Abstract
Herein we present Gene-Collector, a method for multiplex amplification of nucleic acids. The procedure has been employed to successfully amplify the coding sequence of 10 human cancer genes in one assay with uniform abundance of the final products. Amplification is initiated by a multiplex PCR in this case with 170 primer pairs. Each PCR product is then specifically circularized by ligation on a Collector probe capable of juxtapositioning only the perfectly matched cognate primer pairs. Any amplification artifacts typically associated with multiplex PCR derived from the use of many primer pairs such as false amplicons, primer-dimers etc. are not circularized and degraded by exonuclease treatment. Circular DNA molecules are then further enriched by randomly primed rolling circle replication. Amplification was successful for 90% of the targeted amplicons as seen by hybridization to a custom resequencing DNA micro-array. Real-time quantitative PCR revealed that 96% of the amplification products were all within 4-fold of the average abundance. Gene-Collector has utility for numerous applications such as high throughput resequencing, SNP analyses, and pathogen detection.
INTRODUCTION
DNA analysis instruments are becoming increasingly more powerful in the capacity of sequence analysis. DNA resequencing microarrays (1,2) and high throughput parallel sequencing instruments (3,4) are currently used for whole genome analyses of low complexity genomes down to single nucleotide resolution. However, the human genome remains too large to access without complexity reduction by directed amplification of specific sequences. To match the throughput of these instruments, the amplification bottleneck needs to be addressed with more efficient technologies.
To increase assay throughput and allow for more efficient use of precious DNA samples, simultaneous amplification of many targets can be carried out by combining many specific primer pairs in individual PCRs (5,6). However, it is one of the crucial problems with PCR that when large numbers of specific primer pairs are added to the same reaction, both correct and incorrect amplicons are formed. At a later stage, this skews the uniformity of the products to the point where many amplicons drop out in favor of artifacts. Even with careful attention paid to the design of the primers, PCR is usually limited to 10–20 simultaneous reactions before yield and evenness is compromised by the accumulation of irrelevant amplification products (7,8). Therefore, large numbers of separate PCRs are typically performed whenever many genomic sequences need to be analyzed.
The correct amplicons in a multiplex PCR have a unique feature compared to the false ones in that their end sequences are composed of a cognate primer pair as apposed to a primer from one pair combined with a primer from another pair. The method we present herein takes advantage of this feature and specifically circularizes only the cognate paired ends through hybridization and ligation on a so-called Collector oligonucleotide probe. After the specific circularization reaction, two measures are used to enrich for circular DNA, exonuclease treatment for selective degradation of linear DNA and by rolling circle amplification. The method is thereby not limited by the primer cross-reaction-based amplification artifacts typically associated with multiplexed PCR.
Gene-Collector is related to the previously published Selector technology (9). Instead of circularizing multiplexed PCR-amplified DNA targets, the Selector technology circularizes specific genomic DNA targets derived from restriction enzyme digestions. As a consequence, the Selector technology requires a unique probe design for every specific set of target sequences (10), which renders it less modular in comparison to Gene-Collector, where new sets of Collector oligonucleotides can be mixed with any previously existing ones, making all Collector probes compatible with each other. We demonstrate the specificity and flexibility of Gene-Collector by multiplex amplification of 170 targets located in the coding regions of 10 human cancer genes: EGFR, AKT1, AKT2, APC, FRAP1, KRAS, MARK3, SMAD4, TGFBR2 and TP53.
MATERIALS AND METHODS
Oligonucleotide probes and target amplicons
All oligonucleotides were synthesized at the Stanford Genome Technology Center, see Supplementary Table 1 for primers, probes and target amplicon sequences. The thymidines were substituted with uracil bases in the Collector probes for degradation purposes by uracil-DNA glycosylase. However, this enzymatic procedure was later found not to be necessary and removed from the protocol.
Table 1.
Analysis of failed amplifications. One primer pair was incorrectly designed through human error and two target sequences lacked Collector probes as a negative control leaving a total of 167 amplicons with a chance of successful amplification. Quantitative PCR revealed at what stage the Gene-Collector protocol failed. The failure reason for the final four amplicons still remains unknown as no successful quantitative PCR primers could be designed
| Failures | Total | % Success | Fraction | |
|---|---|---|---|---|
| Targeted amplicons | 170 | |||
| Human design error | 1 | 169 | ||
| No collector probe | 2 | 167 | ||
| negative control | ||||
| Failed at Mux-PCR | 5 | 162 | 97% | 162/167 |
| Failed at ligation | 3 | 159 | 95% | 159/167 |
| Failed at final amplification | 5 | 154 | 92% | 154/167 |
| Unknown failures (no qPCR) | 4 | 150 | 90% | 150/167 |
| 2 with 75% GC |
Gene-Collector protocol
First, multiplex PCR was run in 50 μl with all 340 primers (170 pairs) at 100 nM concentration each using 10 units pfu polymerase in 1 × pfu buffer (Stratagene), 200 μM each dNTP and 200 ng human genomic DNA, at 95°C for 5 min—[(95°C for 30 s; 55°C for 2 min; 72°C for 8 min) × 8] followed by 72°C for 10 min. Excess primers were removed by the addition of exonuclease I and incubated for 30 min at 30°C, followed by removal of enzymes by a Qiagen PCR purification column. Amplicon circularization by ligation was performed on 20 nM of each collector probe in 1× Ampligase buffer (Epicentre), 5 units Ampligase, 5 units OptiKinase (USB), 1 mM ATP, 1 mM DTT at 37°C for 30 min—[(95°C for 30 s; 65°C for 2 min; 55°C for 1 min, 60°C for 5 min) × 10] in 50 μl. A combination of exonuclease I, exonuclease T7 gene 6 and λ-exonuclease reduced the amount of linear DNA during 45 min at 37°C and then stopped when heated for 20 min at 80°C. The circular DNA was concentrated by a second Qiagen PCR purification column eluted in the supplied elution buffer and set to evaporate for ∼45 min at 65°C. One microliter of the 10-fold concentrated circles were added to a 10 μl TempliPhi reaction (GE) supplemented with 10% DMSO and run at 30°C for 16 h, then inactivated at 65°C for 10 min.
Resequencing by hybridization
A 50-kb high-density DNA array was designed by Affymetrix to match the 10-gene reference sequence. The collector amplified product was purified in a PCR purification column (Qiagen). One hundred and fifty nanograms of purified product was fragmented, labeled and finally hybridized according to the protocol provided by Affymetrix (GeneChip CustomSeq Resequencing Array Protocol). The array was washed and stained using the Affymetrix GeneChip Fluidics Station 450 and scanned using GeneChip Scanner 3000 according to the protocol. The scanned probe array image was analyzed using Affymetrix GeneChip Sequence Analysis Software.
Quantitative PCR of amplicons
Ten microliter reactions containing 400 nM of qPCR primers specific for the individual amplicons with 2 μl of the TempliPhi reaction diluted 1000-fold in TE buffer were performed to assay their relative abundance. Bio-Rad Sybr Green master mix (1×) was used on an ABI 7900 instrument, see Supplementary Table 1 for primers.
RESULTS
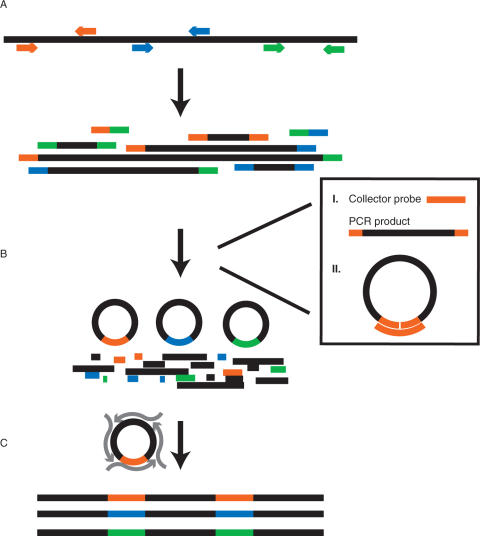
Coding-sequence-specific PCR primer pairs were designed using ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html) for 10 cancer genes, see Supplementary Table 1. The resulting 170 primer pairs were synthesized and pooled into one tube. A multiplexed PCR was then run for eight cycles using pfu polymerase which generates blunt-end PCR products suitable for circularization by ligation (11). Excess primers were then removed using a single strand-specific exonuclease followed by a Qiagen PCR product purification column. A pool of Collector probes, each specific to one correct amplicon then guided a circularization reaction of matched PCR primer pair ends and closed circles were formed by a DNA ligase enzyme. The ligation reaction also involved a pre-step at 37°C for phosphorylation of 5′-ends by a kinase enzyme prior to ligation. Circularization was then followed by the addition of an exonuclease cocktail to degrade linear DNA such as amplification artifacts, genomic DNA and excess Collector probes. The circularized sequences were finally amplified using hyper-branched rolling circle amplification with random hexamers and phi-29 polymerase, TempliPhi (12). An outline of the Gene-Collector procedure is displayed in Figure 1.
Figure 1.
Principles of Gene-Collector. (A) A multiplex PCR is carried out using target specific primer pairs, generating both correct and incorrect products. For clarity, only three of the 170 primer pairs are shown and are color coded. (B) Guided by the collector probe, targets that contain matched primer pairs are circularized, leaving non-cognate products linear and thus susceptible to exonuclease degradation. In detail, (I) a collector probe contains complementary sequences to a cognate primer pair (orange). (II) The collector probe and the DNA ligase enable circularization of correctly amplified targets. (C) A universal amplification is then carried out using a randomly primed rolling circle amplification, generating a final product of concatemers of correct target sequences.
The success rate of the amplification was assessed by hybridizing the final product on an Affymetrix custom-designed resequencing array containing probes scanning the coding sequence of these 10 genes with four variant probes for each nucleotide position, A, T, G and C. The array revealed that 90% of the target sequences had been successfully amplified as assessed by providing accurately read sequence for at least 30% of the nucleotides in each individual amplicon located in continuous stretches of sequence. The performance of the resequencing array itself will be reported elsewhere (Dahl et al. in preparation). Using real-time PCR with primers specific to the individual amplicons, we evaluated the failed amplifications and at which stage of the Collector protocol they had dropped out, see Table 1. Several sequences could probably be recovered through re-design of the initial multiplex PCR primers or by using prevalidated primer sets.
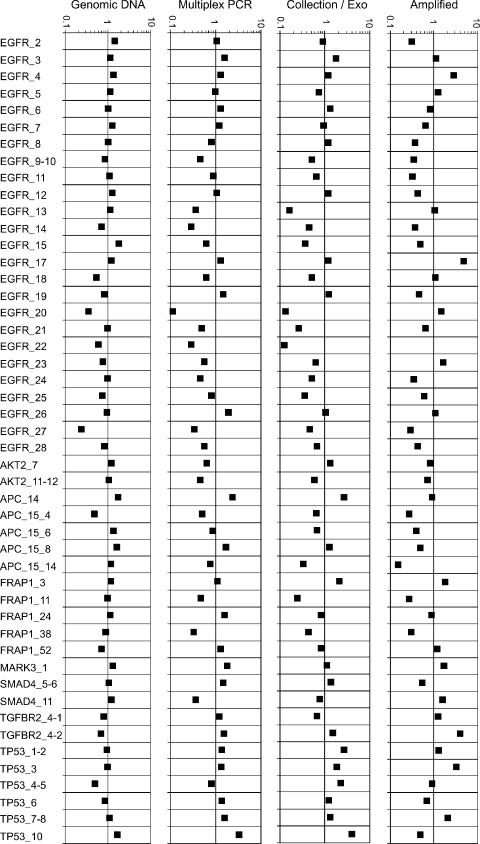
Uniform abundance of each product is an important feature of any multiplex amplification protocol, especially when used as a sample preparation step for the next generation high-throughput sequencing instruments, to avoid over- or under-sampling of target amplicons. The initial multiplex PCR is conducted under very non-stringent conditions in order to give all target sequences the best chance of efficient amplification. This would normally generate many amplification artifacts but these are efficiently removed by circularization and exonuclease degradation. To ensure uniformity of the multiplex-PCR, extension times were required to be long at 8 min, with primer hybridizations conducted at 55°C for 2 min. Each stage of the reaction was analyzed for evenness by quantitative PCR, see Figure 2. Surprisingly, some primer pairs which did not work in individual PCRs under standard conditions as analyzed by agarose gel, did produce the correct product with the Gene-Collector procedure (data not shown). The final amplification by TempliPhi was supplemented with a 10% final DMSO concentration to reduce the skewing effects of varied amplicon GC content. The average abundance of each final product was estimated to be at ∼10 nM in a 10 μl reaction volume with 96% of all amplicons having no less than one-fourth of the average abundance.
Figure 2.
Evenness measurements of the various stages of the Gene-Collector process assessed by quantitative PCR. A subset of 48 targets, all successfully amplified according to the resequencing array, was chosen to represent the overall variation in amplification efficiency. The starting material of human genomic DNA, assumed to be perfectly uniform, is compared to the evenness after the multiplex PCR, the ligation and exonuclease treatment and finally the rolling circle amplified material. The Y-axis represents a log-scale with deviations from 1 being relative differences from the average abundance. No compensation for differences in real-time PCR efficiency between reactions was used. However, the genomic DNA starting material represents a measure of this variation and the general imprecision of the real-time PCRs. Here, 96% of the final amplicons analyzed was no less than one-fourth of the average abundance.
In order to measure the levels of false amplification products generated by the Gene-Collector protocol, the final product was cloned and sequenced. The TempliPhi reaction produces concatemeric products of ∼10 kb each, which were fragmented by sonication, gel purified and cloned into a sequencing vector. When 96 colonies were picked and Sanger sequenced, 93 reads showed that 58% of the reads were of expected products, see Table 2. As cloning selects the sequence representation randomly, it provides an additional measure of frequency distribution. Most amplicons appeared only once showing even representation. Nine amplicons appeared twice and two of the targets three times. No non-specific products appeared more than once. The fraction of paired matched primers found among the non-specific products was much lower than for the specific ones. As can be seen in Table 2, few non-specific products were formed by two matched primer pairs amplifying a non-target sequence. This type of false product would still become circularized by the Collector probe but are not the main source of errors. A complete list of sequences is available in Supplementary Table 2. As expected from cloned rolling-circle-amplified material, many sequencing reactions produced concatemeric reads of repeated elements. Interestingly, this provided redundant sequencing within one and the same read with up to 3-fold coverage.
Table 2.
Analysis of amplification product by cloning and sequencing. From the 93 total reads produced, 58% of these were of the expected products. Primer sequences were only rarely found within the non-specific products either as single primers, non-matched pairs or as matched pairs suggesting that the TempliPhi reaction produced the majority of the artifacts or that they were simply caused by remaining genomic DNA
| Reads | % of total | Fraction | |
|---|---|---|---|
| Total sequence reads | 93 | ||
| Correct products | 54 | 58% | 54/93 |
| two matched primers | 52 | ||
| one primer | 52 | ||
| Non-specific products | 39 | 42% | 39/93 |
| two matched primers | 4 | ||
| one primer | 8 | ||
| two non matched primers | 2 | ||
| not found in human genome | 1 |
DISCUSSION
We have amplified all the coding sequences located in 10 cancer genes using a multiplexed procedure termed Gene-Collector. Resequencing of large numbers of cancer- related genes has recently shown to provide important biological insights into the disease (13). Even with extensive optimization, standard multiplex PCR is not a feasible approach to large-scale genetic studies as the failure rate is too high due to the many false amplicons out competing the correct ones for the amplification reagents. However, even though these false amplicons do result, the correct products are also present and at uniform abundance early in the amplification. Gene-Collector reduces the presence of false products enabling further amplification of the correct ones.
The presented initial multiplex PCR had very relaxed conditions in order to give all primer pairs the ability to hybridize through the use of low hybridization temperature and long duration. Polymerization of all templates was assured by a long extension time and an ample amount of DNA polymerase. This condition was suitable for all amplicons as the Collector procedure removes artifacts by exonuclease degradation. Primer-dimer artifacts, which are a major problem in traditional multiplexed PCR, are of little concern for Gene-Collector as the circularization process is impossible of such short DNA strands due to the lower limit size constraints of partially double stranded circular DNA (14).
Alternatively, one may use PCR in the final amplification of the circularized amplicons, which then gives distinct bands on standard agarose gel (Baner and Fredriksson in preparation). This version of the Gene-Collector protocol includes a general primer pair motif within the Collector probe and generates a purer product than the randomly primed RCA. This could, for example, be suitable for rapid multiplex pathogen detection using electrophoretic separation.
The relative abundance of products from the rolling circle reaction was very even. The rarely observed unevenness of this final product could be due to various factors. The lengths of amplicons spans from 160 to 800 bp and with varied GC content, possibly resulting in different circularization efficiency and/or final amplification efficiency. As only a few of the amplification artifacts found by cloning and Sanger sequencing contained a primer sequence, we believe these to be mainly associated with the randomly primed RCA which is known to also amplify linear DNA but with a much lower efficiency. The impurities may also be derived from remaining fragments of genomic DNA and if so, their relative presence should decrease with increased levels of multiplexing. Further improvement of the final product purity is desired for certain applications and is under development. One may also note that target sequences could be arrayed if the circularization is performed on immobilized Collector probes.
Gene-Collector should be of great value for a wide range of amplification-based applications, particularly in combination with highly parallel DNA analysis platforms. The level of further multiplexing achievable with the Gene-Collector protocol will probably be more limited by how many primer pairs one can use in the initial multiplex PCR then on the circularization process. One class of parallel DNA analysis is large-scale sequencing and resequencing platforms (15), such as sequencing by hybridization (1,2), sequencing by ligation (4) or sequencing by synthesis (3) systems. The Collector technology also displays promising properties to be combined with PCR-intense genotyping methods (7,16), like mini-sequencing (17,18) and primer extension-based methods in concert with mass spectrometry analysis (19), as well as high throughput pathogen detection. Gene-Collector could also be combined with genetic variation detection techniques that require many single PCRs (20,21) to increase assay throughput.
In summary, the presented multiplexed protocol enables analysis of small and precious sample materials, reduces enzyme consumption and offers higher throughput of DNA amplification.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
This work was supported by the Swedish Research Council, The Swedish Society for Medical Research, The Wenner-Gren Foundations, and the NIH (Center Grant 2P01HG000205). Special thanks to Keith Anderson and Mike Jensen at the Stanford Genome Technology Center for synthesis of oligonucleotides. Funding to pay the Open Access publication charge was provided by NIH (P01HG000205).
Conflict of interest statement. S.F. and F.D. are inventors on a patent application describing the published method.
REFERENCES
- 1.Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, et al. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 2.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 3.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005 doi: 10.1126/science.1117389. 1117389. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shigemori Y, Mikawa T, Shibata T, Oishi M. Multiplex PCR: use of heat-stable Thermus thermophilus RecA protein to minimize non-specific PCR products. Nucleic Acids Res. 2005;33:e126-. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syvanen AC. Toward genome-wide SNP genotyping. Nat. Genet. 2005;37(Suppl):S5–S10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 8.Broude NE, Zhang L, Woodward K, Englert D, Cantor CR. Multiplex allele-specific target amplification based on PCR suppression. Proc. Natl. Acad. Sci. USA. 2001;98:206–211. doi: 10.1073/pnas.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenberg J, Dahl F, Landegren U, Nilsson M. PieceMaker: selection of DNA fragments for selector-guided multiplex amplification. Nucleic Acids Res. 2005;33:e72. doi: 10.1093/nar/gni071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antson DO, Isaksson A, Landegren U, Nilsson M. PCR-generated padlock probes detect single nucleotide variation in genomic DNA. Nucleic Acids Res. 2000;28:E58. doi: 10.1093/nar/28.12.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Baner J, Nilsson M, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat. Rev. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 16.Syvanen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 17.Syvanen AC, Aalto-Setala K, Harju L, Kontula K, Soderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- 18.Pastinen T, Raitio M, Lindroos K, Tainola P, Peltonen L, Syvanen AC. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 2000;10:1031–1042. doi: 10.1101/gr.10.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by mass spectrometry. Mass Spectrom. Rev. 2002;21:388–418. doi: 10.1002/mas.1009. [DOI] [PubMed] [Google Scholar]
- 20.Faham M, Baharloo S, Tomitaka S, DeYoung J, Freimer NB. Mismatch repair detection (MRD): high-throughput scanning for DNA variations. Hum. Mol. Genet. 2001;10:1657–1664. doi: 10.1093/hmg/10.16.1657. [DOI] [PubMed] [Google Scholar]
- 21.Fakhrai-Rad H, Zheng J, Willis TD, Wong K, Suyenaga K, Moorhead M, Eberle J, Thorstenson YR, Jones T, Davis RW, Namsaraev E, Faham M. SNP discovery in pooled samples with mismatch repair detection. Genome Res. 2004;14:1404–1412. doi: 10.1101/gr.2373904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.