Abstract
Restriction endonuclease MvaI recognizes the sequence CC/WGG (W stands for A or T, ‘/’ designates the cleavage site) and generates products with single nucleotide 5′-overhangs. The enzyme has been noted for its tolerance towards DNA modifications. Here, we report a biochemical characterization and crystal structures of MvaI in an apo-form and in a complex with target DNA at 1.5 Å resolution. Our results show that MvaI is a monomer and recognizes its pseudosymmetric target sequence asymmetrically. The enzyme consists of two lobes. The catalytic lobe anchors the active site residues Glu36, Asp50, Glu55 and Lys57 and contacts the bases from the minor grove side. The recognition lobe mediates all major grove interactions with the bases. The enzyme in the crystal is bound to the strand with T at the center of the recognition sequence. The crystal structure with calcium ions and DNA mimics the prereactive state. MvaI shows structural similarities to BcnI, which cleaves the related sequence CC/SGG and to MutH enzyme, which is a component of the DNA repair machinery, and nicks one DNA strand instead of making a double-strand break.
INTRODUCTION
Restriction endonucleases (REases) are usually classified according to their specificity, target site symmetry, oligomeric structure and/or the presence of auxiliary domains that modulate the activity (1). A less widely used criterion for REase classification is the length of sticky ends in cleaved DNA duplexes. Grouping REases by this criterion reveals a very uneven distribution in the frequency of different cleavage patterns. Indeed, 4 nucleotide (nt) 5′-overhangs are most abundant. Moreover, protruding 5′-ends are more common than recessed 5′-ends and overhangs with an even number of nucleotides outnumber those with an odd number (2). It is currently thought that different cleavage patterns are mainly achieved by the changes at the dimer interface between the conserved monomers of restriction enzymes (3,4). However, the crystal structure of Ecl18kI, which cleaves the CCNGG sequence and generates 5 nt 5′-overhangs demonstrated that Ecl18kI achieves its specificity by a flip of the central nucleotides rather than by a rearrangement of the dimer interface (5). This finding suggests that the cleavage pattern may reflect fundamental structural differences and prompts further studies of endonucleases that generate rare cleavage patterns. Here, we focus on MvaI, the first example of a restriction enzyme that recognizes a pseudopalindromic sequence CC/WGG (W stands for A or T) and generates 1 nt 5′-overhangs upon cleavage.
The MvaI REase is a part of the MvaI restriction–modification (RM) system discovered in Micrococcus varians strain RFL19 (now renamed to Kocuria varians). The MvaI REase is accompanied by the cognate methyltransferase (MTase) which methylates the N4-amino group of the inner cytosine residue making the host DNA resistant to MvaI cleavage (6). The modification of the same cytosine to 5mC, which can be catalyzed by several physiologically unrelated MTases, is not protective against MvaI cleavage (7).
The MvaI REase is unusual in many respects. (a) The enzyme generates 1 nt 5′-overhangs (6). (b) MvaI cleaves unmodified DNA duplexes in two single-strand scissions (8). (c) The cleavage rates of the two strands are significantly different (9). (d) Unlike the prototype EcoRII REase, MvaI is very tolerant of modifications in synthetic substrates (10). The enzyme completes the cleavage of substrates with a nick in one strand, irrespective of whether or not a phosphate is present at the nicking site (8). Moreover, selective modification of one DNA strand typically affects the cleavage of either the modified or the unmodified strand, depending on the nature of the modification, but only very drastic modifications impair cleavage of both strands (7,10).
Here, we show that MvaI is a monomer in solution that binds duplex DNA in 1:1 stoichiometric ratio and we present a crystallographic analysis of MvaI in the apo- and DNA-bound forms both at 1.5 Å resolution. The crystallographic results show that MvaI interacts with a complete recognition site and not just with a half-site of the pseudosymmetric target sequence. Therefore, we suggest that the enzyme acts as a ‘nickase’ initially, but may swing around its DNA target to make a second cut that completes the cleavage reaction.
MATERIALS AND METHODS
MvaI REase cloning and expression
Escherichia coli strain ER2566 (F–λ–fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10–TetS)2 R(zgb-210::Tn10)(TetS) endA1 [dcm]) was used as a host for cloning and overexpression experiments. ER2566 strain was shown experimentally to exhibit the dcm− phenotype.
The MvaI REase (mvaIR) gene was amplified from the cloning vector kindly provided by Fermentas (Vilnius, Lithuania). The mvaIR gene was re-cloned into the pBAD24 (Apr) vector and expressed in the ER2566 strain bearing plasmid pACYC184 (Cmr) containing pspGIM gene which was a gift from New England Biolabs (USA). The expression vector for MvaI REase encoded full-length MvaI with three additional MKS amino acids at the N-terminus. To express MvaI REase the ER2566 strain containing the pACYC184_M.PspGI was transformed with pBAD24_R.MvaI. Cells were grown in LB medium with appropriate antibiotics at 37°C to OD600 0.7 and MvaI REase expression induced by addition of arabinose to the final 0.2% concentration. After 4 h of induction the cells were harvested by centrifugation and the pellet was stored at −20°C.
MvaI purification
Frozen cells were thawed and resuspended in 4 volumes of buffer A (0.01 M Tris-HCl, pH 8.0 at 25°C, 1 mM EDTA, 7 mM 2-mercaptoethanol) with 0.1 M NaCl. The cell suspension was sonicated and the cell debris was removed by centrifugation at 18 000 r.p.m for 1 h. The supernatant was loaded on phosphocellulose P11 column (Whatman) equilibrated with buffer A with 0.1 M NaCl. The MvaI protein was not bound to P11 beads, the flow-through was collected and subjected to subsequent chromatography on the Heparin Sepharose, Blue Sepharose (Amersham Biosciences) and phosphocellulose P11 columns (Whatman). The protein was eluted from all columns with a linear NaCl gradient in buffer A. All purification steps were monitored by λ DNA cleavage and SDS-PAGE. Fractions containing MvaI REase activity were pooled and dialyzed against storage buffer (0.01 M Tris-HCl, pH 8.0 at 25°C, 0.4 M KCl, 1 mM EDTA, 1 mM DTT, 50% glycerol) and stored at −20°C. The protein was purified to electrophoretic homogeneity in a Coomassie-blue-stained SDS-gel. The overall yield of MvaI purification was 10 mg from 5 l culture.
Analytical ultracentrifugation
Analytical ultracentrifugation was done in an An-60 rotor with a Coulter-Beckman Model XL-A analytical ultracentrifuge equipped with UV absorption detection using charcoal filled epon centrepieces. Sedimentation velocity was measured at 4°C and 44 000 r.p.m. in a buffer containing 0.01 M Tris-HCl, pH 7.4, 0.4 M KCl, 0.1 mM DTT and 0.1 mM EDTA at a loading concentration of 2.5 µmol. Absorption profiles were recorded at 230 nm to increase the sensitivity of detection. The sedimentation coefficient distribution was analyzed with the program SEDFIT (11). Viscosity and density correction to calculate s20°C,W was done using the data supplied by the program package SEDNTERP (http://www.rasmb.bbri.org). Sedimentation equilibrium was measured in six-channel centrepieces at 18 000 r.p.m. and 20°C. Samples were spun until no change in absorbance profiles could be observed for at least 12 h at which time equilibrium was assumed to have been reached. Molar masses were evaluated from the concentration gradients observed in these last 12 h as described earlier (12).
Analytical gel filtration
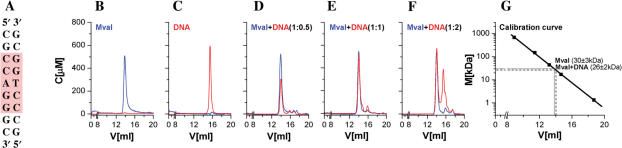
Here, 30 μg (1 nmol) of MvaI protein was mixed with blunt-ended 9-mer cognate oligoduplex 1 (Figure 1A) in buffer B (0.01 M Tris-HCl, pH 8.0, 0.3 M KCl, 5 mM CaCl2, 1 mM DTT, 5% glycerol) at different stoichiometric ratios, and incubated on ice for 1 h. The samples were loaded on a Superose ™ 12 HR 10/30 column (Amersham Biosciences), which was equilibrated in buffer B and run with a flow rate of 0.4 ml/min. The column was calibrated with Biorad protein standards (vitamin B-12, 1.35 kDa; myoglobin, 17 kDa; ovalbumin, 44 kDa; IgG, 150 kDa and thyroglobin, 670 kDa). For the interpolation of unknown molecular mass, a linear dependence of the logarithm of the molecular mass on the elution time was assumed.
Figure 1.
Analytical gel filtration experiments to determine the MvaI oligomeric state and the stoichiometry of DNA binding for the oligoduplex 1 shown in (A). Elution profiles were recorded simultaneously at 260 and 280 nm and deconvoluted to obtain separate curves for the MvaI (blue) and DNA (red) concentration. (B) MvaI alone, (C) DNA alone, (D) mixture with a 2:1 molar excess of MvaI over DNA, (E) stoichiometric mixture, (F) mixture with a 2:1 molar excess of DNA over MvaI, (G) calibration curve for Superose ™ 12 HR 10/30 column (Amersham Biosciences) with standards from Biorad (vitamin B-12, 1.35 kDa; myoglobin, 17 kDa; ovalbumin, 44kDa; IgG, 150 kDa and thyroglobin, 670 kDa).
Elution profiles were monitored by an Ettan (Amersham) two-wavelength detector at 260 and 280 nm. The A(260):A(280) absorbance ratios necessary for profile deconvolution were deduced from the ratios of A(260) and A(280) peak heights after injection of only protein or only DNA. For our system, we determined A(260):A(280) ∼1.9 for DNA and A(260):A(280) ∼0.56 for MvaI without DNA. Absolute absorbance values were calculated as follows: for the double-stranded DNA, an A(260) = 1 cm−1 was taken to correspond to 0.15 mM nucleotides or 8.33 μM of the 9-mer oligoduplex. For MvaI apo-form, an extinction coefficient of 34 380 M−1cm−1 was calculated by the EXPASY server (http://www.expasy.ch) (13). After some elementary algebraic manipulations, it follows that cprotein = −21.3A(260) + 40.8A(280) and cDNA = 11.7A(260) − 6.5A(280), if absorbances and concentrations are measured in units of cm−1 and μM, respectively.
Native gel electrophoresis
Native gel electrophoresis was run in acidic conditions according to a protocol from Dr Lebendiker (The Hebrew University, Jerusalem, http://wolfson.huji.ac.il/purification/Protocols/PAGE_Acidic.html). Electrophoresis was performed at 4°C at 25 mA. Care was taken to reverse the polarity relative to the usual arrangement, because at pH 4.3 the proteins migrate as positively charged species. Here, 5 μg (0.17 nmol) of MvaI was mixed in 10 μl of buffer B with oligoduplex 1 (Figure 1A) in various stoichiometric ratios. Mixtures were incubated on ice overnight, supplemented with 2 μl of loading dye (1.45 ml of 50% glycerol, 0.5 ml of 0.25 M acetate-KOH, traces of methylene green) and immediately loaded on the gel.
Crystallization
Crystallization was done by the vapor diffusion technique at room temperature. Initial high-throughput screens were set up at the 200-nl scale using a Cartesian pipetting robot and 96-well Greiner sitting drop plates. Crystallization trials with larger drop volumes were pipetted in CRYSCHEM plates (Hampton Research).
DNA-free form
MvaI was dialyzed against buffer B and concentrated to 7 mg/ml. Initial screening identified Hampton Research Crystal Screen 2 buffer 34 (0.05 M cadmium sulfate hydrate, 0.1 M HEPES pH 7.5, 1.0 M sodium acetate trihydrate) as promising for crystallization. The best crystals appeared in a drop containing 1.5 µl of protein, 1.5 µl of buffer C (0.05 M cadmium sulfate hydrate, 0.1 M HEPES pH 7.74, 1.2 M sodium acetate trihydrate) and 0.3 µl of 1 M glycine as an additive, which was equilibrated against buffer C in the reservoir. The largest crystal was flash-cryocooled in a drop containing 16 µl buffer C, 2 µl of 1 M glycine and 2 µl of (2R,3R)-(−)-2,3-butandiol. Diffraction improved after a single annealing step.
DNA-bound form MvaI was mixed with oligoduplex 2 (Figure 2)
Figure 2.
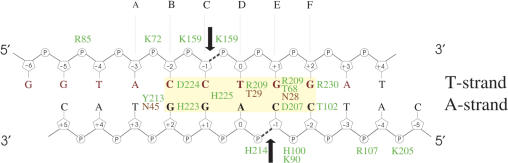
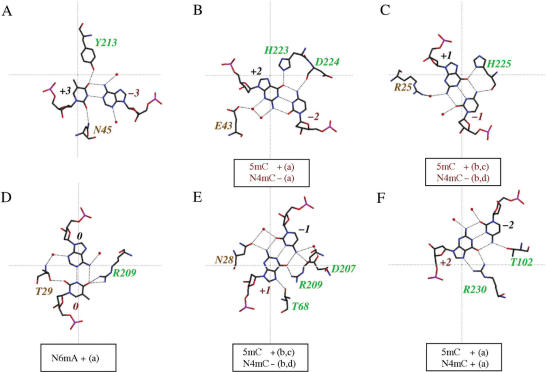
Summary of MvaI–oligoduplex 2 interactions. Residues of the catalytic and recognition lobes are marked in orange and green, respectively. Arginine, lysine and histidine residues of MvaI involved in the interactions with DNA phosphate oxygen atoms are indicated close to the respective phosphates. Direct hydrogen bonding interactions of MvaI with the DNA bases are indicated next to the bases, on their right side (e.g. Y213 and N45 interact with T + 3). Indirect water-mediated hydrogen bonding interactions are not shown. The black arrows indicate the MvaI cleavage sites. The binding mode of the DNA in the MvaI crystals brings only the T-strand close to the active site. Labels A, B, C, D, E and F refer to the panels in Figure 5.
in 1:1 molar ratio and incubated overnight on ice. The protein–DNA complex (final concentration 6.4 mg/ml) was applied on a 24-well plate with appropriate buffers. The crystals appeared in a drop containing 2 µl of protein and DNA solution and 2 µl of buffer D (0.1 M HEPES pH 7.89, 0.2 M CaCl2, 25% PEG4000). Crystals were flash-cryocooled in crystallization buffer D supplemented with 20% of glycerol.
Data collection and structure determination
All diffraction data were collected at 100 K. In-house data were measured on a RUH300 generator with copper anode from MSC/Rigaku equipped with Osmic multilayer optics and an MAR345 image plate. Synchrotron data were collected at beamline BW6 at DESY, Hamburg. All data were processed with MOSFLM (14) and scaled with SCALA (15) (Table 1).
Table 1.
Data collection, phasing and refinement statistics
| MvaI without DNA (20.0–1.5 Å) | MvaI with DNA (30.0–1.5 Å) | |||
|---|---|---|---|---|
| Space group | P2(1) | P2(1) | ||
| a (Å) | 58.5 | 53.9 | ||
| b (Å) | 63.3 | 81.4 | ||
| c (Å) | 71.9 | 71.3 | ||
| β (°) | 100.8 | 90.5 | ||
| Wavelength (Å) | 1.54 (in house) | 1.05 (BW6, DESY) | 1.05 (BW6, DESY) | |
| Resolution range (Å) | 12.0–1.7 | 20.0–1.5 | 30.0–1.5 | |
| Total reflections | 243 586 | 233 261 | 298 191 | |
| Unique reflections | 52 083 | 75 246 | 94 979 | |
| Completeness (%) (last shell) | 91.6 (99.0) | 91.2 (99.6) | 96.6 (95.3) | |
| I/σ (last shell) | 10.8 (2.9) | 30.8 (4.1) | 6.2 (2.5) | |
| R(sym) (%) (last shell) | 5.2 (25.0) | 6.1 (23.2) | 7.0 (28.8) | |
| B(iso) from Wilson (Å2) | 17.3 | 19.8 | 14.5 | |
| FOM (SHARP) | 0.375 | – | – | |
| Reflections work/test | 68 906/3629 | 90 188/4789 | ||
| Protein atoms (excluding H) | 3847 | 4081 | ||
| DNA atoms (excluding H) | – | 868 | ||
| Solvent molecules | 313 | 583 | ||
| R-factor (%) | 18.4 | 17.7 | ||
| R-free (%) | 21.2 | 19.5 | ||
| RMSD bond lengths (Å) | 0.014 | 0.008 | ||
| RMSD angles (°) | 1.5 | 1.2 | ||
| Ramachandran core region (%) | 92.8 | 90.5 | ||
| Ramachandran allowed region (%) | 7.2 | 9.1 | ||
| Ramachandran additionally allowed region (%) | 0.0 | 0.0 | ||
| Ramachandran disallowed region (%) | 0.0 | 0.4 | ||
DNA-free form
This crystal form contained two MvaI molecules in the asymmetric unit. It was solved by the SAD method, taking advantage of the presence of Cd2+ ions in the crystallization buffer, which bound to some defined sites on the protein surface. Five Cd2+ sites were identified with reasonable statistics (CCall 28.7, CCweak 17.3) by the SHELXD program (16). Initial phasing with these sites in SHELXE (17) resulted in a significant contrast difference (0.61 for the correct hand versus 0.45 for the incorrect hand assuming 50% solvent content). Optimal SAD phases were calculated by the SHARP program and then extended to the full resolution of the synchrotron dataset. The extended phases were of sufficient quality for ARP/wARP (18) to automatically build 333 amino acids in 19 chains or 67% of the total number of residues that are chemically present in the MvaI crystals. The partial model was sufficient to derive the non-crystallographic (NCS) symmetry, which could then be used to map fragments between monomers. The ‘symmetrized’ model was then used as the starting point for a further round of model building, and then polished manually.
DNA-bound form
This crystal form contained two complexes of MvaI bound to duplex DNA in the asymmetric unit. Molecular replacement was unsuccessful with the complete MvaI model from the DNA-free crystal form, but two copies of the recognition domain could be placed using the MOLREP program. The initially weak model phases were improved using the protocol for NCS-phased refinement from the CCP4i interface. The resulting phases were of sufficient quality for automatic model building using ARP/wARP (18). DNA was built manually starting from a model of B-DNA of the proper sequence generated with the modeling program 3DNA (19).
Manual corrections to the models were done with the programs O (20) and XtalView (21). All structures were refined with the maximum likelihood program REFMAC (22) treating each lobe of MvaI and each DNA duplex as separate TLS entities. Data collection and refinement statistics are summarized in Table 1.
RESULTS
MvaI is a monomer according to analytical ultracentrifugation
At the sequence level, MvaI shows weak similarities to the monomeric DNA repair protein MutH (23). This result prompted us to check the oligomerization state of MvaI in solution. Analytical ultracentrifugation shows MvaI to be mostly monomeric. Sedimentation velocity runs in the analytical ultracentrifuge with MvaI gave a sedimentation constant of s20°C,W = 2.62 S. Using a mass for the monomeric protein of 28.6 kg/mol this corresponds to a frictional ratio of 1.27. For spherical hydrated proteins, a frictional ratio of 1.1–1.2 is expected (24) and thus MvaI can be viewed as a mostly globular, monomeric particle. Sedimentation equilibrium gave a molar mass of 35 kg/mol indicating the protein to show some aggregation. This aggregation could be suppressed by the addition of 0.8 M GuaHCl where a molar mass of 27.5 kg/mol is observed.
MvaI is a monomer according to analytical gel filtration
The monomeric state of MvaI in the apo-form and in complex with the cognate, blunt-ended 9-bp oligoduplex 1 (Figure 1A) was independently analyzed by analytical gel filtration. Experiments were run in the presence of calcium ions, which support DNA binding, but not hydrolysis (data not shown). Although MvaI and MvaI–DNA complexes are not resolved on the column, the amounts of MvaI and DNA in the elution peaks could be separately quantified at 260 and 280 nm, respectively, and deconvoluted based on the known A(260):A(280) absorbance ratios for MvaI and DNA (see Materials and methods). For consistency, the deconvolution procedure was applied to all profiles, even if only protein or only DNA was injected (Figure 1B–F).
In the experimental conditions (see Materials and methods), MvaI alone elutes from the column at 13.82 ml, which translates into a molecular mass of 30 ± 3 kDa, in agreement with the calculated monomer mass 28.6 kDa (Figure 1B,G). The cognate 9-bp oligoduplex 1 (Figure 1A) alone elutes much later, at 15.47 ml (Figure 1C), but it coelutes with MvaI up to a stoichiometric ratio of 1 oligoduplex per 1 MvaI monomer (Figure 1D and E). If oligoduplex is present in excess over the MvaI monomer, two peaks result. One equivalent of DNA coelutes with the protein, and the rest elutes as free DNA (Figure 1F). The retention volume for the MvaI–DNA complex is 14.01 ml and corresponds to an apparent molecular mass value of 26 ± 2 kDa (Figure 1G) which is slightly lower than the apparent mass of MvaI alone, probably because the more compact shape of the complex nearly balances the increase in molecular mass due to DNA binding. We conclude that MvaI remains monomeric in the presence of cognate DNA, at least in our experimental conditions. Our findings are inconsistent with a prior report of MvaI dimerization in the presence of both cognate and non-cognate DNA (25).
MvaI binds duplex DNA in 1:1 stoichiometric ratio
The gel filtration experiments suggest that MvaI is a monomer that binds target duplex DNA in 1:1 stoichiometric ratio. This was independently confirmed by gel electrophoresis under acidic conditions (see, Materials and methods). In these conditions, free DNA does not enter the gel, but MvaI alone (predicted isoelectric point 6.8) migrates into the gel (Supplementary Figure 1, lane 1). The addition of substoichiometric amounts of cognate oligoduplex leads to the appearance of a second, slower migrating band, the MvaI–DNA complex (Supplementary Figure 1, lane 2). If oligoduplex is added in stoichiometric ratio, the band from MvaI alone almost disappears, and essentially only the MvaI–DNA band is present (Supplementary Figure 1, lane 3). Still further increase of the DNA concentration has no effect on the band pattern (Supplementary Figure 1, lane 4).
MvaI structure determination
The available biochemical data on MvaI and our findings about its oligomeric state and DNA-binding stoichiometry suggested that MvaI is a highly unusual restriction enzyme and prompted us to determine its structure. MvaI was crystallized in the apo-form and in the presence of oligoduplex 2 (Figure 2). The crystals with and without DNA happened to be monoclinic and diffract to 1.5 Å resolution. The apo-form was solved by the SAD method, and the form with DNA by molecular replacement, using the previously determined model of the DNA-free form (details in Materials and methods and Table 1).
Both crystal forms of MvaI contained two monomers in the asymmetric unit. In the apo-structure, the interaction of the monomers, which are related by a curious 2-fold symmetry, buries ∼1800 Å2 of solvent-exposed surface. Despite this extensive contact, the interaction cannot be physiological, because (a) it is not observed in solution, (b) the interface is not conserved and (c) it locks MvaI in an open conformation that is not compatible with DNA binding (see below). In the crystals of the MvaI–DNA complex, the largest interfaces between adjacent molecules in the crystal bury only ∼900 and ∼600 Å2. Moreover, these contacts relate molecules that cannot be mapped on each other by a simple 2-fold rotation, and they differ from the extensive contact in the apo-MvaI structure. Therefore, we conclude that MvaI crystallized as a monomer in all cases. The two molecules in the apo-MvaI structure are very similar, probably because the local 2-fold axis enforces it. Likewise, the two MvaI molecules in the asymmetric unit of the crystals with DNA overlap almost perfectly, probably because the complexes with DNA are very compact and therefore rigid. The same is true for the bound DNA duplexes, which can be described by very similar conformational parameters according to the 3DNA program (19) (Supplementary Table 1), even though no restraints or constraints were applied during refinement. For simplicity, we will not distinguish between the two monomers in each crystal form in the following text.
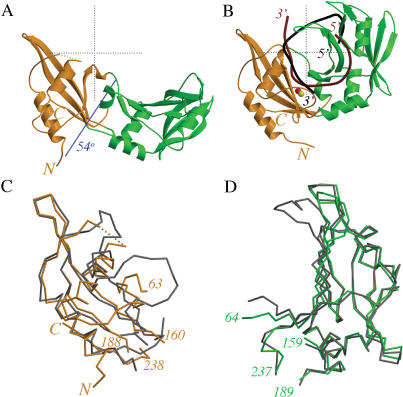
MvaI consists of two lobes
MvaI is organized into two lobes which we term the catalytic lobe (residues 1–63, 160–188 and 238–246, orange in Figure 3) and the recognition lobe (residues 64–159 and residues 189–237, green in Figure 3) based on their mechanistic roles discussed below. The orientation of the two lobes differs radically depending on whether or not DNA is bound: in the absence of DNA, the molecule has an almost flat appearance (Figure 3A), but in the presence of DNA, it forms a tight ‘clamp’ around it (Figure 3B). Despite the drastic change in hinge angle (∼54°), there is no major conformational difference within the lobes, except in loops (Figure 3C and D and Supplementary Figure 2). The rearrangement of residues 40–57 is significant, because this region includes catalytic residues.
Figure 3.
Overall view of the MvaI structure. The catalytic lobe of MvaI is shown in orange and the recognition lobe is presented in green. (A) Ribbon representation of the open conformation in the apo-form of MvaI. Rotation of the recognition lobe around the blue axis by 54° brings this lobe into a similar orientation relative to the catalytic lobe as in the MvaI–DNA complex. (B) Ribbon representation of the closed conformation in the MvaI–DNA complex. The DNA strand that comes close to the active site and would be cleaved in the presence of Mg2+ ions is shown in dark red. The complementary strand is presented in black. (C) Superposition of the Cα-traces of the catalytic lobes taken from the structures without (orange) and with (gray) DNA. (D) Superposition of the Cα-traces of the recognition lobes from the structures without (green) and with (gray) DNA. All panels were prepared with the MOLSCRIPT program (44).
Catalytic lobe
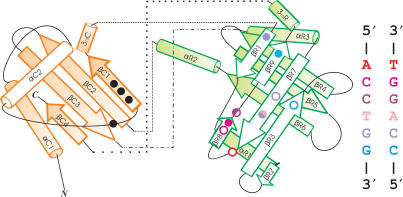
The catalytic lobe is organized around a four-stranded mixed β-sheet which is flanked by two α-helices and a 310-helix (orange part of Figure 4). The topology of the sheet is +1x, +1, +1, +1 according to the Richardson nomenclature (26). Note that elements of the recognition lobe are interspersed between strands βC1 and βC2 and also between strands βC3 and βC4. Strands βC2 and βC3 are connected by a simple hairpin. The fold of the catalytic lobe is fairly similar to the fold of its counterpart in MutH, which has been termed the ‘N-arm’ of this enzyme (27). There are also other, more distant similarities to the cores of other REases of the PD … EXK family (data not shown).
Figure 4.
Schematic representation of the MvaI fold. The catalytic domain is in orange and the recognition domain is in green. Catalytic residues are marked by black dots, and residues that are involved in hydrogen-bonding interactions are marked by colored circles. Colors are ramped from red to blue following the T-strand in 5′ → 3′ direction. Filled circles mark residues that interact with the T-strand and open circles indicate residues that interact with the A-strand in the crystal structure. The secondary structure assignment was done for the structure with DNA using the DSSP program (45): αC1 (L5-V16), αC2 (G30-L38), βC1 (I54-E60), 310C (T61-A63), βR1 (L67-T71), αR1 (A80-Y88), βR2 (K90-K91), βR3 (N97-V103), βR4 (Y116-D123), βR5 (V128-D135), βR6 (M140-S148), αR2 (F149-K159), βC2 (Y162-E173), βC3 (K176-T188), αR3 (V192-N201), βR7 (I204-A212), βR8 (T222-D224), βR9 (A228-I231), 310R (M233-E238) and βC4 (E241-V244).
The term ‘catalytic’ lobe was chosen because this part of MvaI anchors all catalytic residues of the enzyme. In addition, it also contributes some of the minor grove interactions with DNA, which are unlikely to play a major role in sequence discrimination (see below).
Recognition lobe
In contrast to the catalytic lobe, the recognition lobe is characteristic of MvaI. The fold is organized around two antiparallel β-sheets (green part of Figure 4). The larger of the two sheets consists of strands βR1, βR2, βR3, βR7 and βR8/βR9, which are connected in +4x, −1, −1, −1 topology. This would correspond to the Greek key motif if the small βR2 is not taken into account. The smaller sheet is built from strands βR4, βR5 and βR6 and is a β-meander. Protein architecture dictates two preferred ways for β-sheets to stack against each other: aligned, with an angle between the strands in the two sheets ∼30°, and perpendicular, with an angle between the strands ∼90° (28,29). The MvaI recognition lobe clearly belongs to the latter group and therefore resembles a half-barrel or barrel (30,31) except for the lack of hydrogen bonds to connect the sheets.
The term ‘recognition’ lobe was chosen because this lobe anchors the residues that interact specifically with the major grove of DNA and likely mediate sequence discrimination. We also note that nearly all basic residues (Lys64, Lys72, Arg85, Lys90, His100, Arg107, Lys159, Lys205, His214) that interact with the phosphodiester backbone of DNA are located in the recognition lobe.
One active site, asymmetric recognition of the pseudosymmetric target sequence
MvaI acts as a monomer and recognizes its pseudosymmetric target sequence asymmetrically (Figures 2 and 3B). As the enzyme has only one active site, this implies that only one strand can come into proximity of the active site. Although MvaI can bind target DNA in two orientations, the strand with the central T (‘T-strand’) binds exclusively or predominantly close to the active site in our crystal form of MvaI with DNA.
Sequence readout
As expected, MvaI contacts the specifically recognized target bases, but in addition it also engages in hydrogen bonds with a flanking base pair. In all positions, the catalytic lobe approaches the bases exclusively from the minor grove side, and the recognition lobe interacts with the bases exclusively from the major grove side. For the detailed discussion, we follow the T-strand from the −3 to the +2 position (from left to right according to the scheme in Figure 2).
A − 3 T + 3: This A-T base pair is not part of the recognition sequence, but it nevertheless makes two direct hydrogen bonds with MvaI. The O2 and O4 atoms of thymine accept hydrogen bonds from the side chain amide group of Asn45 and the OH of Tyr213, respectively. From the structural perspective, it can be expected that these interactions contribute little to sequence specificity: the position of the O2 atom, the so-called ‘outer minor grove’ is taken by a hydrogen bond acceptor for all four possible base pairs (32). On the major grove side, the side chain oxygen atom of Tyr213 can act as a hydrogen bond donor as in the crystallographically observed complex, but might also act as a hydrogen bond acceptor, if other base pairs are present in this position (Figure 5A).
Figure 5.
Hydrogen-bonding interactions between MvaI and DNA. Panels are ordered following the T-strand in 5′ → 3′ direction as indicated in Figure 2. Residues of the catalytic lobe are labeled in orange, and residues of the recognition lobe are in green. Hydrogen bonds are indicated by dotted lines. Prior experimental results on the effects of single-strand methylations are indicated below the interaction diagrams. A ‘+’ sign indicates that the methylation is compatible with T-strand cleavage, and a ‘−’ indicates that the corresponding methylation reduces T-strand cleavage by at least 50%. The abbreviations (a), (b), (c), (d) refer to the following publications. (a) Gromova et al. (1991) (10) (b) Butkus et al. (1985) (6) (c) Kubareva et al. (1988) (7) (d) Kubareva et al. (1990) (46).
C − 2 G + 2: This C-G base pair forms only indirect hydrogen bonds with MvaI on the minor grove side, but is involved in two direct hydrogen-bonding interactions with the enzyme on the major grove side. The N4 atom of cytosine donates a hydrogen bond to the main chain carbonyl oxygen atom of Asp224 and the O6 atom of guanine accepts a hydrogen bond from the Nε atom of His223. This interpretation requires that His223 is either charged or is in the tautomeric state with the proton on the Nɛ atom (Figure 5B and data not shown).
C − 1 G + 1: This C-G base pair makes an indirect hydrogen bond to Arg25 on the minor grove side and interacts with His225 on the major grove side. The main chain carbonyl oxygen atom of this residue accepts a hydrogen bond from the N4 atom of cytosine, and the Nδ atom of its imidazole ring donates a hydrogen bond to the O6 atom of guanine. For the interaction between the imidazole ring and the base to be sequence selective, the tautomerization state of His225 should be locked by interactions within MvaI, and not just by the hydrogen bond to the base. As the Nε atom of His225 interacts with a water molecule that could be either a donor or an acceptor, if and how this ‘lock’ is provided remains unclear (Figure 5C and data not shown).
T0 A0: This T-A base pair accepts a direct hydrogen bond from Thr29 Oγ to the thymine O2 atom and an indirect hydrogen bond to the adenine N3 atom. As the ‘outer minor grove’ positions are taken by hydrogen bond acceptors for all possible base pairs, this interaction probably contributes little to specificity. On the major grove side, Arg209 donates a hydrogen bond to the O4 oxygen atom of the T-strand thymine in the crystallized MvaI–DNA complex. There is no trace of the alternative binding mode, which swaps purine and pyrimidine and must occur as well in solution (Figure 5D).
G + 1 C − 1: This G-C base pair is involved in extensive hydrogen-bonding interactions. On the minor grove side, Asn28 accepts a direct hydrogen bond from the N2 atom of guanine and anchors a water molecule that donates a hydrogen bond to the O2 atom of cytosine. On the major grove side, the carboxylate of Asp207 accepts a direct and a water-mediated hydrogen bond from the cytosine N4 atom. The guanidino group of Arg209 and Thr68 Oγ atoms donate hydrogen bonds to the O6 and N7 atoms of guanine, respectively (Figure 5E).
G + 2 C − 2: This G-C base pair makes only major grove interactions with MvaI. The guanidino group of Arg230 donates hydrogen bonds to the guanine atoms O6 and N7, and the Thr102 Oγ atom accepts a hydrogen bond from the cytosine N4 atom (Figure 5F).
Methylation sensitivity
The methylation sensitivity of MvaI has been extensively studied with special emphasis on the differential effects of methylation on the hydrolysis of the two DNA strands. Experimentally, it was found that 5mC can replace cytosine in all positions (Supplementary Table 2), but this would have been difficult to predict from the crystallographic results. In silico introduction of 5mC instead of the outer and inner cytosines of the T-strand brings the extra methyl groups within 2.9 Å of Gly226 Cα and within 3.0 Å of His225 O, respectively, if base positions are not adjusted and the enzyme is kept rigid. Similarly, in silico conversion of the cytosines to 5mC in the A-strand would introduce methyl groups 2.3 Å away from Thr102 Oγ and 3.3 Å away from His100 Cβ, respectively, and would additionally require the displacement of a water molecule. Therefore, the experimentally observed tolerance of MvaI to 5mC has to be attributed either to flexibility of the enzyme, or alternatively to the adjustability of the exact positions of the bases (data not shown).
In contrast to the effects of C5 methylation, the consequences of N4 methylation on T-strand cleavage can be readily explained by the crystal structure. N4mC cannot replace the inner cytosine in either strand, because the extra methyl group would clash with a hydrogen bond acceptor of the protein (His225 O, Figure 5C and Asp207 Oδ, Figure 5E). Likewise, N4mC in the outer position of the T-strand is not tolerated, again because the methyl group would clash with a hydrogen bond acceptor on the protein (Asp224 O in this case, Figure 5B). Experimentally, substitution of the A-strand outer cytosine with N4mC has been reported not to interfere with cleavage: apparently the methyl group can displace the side chain of Thr102 (Figure 5F).
Substitution of the central adenine with N6mA interferes with A-strand cleavage, but does not affect T-strand cleavage. The latter result is consistent with the crystal structure, because the methyl group of N6mA only needs to displace a water molecule to fit in (Figure 5D).
The rules for T-strand cleavage have direct implications for A-strand cleavage, and therefore the above results can be summarized in four simple rules: (a) substitution of cytosine with 5mC has no effect. (b) Substitution of the inner cytosine with N4mC in one strand blocks cleavage of both strands. (c) Substitution of an outer cytosine in one strand by N4mC abolishes cleavage of this strand, but does not interfere with cleavage of the complementary strand. (d) Substitution of the central adenine with N6mA affects A-strand, but not T-strand cleavage. Together, these four rules correctly predict the outcome of a large number of experiments on the methylation sensitivity of MvaI (Supplementary Table 2).
Note that the MvaI MTase methylates the N4 atoms of the inner cytosines. Rule (b) predicts correctly that this modification interferes with DNA cleavage. Conversely, the Dcm MTase converts the same cytosine to 5mC, which should not have an effect on DNA cleavage. This is consistent with the experimental observation that DNA from dcm+ strains can be cleaved by MvaI (Supplementary Table 2).
MvaI active site
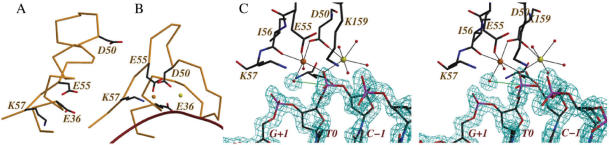
MvaI crystals with and without DNA were grown in the absence of Mg2+, but in the presence of Ca2+ ions, which support DNA binding (see Figure 1), but not DNA hydrolysis (data not shown). In the DNA-free form, Cd2+ ions were present in addition to the Ca2+ ions, but no metal ions were found in the vicinity of the active site residues. This unexpected result is due to the arrangement of residues 40–57, which are present in radically different conformations in the apo-MvaI structure (Figure 6A) and in the complex with DNA (Figure 6B).
Figure 6.
MvaI active site: (A) Conformation in the crystal of the apo-form. (B) Conformation in the cocrystals with DNA and Ca2+ ions. In (B), the yellow and orange balls represent the two Ca2+ ions in the structure, and the dark red curve is the T-strand of DNA in ribbon representation. (C) Stereo representation of the active site of MvaI in the form with bound DNA. The 2fofc density has been contoured at 1.2σ and is shown around the DNA and the catalytic water molecule or hydroxide ion. A productive nucleophilic attack, which does not take place in the crystals would require an approach of the catalytic water molecule or hydroxide ion towards the phosphorus atom of the scissile phosphate along the green line. Glu36 at the back of the figure has not been labeled to avoid overlap.
In the productive orientation two metal-binding sites are formed, which are occupied by Ca2+ ions from the buffer (the identification of the metals is supported by the ligand distances and the X-ray anomalous signal). In both standard electron density maps and anomalous difference Fourier maps, the peak heights for the two metals are very different. The weaker peak (yellow ball in Figure 6C) corresponds to a Ca2+ liganded to an oxygen atom of the scissile phosphate, one Asp50 Oδ atom and three or four water molecules (depending on which molecule in the asymmetric unit is used for the analysis). The stronger peak corresponds to a hexa-coordinated Ca2+ ion with an almost perfect octahedral coordination sphere. The ligands to this metal ion are the other Oδ oxygen atom of Asp50, the Oε atom of Glu55, the main chain carbonyl oxygen atom of Ile56, an oxygen atom of the scissile phosphate, which acts as a bridge between the two metals, and two solvent molecules (Figure 6C).
One of these solvent molecules, which could either be a water molecule or a hydroxide ion (shown with its electron density in Figure 6C) is positioned exactly on the line that links the O3′ oxygen atom of the C−1 residue to the phosphorus atom of the scissile phosphate. This solvent molecule is ideally positioned for an in-line attack on the phosphate atom, which might proceed via a bipyramidal transition state and would correctly predict reaction products with a free 3′-end and a phosphorylated 5′-end.
Although the above catalytic mechanism appears plausible, the reaction does not proceed in the crystals, as evidenced by several detailed features of the crystal structure. (a) There is robust electron density for the potentially scissile phosphorus oxygen bond, suggesting that this bond is predominantly not cleaved. (b) At least at the present resolution, there is no significant deformation of the tetrahedral geometry at the phosphorus atom towards a trigonal bipyramidal arrangement. (c) The putative ‘catalytic’ water molecule is 3.3 Å away from the phosphorus atom. This distance is not significantly smaller than the sum of the van der Waals radii, if coordinate errors of the crystal structure are taken into account, but it is significantly larger than a typical phosphorus oxygen bond in DNA (∼1.6 Å). Therefore, it seems that the reaction has either not started in the crystal structure, or alternatively that it is trapped very early in the trajectory, which is consistent with the biochemical observation that Ca2+ ions in the active site support DNA binding, but not DNA cleavage. Why is the reaction blocked with Ca2+ ions in the active site and would proceed with Mg2+ ions in the active site? Typical oxygen ligand distances are much shorter for Mg2+ than for Ca2+ (2.1 Å versus 2.4 Å) (33), which might lead to a slight mispositioning of the nucleophilic water molecule in the complex with Ca2+. However, more sophisticated simulation studies attribute the different efficiencies of the Mg2+ and Ca2+ forms of REases to kinetic effects and not to the properties of the prereactive states of these enzymes (34,35).
DISCUSSION
Monomeric REases
Nearly all restriction enzymes that recognize palindromic or pseudopalindromic sequences form functional dimers or higher order assemblies of dimers that match the 2-fold symmetry or pseudosymmetry of their target sequence (35). Nevertheless, our suggestion that MvaI interacts with its (pseudo)palindromic target sequence as a monomer already has precedence among REases. MspI and HinP1I, which are specific for the palindromic sequences C/CGG and G/CGC, respectively, have both been reported to be monomers in solution (36–38).
In the case of MspI, the authors of the crystal structure have proposed several possible explanations for how a monomeric restriction enzyme can generate double-strand breaks in DNA. (a) Although monomers are present in the absence of DNA, there might be a monomer–dimer equilibrium in the presence of DNA. Therefore, an initial monomer–DNA complex might recruit another monomer for the cleavage reaction. (b) Alternatively, MspI might achieve double-strand DNA cleavage by bringing together two monomers bound on two separate recognition sites. (c) MspI might cut strands sequentially, possibly in a concerted manner by a flip of the enzyme around the DNA after the first cleavage reaction (36).
In the case of HinP1I, a conserved dimer is present in crystals of the apo-form and of the DNA complex, despite the predominantly monomeric nature of the enzyme in solution (37). This unusual back-to-back dimer can bind two copies of DNA, which are recognized asymmetrically and are in contact with one active site (38). As expected, only one strand was cleaved if these crystals were soaked with Mg2+ ions. In solution, HinP1I treatment of circular DNA with multiple recognition sites leads first to a nicked product, but linear DNA appears earlier than one might expect from a completely random nicking reaction, suggesting either that the likelihood of strand hydrolysis at any HinP1I site increases when the other strand is cleaved or that the enzyme remains in the vicinity of its site after hydrolysis and can rebind (38).
MvaI binds pseudosymmetric DNA asymmetrically
The results reported here show that MvaI is a monomer that recognizes its target DNA asymmetrically. The data explain the cleavage rate differences for the two DNA strands (8,9) and are consistent with a possible sequential cleavage model. As MvaI forms a ‘clamp’ around the DNA, rotation of MvaI around the long axis of DNA seems plausible (Figure 3B). However, such a rotation alone can never lead to a second productive binding event, simply because the two DNA strands run in opposite direction. To enable cleavage of the second strand by the same active site, a rotation around the 2-fold axis of pseudosymmetry perpendicular to the DNA is required to position the enzyme in a second catalytically productive orientation. The crystallographically observed hinge mobility might be important to straighten out the protein clamp, so that this rotation can proceed without clashes with the DNA. However, alternative models to explain the endonuclease activity of MvaI such as hairpin formation (39,40) cannot yet be ruled out.
Our biochemical and crystallographic results indicate that MvaI is monomeric in the presence and absence of DNA and conflict with a prior report that MvaI dimerizes in the presence of DNA (25). The crystal structure of the MvaI-DNA complex shows that two monomers of MvaI cannot simultaneously interact specifically with the target DNA, because they would compete for interactions with the same bases. Moreover, two MvaI monomers which would be related by the (pseudo) 2-fold symmetry of the target sequence would also clash in various other places (not shown). Nonetheless, if MvaI dimerization on the DNA is indeed relevant in some conditions, then hinge mobility of MvaI could explain how one MvaI monomer could recruit another monomer in a second productive orientation without clashes or competition for target bases.
MvaI generates a cleavage pattern with single nucleotide 5′-overhangs
To our knowledge, MvaI is the first crystallographically studied REase that generates products with single nucleotide 5′-overhangs. The overhangs are 1 nt shorter than the 2 nt overhangs generated by MspI and HinP1I. This emphasizes the versatility of monomeric REases with respect to the cleavage pattern. In principle, such monomeric REases could generate overhangs of any length, simply by asymmetrically binding a palindromic or pseudopalindromic recognition sequence with the center of symmetry or pseudosymmetry placed in the appropriate distance from the scissile phosphate bond. Note that although DNA recognition is asymmetric, the target sequence has to be symmetric or pseudosymmetric, simply because for double-strand breaks to occur, the DNA must be recognized in both orientations by the enzyme. The requirement for symmetry or pseudosymmetry of the target might be relaxed in cases with degenerate sequence recognition.
MvaI is barely similar to the neoschizomers EcoRII and PspGI
MvaI recognizes the same sequence as EcoRII, but generates single nucleotide 5′-protruding ends while EcoRII and PspGI produce five nucleotide 5′-overhangs. A comparison of the MvaI and EcoRII (41) protomers shows that these two enzymes are barely related [the DALI score 0.9 for the pairwise superposition is insignificant (42)] The similarity is restricted to the catalytic core, which is essentially conserved in all restriction enzymes. Moreover, PspGI and EcoRII are dimers and show similarities to the Ecl18kI enzyme specific for the /CCNGG sequence (5) rather than MvaI. This example indicates once more that cleavage stagger and not only the target sequence should be considered as the ‘phenotype’ of a restriction enzyme for the statement ‘phenotype predicts genotype’ (3) to hold true.
The catalytic domain of MvaI is similar to the catalytic domain of the DNA repair protein MutH
Weak sequence similarity at the PSI-BLAST level, which has been noted before (23), suggests that MvaI is related to MutH, a component of the DNA-repair machinery in bacteria, which acts as a nickase on hemi-methylated DNA (43). Despite the functional differences, there are many parallels between MvaI and MutH. Both proteins are two-lobed monomers and recognize almost symmetric target sequences asymmetrically. A comparison of the X-ray structures confirms the similarities, particularly in the vicinity of the active sites, but also reveals substantial differences in the recognition domains, which is not surprising because the two enzymes recognize unrelated target sequences (CC /WGG for MvaI versus /GATC for MutH). At the sequence level, MvaI is strikingly similar to BcnI, which recognizes the related sequence CC /SGG (S stands for C or G) and cleaves it like MvaI. In accompanying manuscripts, we present the crystal structures of BcnI with and without target DNA and compare them in detail with the corresponding structures for MvaI and MutH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Prof. Hans Bartunik for generous allocation of beamtime on BW6 (DESY, Hamburg) and Dr Gleb Bourenkov and Dr Galina Kachalova for assistance with data collection. We are grateful to AB ‘Fermentas’ for the MvaI RM system clone and NEB for the PspGI methylase clone. Research in the V.S. laboratory was supported by the Howard Hughes Medical Institute (HHMI) International Research Scholar grant # 55000336. M.B. thanks the European Molecular Biology Organization (EMBO) and HHMI for the Young Investigator award. H.C. is grateful to the Polish Ministry of Science and Higher Education for a POL-POSTDOC grant (PBZ/MEiN/01/2006/24). This work was supported by the European Commission 5th Framework Programme project ‘Center of Excellence in Molecular Bio-Medicine Contract no: QLK6-CT-2002-90363’ (Warsaw) and ‘Center of Exellence – Biocell’ Contract no: QLK2-CT-2002-30575 (Vilnius). The funds from the Polish Ministry of Science and Information Technology for the purchase of a crystallization robot (Decision Nr. 5210/IA/1789/2005) are gratefully acknowledged. The apo-MvaI and the MvaI–DNA structures will be available upon acceptance of the article from the Protein Data Bank (http://www.rcsb.org) under accession codes 2OA9 and 2OAA, respectively. Funding to pay the Open Access publication charge was provided by UNESCO/Polish Academy of Sciences Cellular and Molecular Biology Network.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman M, Lunnen K, Wilson G, Greci J, Schildkraut I, Phillips SE. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. Embo J. 1998;17:5466–5476. doi: 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. Embo J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butkus VV, Klimasauskas SJ, Janulaitis AA. Analysis of products of DNA modification by methylases: a procedure for the determination of 5- and N4-methylcytosines in DNA. Anal Biochem. 1985;148:194–198. doi: 10.1016/0003-2697(85)90645-1. [DOI] [PubMed] [Google Scholar]
- 7.Kubareva EA, Pein CD, Gromova ES, Kuznezova SA, Tashlitzki VN, Cech D, Shabarova ZA. The role of modifications in oligonucleotides in sequence recognition by MvaI restriction endonuclease. Eur J Biochem. 1988;175:615–618. doi: 10.1111/j.1432-1033.1988.tb14236.x. [DOI] [PubMed] [Google Scholar]
- 8.Pein CD, Cech D, Gromova ES, Orezkaya TS, Shabarova ZA, Kubareva EA. Interaction of the MvaI restriction enzyme with synthetic DNA fragments. Nucleic Acids Symp Ser. 1987;18:225–228. [PubMed] [Google Scholar]
- 9.Kubareva EA, Gromova ES, Pein CD, Krug A, Oretskaya TS, Cech D, Shabarova ZA. Oligonucleotide cleavage by restriction endonucleases MvaI and EcoRII: a comprehensive study on the influence of structural parameters on the enzyme-substrate interaction. Biochim Biophys Acta. 1991;1088:395–400. doi: 10.1016/0167-4781(91)90131-5. [DOI] [PubMed] [Google Scholar]
- 10.Gromova ES, Kubareva EA, Vinogradova MN, Oretskaya TS, Shabarova ZA. Peculiarities of recognition of CCA/TGG sequences in DNA by restriction endonucleases MvaI and EcoRII. J Mol Recognit. 1991;4:133–141. doi: 10.1002/jmr.300040405. [DOI] [PubMed] [Google Scholar]
- 11.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J Mol Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 13.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 14.Leslie AWG. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 http://www.esrf.eu/info/science/newsletter/jun01/autopx.html. [Google Scholar]
- 15.Collaborative Computational Project Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 17.Sheldrick GM. Macromolecular phasing with SHELXE. Zeitschrift für Kristallographie. 2002;217:644–650. [Google Scholar]
- 18.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 19.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones TA, Zou JY, Cowan SW, Kjeldgaard. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Bujnicki JM. In: Restriction Endonucleases: Structure, Function, and Evolution. Springer, Berlin Heidelberg. Pingoud AM, editor. Vol. 14. Berlin Heidelberg: Springer; 2004. pp. 63–94. [Google Scholar]
- 24.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ovetchkina LG, Popova SR, Zinoviev VV, Vaitkevicius D, Janulaitis A, Gorbunov JA, Malygin EG. Endonuclease MvaI dimerization induced by the oligonucleotide substrate. Biopolymery Kletka (Russ.) 1988;4:239–242. [Google Scholar]
- 26.Richardson JS. beta-Sheet topology and the relatedness of proteins. Nature. 1977;268:495–500. doi: 10.1038/268495a0. [DOI] [PubMed] [Google Scholar]
- 27.Ban C, Yang W. Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. Embo J. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 29.Chothia C, Janin J. Orthogonal packing of beta-pleated sheets in proteins. Biochemistry. 1982;21:3955–3965. doi: 10.1021/bi00260a009. [DOI] [PubMed] [Google Scholar]
- 30.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. II. The observed structures. J Mol Biol. 1994;236:1382–1400. doi: 10.1016/0022-2836(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 31.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 32.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding MM. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 8):1432–1443. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 34.Mordasini T, Curioni A, Andreoni W. Why do divalent metal ions either promote or inhibit enzymatic reactions? The case of BamHI restriction endonuclease from combined quantum-classical simulations. J Biol Chem. 2003;278:4381–4384. doi: 10.1074/jbc.C200664200. [DOI] [PubMed] [Google Scholar]
- 35.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Horton JR, Maunus R, Wilson GG, Roberts RJ, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 40.Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 41.Zhou XE, Wang Y, Reuter M, Mucke M, Kruger DH, Meehan EJ, Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J Mol Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 42.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 43.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 44.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Applied Crystallography. 1991;24:946–950. [Google Scholar]
- 45.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Kubareva EA, Gromova ES, Romanova EA, Oretskaia TS, Shabarova ZA. [Cleavage by restriction endonucleases MvaI and EcoRII of substrates modified in amino groups of heterocyclic bases] Bioorg Khim. 1990;16:501–506. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.