S, Berrow NS, Alderton D, Nettleship JE, Stammers DK, Saunders NJ, Owens RJ. Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis. BMC Struct. Biol. 2005;5:13. doi: 10.1186/1472-6807-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 14.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum. Mol. Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 15.Albeck S, Alzari P, Andreini C, Banci L, Berry IM, Bertini I, Cambillau C, Canard B, Carter L, et al. SPINE bioinformatics and data-management aspects of high-throughput structural biology. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1184–1195. doi: 10.1107/S090744490602991X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 17.Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benoit RM, Wilhelm RN, Scherer-Becker D, Ostermeier C. An improved method for fast, robust, and seamless integration of DNA fragments into multiple plasmids. Protein Expr. Purif. 2006;45:66–71. doi: 10.1016/j.pep.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 20.Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J. Struct. Funct. Genomics. 2004;5:267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 21.Chambers SP, Austen DA, Fulghum JR, Kim WM. High-throughput screening for soluble recombinant expressed kinases in Escherichia coli and insect cells. Protein Expr. Purif. 2004;36:40–47. doi: 10.1016/j.pep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Jones IM. Rapid parallel expression in E. Coli and insect cells: analysis of five lef gene products of the Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) Virus Genes. 2004;29:191–197. doi: 10.1023/B:VIRU.0000036379.15968.4a. [DOI] [PubMed] [Google Scholar]
- 23.Pham PL, Perret S, Cass B, Carpentier E, St-Laurent G, Bisson L, Kamen A, Durocher Y. Transient gene expression in HEK293 cells: peptone addition posttransfection improves recombinant protein synthesis. Biotechnol. Bioeng. 2005;90:332–344. doi: 10.1002/bit.20428. [DOI] [PubMed] [Google Scholar]
- 24.Geisse S, Henke M. Large-scale transient transfection of mammalian cells: a newly emerging attractive option for recombinant protein production. J. Struct. Funct. Genomics. 2005;6:165–170. doi: 10.1007/s10969-005-2826-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.