Abstract
A general consequence of pre-mRNA splicing is the stable deposition of several proteins 20–24 nucleotides (nt) upstream of exon–exon junctions on spliced mRNAs. This exon junction complex (EJC) contains factors involved in mRNA export, cytoplasmic localization, and nonsense-mediated mRNA decay. Here we probed the mechanism and timing of EJC assembly. Over the course of splicing, the 5′ exon is subject to numerous dynamic protein–RNA interactions involving at least nine distinct polypeptides. Within the fully assembled spliceosome, these interactions afford protection to the last 25–27 nt of the 5′ exon intermediate. Coincident with exon ligation, interactions at the 3′ end of the 5′ exon disappear, and new species associate with position −24. Mass spectrometry and Western blotting of purified H, C, and mRNP complexes revealed that at least one EJC component, REF/Aly, can interact with pre-mRNA prior to spliceosome assembly, whereas Y14, Magoh, RNPS1, UAP56, and SRm160 are found in intermediate-containing spliceosomes. Upon exon ligation, association of RNPS1, UAP56, and SRm160 is destabilized. In contrast, REF/Aly, Y14, and Magoh remain stably bound to spliced mRNA, indicating that these three proteins are components of the EJC core.
Keywords: Pre-mRNA splicing, spliceosome, exon junction complex, mRNP
Throughout their lifetimes in vivo, mRNAs exist as mRNA–protein particles (mRNPs). The associated proteins control every aspect of mRNA metabolism, from subcellular transport to translational efficiency to rate of decay. Exactly which proteins associate with a particular mRNA depends on its sequence, its subcellular localization, and its synthetic history. Furthermore, the complement of mRNP proteins evolves as the mRNA moves to different locations and is acted on by such processes as nuclear export and translation. Many proteins join the mRNP in the nucleus and then accompany it to the cytoplasm, where they influence subsequent mRNA metabolism (Shyu and Wilkinson 2000). A subset of such proteins are specifically loaded as a consequence of pre-mRNA splicing (Blencowe et al. 1998; Mayeda et al. 1999; Kataoka et al. 2000; Le Hir et al. 2000a,b; McGarvey et al. 2000; Zhou et al. 2000; Luo et al. 2001). These splicing-specific mRNP proteins constitute the exon junction complex (EJC), which associates with spliced mRNAs in a sequence-independent manner a fixed distance upstream of exon–exon junctions (Le Hir et al. 2000b). Using coimmunoprecipitation strategies, we and others recently reported that the EJC assembled in HeLa nuclear extract contains numerous epitopes, including those recognized by antibodies against REF/Aly, Y14, SRm160, DEK, RNPS1, UAP56, and Magoh (Blencowe et al. 1998; Mayeda et al. 1999; Kataoka et al. 2000, 2001; Le Hir et al. 2000a,b, 2001b; McGarvey et al. 2000; Zhou et al. 2000; Gatfield et al. 2001; Luo et al. 2001). When assembled in vivo, the EJC is additionally precipitated by antibodies against Upf2, Upf3, and the TAP:p15 heterodimer (Kim et al. 2001b; Le Hir et al. 2001a; Lykke-Andersen et al. 2001). Finally, REF/Aly, Y14, SRm160, RNPS1, Upf2, Upf3, and TAP were all present in mRNPs immunopurified from the nuclear fraction of COS cells with antibodies against a subunit of the nuclear cap binding complex (Lejeune et al. 2002).
The known EJC components function at multiple levels of mRNA metabolism. SRm160 and RNPS1 were originally characterized as activators of pre-mRNA splicing (Blencowe et al. 1998; Mayeda et al. 1999). REF/Aly and TAP:p15 are mRNA export factors, and their association with the EJC can facilitate nuclear export of spliced RNAs (Luo and Reed 1999; Zhou et al. 2000; Le Hir et al. 2001a). REF/Aly and TAP:p15 are recruited to spliced mRNAs through physical interactions with UAP56 (Luo et al. 2001; Strasser and Hurt 2001), a putative RNA helicase also implicated as a splicing factor required for early spliceosome assembly (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). Upf2 and Upf3 are factors required for nonsense-mediated mRNA decay (NMD), the process by which cells eliminate mRNAs containing incomplete open reading frames. In higher eukaryotes, termination codons are recognized as premature if they occur upstream of at least one exon–exon junction. By marking the positions of exon–exon junctions, the EJC serves as a conduit for communication between the splicing, translational, and degradative machineries (Kim et al. 2001b; Le Hir et al. 2001a; Lykke-Andersen et al. 2001). Finally, Magoh and Y14 (Mago Nashi and Tsunagi in Drosophila), form a tight heterodimer in vivo and are both required for the proper localization of oskar mRNA during Drosophila development (Hachet and Ephrussi 2001; Mohr et al. 2001). Because Magoh and Y14 are recruited to mRNAs by splicing and then travel to the cytoplasm as mRNP components (Kataoka et al. 2001; Le Hir et al. 2001b), information necessary for proper cytoplasmic mRNA localization is likely imprinted by splicing in the nucleus.
Because of its diverse constituents, the EJC is undoubtedly a central player in the metabolism of spliced mRNAs. It is therefore of interest to study both its structure and assembly. Here we used a variety of techniques to investigate RNA–protein interactions with the 5′ exon in both spliceosomes and spliced mRNPs. Unlike spliced mRNA, where the EJC protects only 8–10 nucleotides (nt) from RNase digestion (Le Hir et al. 2000b), protection of the 5′ exon intermediate within the C complex, the catalytically active form of the spliceosome, spans 25–27 nt. Photo-cross-linking experiments indicated that this entire region undergoes a series of dynamic protein–RNA interactions over the course of splicing that involves at least nine distinct polypeptides. Two of these cross-linked proteins were identified as SRp20 and REF/Aly. Finally, mass spectrometry and Western blotting of purified H, C, and mRNP complexes revealed that at least one EJC component, REF/Aly, joins the pre-mRNA as early as the H complex, a heterogeneous complex that forms on any largely single-stranded RNA, whereas others join prior to exon ligation. These results show that contacts between the spliceosome and the 5′ exon are much more extensive than previously thought. Furthermore, they provide a framework on which to build a more complete understanding of spliceosome–5′ exon interactions and EJC assembly.
Results
Extensive RNase protection of the 5′ exon intermediate
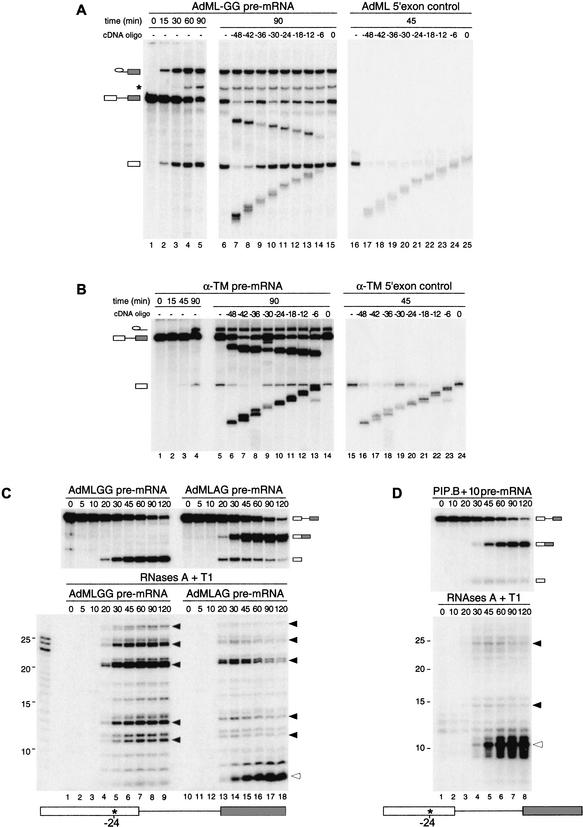
Using comparative RNase H and RNase A/T1 protection analysis of spliced and intronless control mRNAs, we previously demonstrated that 8 nt, including positions −20 to −24 relative to the exon–exon junction, become physically protected upon the completion of splicing (Le Hir et al. 2000b). To determine whether this protection is established prior or subsequent to exon ligation, we now report RNase H analysis of two pre-mRNAs that accumulate splicing intermediates. One RNA derived from the standard AdML splicing substrate (AdML-AG), but contains an AG → GG mutation at the 3′ splice site (AdML-GG; Gozani et al. 1996; Anderson and Moore 1997). The other (α-TM) derived from E2-I2-E3 of rat α-tropomyosin (Smith et al. 1989) and was truncated upstream of the 3′ splice site. Both alterations effectively block the second step of splicing (Fig. 1A, lanes 1–5, 1B, lanes 1–4), allowing accumulation of spliceosomes containing lariat intermediates and free 5′ exons.
Figure 1.
Splicing-dependent protection of 5′ exon intermediates from RNase cleavage. (A) Uniformly labeled AdML-GG pre-mRNA (lanes 1–15) or control 5′ exon RNA (lanes 16–25) was incubated under standard splicing conditions in HeLa cell nuclear extracts for the times indicated. Aliquots of these reactions were further incubated with the cDNA oligonucleotides indicated (lanes 7–15,17–25). RNAs were then separated by 15% denaturing PAGE. Each oligonucleotide was named according to its center position relative to the 5′ splice site, which was defined as 0. Splicing substrates, intermediates, and products are indicated on the left. The asterisk (*) denotes a lariat degradation product often observed in these reactions, which results from 3′–5′ exonucleolytic decay of the accumulated lariat intermediate (Moore and Sharp 1992). Note that in splicing reactions, the RNase H cleavage products migrating below the free 5′ exon derive from both pre-mRNA and free 5′ exons; therefore, 5′ exon intermediate protection is best monitored in splicing reactions by the proportion of intact free 5′ exon remaining after digestion. (B) Similar RNase H analysis using control 5′ exon and spliced α-TM RNAs. (C) Complete RNase digestion of singly labeled AdML RNAs. (Upper panel) Splicing time courses for AdML pre-mRNAs containing a single labeled phosphate at position −24 relative to the 5′ splice site (15% denaturing PAGE). Splicing substrates, intermediates, and products are indicated to the right. (Lower panel) Separation of protected fragments by denaturing PAGE (20%) after RNase A + T1 digestion. Closed arrows indicate fragments corresponding to the 5′ exon intermediate, whereas the open arrow denotes the fragment corresponding to spliced mRNA. (D) Same as C except using PIP.B+10 pre-mRNA containing a single labeled phosphate at position −24 relative to the 5′ splice site.
When incubated under splicing conditions and then probed with a series of 12-nt cDNA oligonucleotides, RNAs corresponding to both 5′ exons not produced by splicing (5′ exon controls) were substantially digested at every position tested (Fig. 1A,B, right panels). In contrast, both 5′ exon intermediates produced by splicing were fully protected from RNase H degradation by all oligonucleotides centered from positions −6 to −24 (middle panels). The AdML-GG 5′ exon intermediate was further protected from digestion by oligonucleotides centered at positions −30 and −36. Unspliced pre-mRNAs in the same reactions also showed some resistance to the same oligonucleotides, but in no case was this protection as significant as for the 5′ exon intermediate. These results are consistent with Figure 1A of Le Hir et al. (2000b), where it can be seen that unspliced TPI pre-mRNA showed no specific resistance to RNase H cleavage. Yet, pre-mRNA molecules that had undergone the first step of splicing exhibited extensive protection, with the last 24 nt of the 5′ exon intermediate (positions −1 to −24) being fully resistant to digestion. Thus, by the time of splicing intermediate formation, stable contacts are established both to the region protected by the EJC in spliced mRNA and along the entire downstream length of the 5′ exon. Furthermore, this extensive 5′ exon intermediate protection is a general phenomenon exhibited by multiple splicing substrates.
To map the extent of 5′ exon intermediate protection more precisely, we next performed RNase A and T1 digests using RNAs site-specifically labeled at position −24 (Fig. 1C). A similar experiment previously revealed that the splicing-dependent, 8-nt protection of AdML mRNA can withstand quite stringent RNase digestion (Le Hir et al. 2000b). Digestion of reactions containing singly labeled AdML-GG pre-mRNA yielded no protected fragments prior to the onset of splicing (Fig. 1C, lanes 1–3,10–12). However, as the 5′ exon intermediate accumulated in undigested samples (Fig. 1C, upper left panel), multiple RNase-resistant fragments accumulated concomitantly in the digested samples (Fig. 1C, lower left panel). The major species were ∼11, ∼13, ∼21, and ∼24 nt, although a protected fragment of ∼27 nt was also clearly visible. These same species were also present in reactions containing AdML-AG pre-mRNA, which undergoes both steps of splicing (Fig. 1C, right panel). In that case, the bands appeared coincident with the 5′ exon intermediate and subsequently disappeared as the 5′ exon was incorporated into mRNA. At the same time, the 8-nt fragment characteristic of spliced mRNA appeared. When the same analysis was performed on a different pre-mRNA (PIP.B+10), we observed two major digestion fragments of ∼15 and ∼25 nt that followed the appearance and disappearance of the 5′ exon intermediate, and a 10-nt fragment that accumulated concomitant with the appearance of spliced mRNA (Fig. 1D).
Several conclusions can be drawn from the above data. First, because no fragments resistant to RNases A and T1 were observed prior to appearance of the 5′ exon intermediate, stable interactions with the −24 region likely form coincident with the first step of splicing. Second, at the splicing intermediate stage, at least 25–27 nt at the 3′ end of the 5′ exon are engaged in these interactions. Third, because protection of this region was observed with several different splicing substrates, these interactions are both a general consequence of the first step of splicing and independent of the 5′ exon sequence.
Multiple proteins cross-link along the 5′ exon during the course of splicing
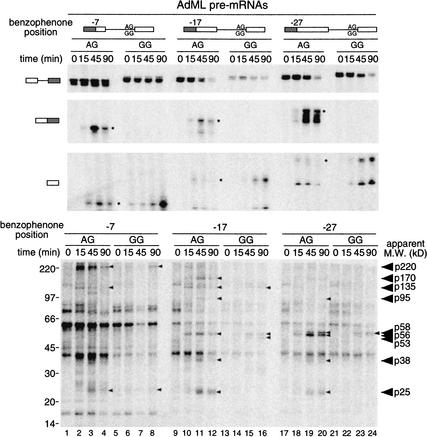
Having demonstrated that 25–27 nt of the 5′ exon intermediate is resistant to RNase degradation (Fig. 1), we next undertook a comprehensive cross-linking strategy to identify the species responsible. We constructed a series of AdML substrates containing a single 32P-label and adjacent photoreactive benzophenone group at three different positions (−7, −17, and −27) within this region. This was accomplished by ligating the same 10-nt oligonucleotide containing a single convertible adenosine as the penultimate nucleotide to three different transcripts initiating at positions −5, −15, and −25 (see Materials and Methods). These modifications were introduced into both the AdML-AG and AdML-GG constructs, allowing us to detect species that cross-link both before and after the second step of splicing. Each substrate was incubated under standard splicing conditions for the times specified, irradiated on ice at 302 nm (the appropriate wavelength for benzophenone activation) and subjected to RNase A degradation followed by SDS-PAGE (Fig. 2). No cross-linked bands were observed when the RNAs lacked a benzophenone moiety or when reactions were treated with proteinase K (data not shown). Therefore, all bands in the lower panel of Figure 2 represented proteins that cross-linked to the photoreactive group at the position indicated.
Figure 2.
Splicing and cross-linking of benzophenone-containing AdML RNAs. (Upper panels) Splicing time courses. RNAs were incubated for the times indicated and separated by denaturing PAGE (7%). Substrate and product structures are indicated above and to the left of each panel. Because pre-mRNAs contained only one 32P label, only a subset of splicing products are detectable. The appearance of doublets for each substrate and product reflects the presence of unligated 3′ RNAs. (Lower panel) Protein cross-linking. After incubation on ice for times indicated, samples were irradiated at 302 nm and digested with RNase A. Molecular mass standards and apparent molecular masses of proteins that cross-linked in a splicing-dependent manner are indicated to the left and right, respectively. Note that lanes 13–16 contained somewhat less input RNA, and thus exhibit reduced cross-linking intensities compared to other samples.
Comparison of the cross-linking time courses revealed numerous labeled bands that appeared with kinetics similar to those of splicing intermediates or products, but were absent prior to spliceosome assembly (Fig. 2, cf. 15-, 45-, and 90-min time points to 0 time point). The nine most distinct such bands had apparent molecular masses of ∼220, ∼170, ∼135, ∼95, ∼58, ∼56, ∼53, ∼38, and ∼25 kD. Other possible splicing-specific cross-links were either less intense or not consistently observed in all experiments (data not shown). It should be noted that cross-links to the GG constructs were generally less intense than those to the AG constructs—except as noted below and in the figure legend, these intensity differences can be explained by the somewhat lower splicing efficiencies of the GG constructs. Three prominent bands with apparent molecular masses of 60, 40, and 18 kD were detectable at all incubation times, suggesting that these proteins interact with the 5′ exon independent of spliceosome assembly.
This cross-linking analysis yielded information as to the location of each protein–RNA interaction along the 5′ exon, as well as its timing relative to the two steps of splicing. According to the intensity of the observed cross-links, p220 likely resides closest to position −7, as it did not yield significant cross-links at either −17 or −27. In contrast, p170 and p53 cross-linked most strongly at position −17, whereas the signal for p58 was most intense at position −27. Cross-linking of p56 was strongest at −27, but also readily apparent at −17. Finally, several proteins (p135, p95, p38, and p25) cross-linked most strongly at position −17, but also significantly at one or both of the other positions.
With regard to timing, the splicing-dependent cross-links fell into three distinct classes. The first class (p220 and p135) consisted of cross-links that appeared coincident with the first step of splicing in AdML-AG reactions (cross-links strongest at 15 min), but disappeared as the second step proceeded. As expected, the intensities of p220 and p135 cross-links increased as the 5′ exon intermediate accumulated with the AdML-GG substrate. Cross-links in the second class (p58, p56, and p25) appeared coincident with the first step of splicing and remained associated with spliced mRNA. These cross-links accumulated on both the AdML-AG and AdML-GG substrates. This class also likely contains p53, although p53 cross-linking to AdML-AG was not clearly detectable. The third class (p170, p95, and p38) comprised cross-links that appeared coincident with the second step of splicing only. These species accumulated on AdML-AG at later time points, but were not apparent with AdML-GG.
Because their migrations in SDS-PAGE were so similar, we thought it possible that p53, p56, and p58 might represent different phosphorylation states of the same protein. To test this, we treated cross-linking reactions with calf intestinal phosphatase prior to gel electrophoresis. Although this treatment did alter the migration of several other bands, no effects were observed on any cross-linked species in the 50–60-kD range (data not shown). Therefore, we conclude that p53, p56, and p58 are likely different polypeptides.
In summary, this analysis revealed that at least nine proteins interact with the 25–27-nt protected region of the 5′ exon over the course of splicing. Each protein appears to associate preferentially with a particular position on the 5′ exon. Furthermore, the interactions are quite dynamic, with the complement of proteins changing significantly as splicing proceeds.
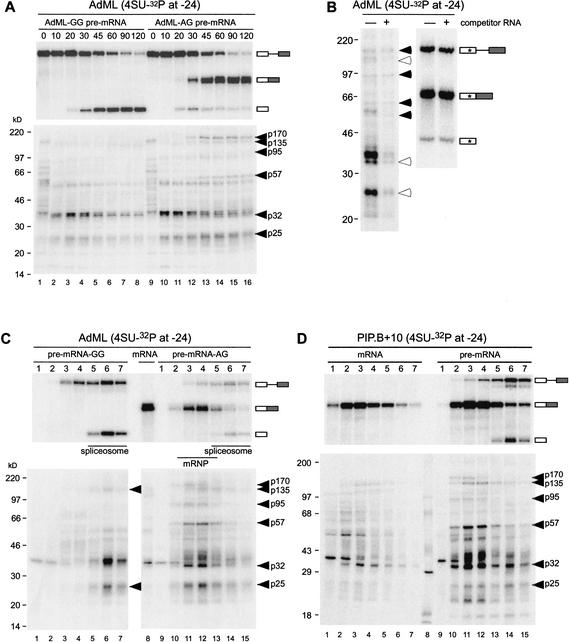
Because many of the above splicing-dependent cross-links were most intense at either position −17 or −27, we chose to extend our analysis by incorporating a different photoactivable group (4-thioU) at position −24 of the AdML-AG, AdML-GG, and PIP.B+10 substrates. In comparison to benzophenone, which can sweep out a radius of up to 15 Å, 4-thioU should only cross-link to those species most closely associated with the RNA (Moore and Query 1998). Paralleling the benzophenone experiments, we first performed splicing and cross-linking time courses for the 4-thioU modified AdML-GG and AdML-AG substrates to reveal proteins that cross-link before and after exon ligation, respectively (Fig. 3A). As a further measure of specificity, we performed cross-linking with AdML-AG–24S in the absence or presence of a 100-fold molar excess of cold competitor (Fig. 3B). To determine which cross-linked bands represented proteins associated with spliceosomes and/or mRNP, we subjected all cross-linking reactions to glycerol gradient fractionation (Fig. 3C,D). In the case of PIP.B+10 (Fig. 3D), we compared the proteins that cross-linked as a consequence of splicing (Fig. 3D, right panel) to those that cross-linked under identical conditions to an intronless control mRNA (Fig. 3D, left panel).
Figure 3.
Splicing and cross-linking of 4-thioU-containing RNAs. (A, upper panel) Splicing time courses of AdML-GG and AdML-AG RNAs containing single 4-thioU and 32P labels at position −24. Splicing precursors, intermediates, and products are indicated to the right. (Lower panel) Pattern of cross-linked proteins for each time point in upper panel. Molecular mass standards and apparent molecular weights of proteins cross-linking in a splicing-specific manner are indicated to the left and right, respectively. (B) Effects of nonspecific competitor RNA on cross-linking pattern for AdML-AG RNA. (Left panel) Following a 60-min incubation under splicing conditions, excess cold competitor RNA was added and reactions incubated for an additional 10 min before UV irradiation and RNase digestion. Open and closed arrows indicate bands that were sensitive or not, respectively, to addition of cold competitor RNA. (Right panel) Denaturing PAGE (15%) of splicing precursors and products in the presence (+) and absence (−) of cold competitor RNA showing that cross-linking differences in the left panel do not simply reflect a difference in the extent of splicing. (C) Glycerol gradient fractionation of splicing reactions containing site-specifically modified AdML-GG (lanes 1–7) or AdML-AG (lanes 9–15) substrates. Substrates were incubated for 90 min (AdML-GG) or 120 min (AdML-AG) and irradiated on ice prior to being loaded onto a 10%–30% glycerol gradient. Following glycerol gradient fractionation, fractions (numbers at top) were divided and either resolved by denaturing PAGE (upper panels) or RNase A digested and separated by SDS-PAGE (lower panels). Lane 8 corresponds to AdML control mRNA incubated for 45 min under splicing conditions, irradiated on ice, and directly resolved by denaturing PAGE (upper panels) or RNase A digested and separated by SDS-PAGE (lower panels), without prior glycerol gradient fractionation. Molecular mass markers and apparent molecular masses of splicing specific cross-linked proteins are indicated to the left and right of each gel, respectively. (D) Same as C except with site-specifically modified PIP.B control mRNA (lanes 1–7) and pre-mRNA (lanes 9–15).
Many of the proteins cross-linking to 4-thioU (Fig. 3) exhibited similarities in terms of their apparent molecular weights and timing of appearance/disappearance to the classes of cross-links observed with the benzophenone substrates (Fig. 2). For instance, as in the benzophenone reactions, two proteins with apparent molecular masses of 170 and 95 kD cross-linked only to the 4-thioU RNAs capable of undergoing the second step of splicing and not to AdML-GG or control PIP.B+10 mRNA. Also in this class (the third class above) for the 4-thioU reactions was a protein of 57 kD apparent molecular mass. The cross-linking intensity of these three bands was unaffected by the addition of cold competitor RNA (Fig. 3B), suggesting that p170, p95, and p57 are all stably associated with the spliced mRNA. Because p57 behaved differently (i.e., it cross-linked only after exon ligation) from the p53, p56, and p58 observed in the benzophenone reactions, we suspect that p57 is distinct from the other 50–60-kD proteins.
In contrast to the above proteins, addition of cold competitor did decrease the intensity of p135 cross-linking (Fig. 3B). Furthermore, p135 appeared to cross-link to the unspliced PIP.B+10 control mRNA (Fig. 3D, left panel), and the fractionation pattern of p135 cross-links did not exactly parallel that of spliced mRNA (Fig. 3C,D, right panels). Taken together with the benzophenone data, these results suggest that p135 interacts with a sequence element present in the 5′ exons of both AdML and PIP.B+10, but it does not bind specifically to mRNA as part of the EJC. The cross-linking patterns of two other bands, p32 and p25, are discussed below.
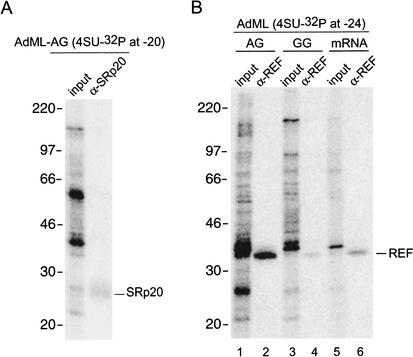
Identification of two cross-linked species as SRp20 and REF/Aly
To identify the cross-linked proteins, we undertook a series of immunoprecipitation experiments with antibodies against known splicing factors and proteins previously reported to be EJC components. Immunoprecipitations were carried out after cross-linked samples had been treated with both heat and SDS to promote protein denaturation (see Materials and Methods). Of the numerous antibodies we tested (against Y14, SRp20, REF/Aly, UAP56, SRm160, DEK, and RNPS1), only those recognizing Y14, SRp20, and REF/Aly clearly precipitated their cognate proteins under the conditions used in Figure 4. Contrary to expectations, although a polyclonal α-Y14 antiserum efficiently precipitated denatured, in vitro translated Y14, this antiserum failed to precipitate any species in the appropriate molecular-mass range (20–26 kD) cross-linked to either benzophenone- or 4-thioU-containing RNAs (data not shown). Instead, we were able to identify the prominent band of ∼25 kDa (p25) that cross-linked to 4-thioU-containing RNAs as SRp20 (Fig. 4A; data not shown). Consistent with the benzophenone data showing that p25 cross-linked at positions −7, −17, and −27 (Fig. 2), this protein also cross-linked to 4-thioU at both positions −24 and −20 (Figs. 3, 4A). In the benzophenone reactions (Fig. 2), p25 cross-links appeared to be splicing-dependent. However, in the 4-thioU time courses (Fig. 3A), cross-linked p25 was detectable (although weakly) even at 0 min, prior to spliceosome assembly. Consistent with the known ability of SRp20 to interact with RNAs independent of splicing (Cavaloc et al. 1999; Schaal and Maniatis 1999), p25 cross-links were also weakly detectable in the PIP.B+10 intronless control mRNA fractionations (Fig. 3D, left). Because the intensity of p25 cross-links increased upon splicing (Figs. 2, 3A,D), it seems reasonable to conclude that although SRp20 can bind the 5′ exon independent of splicing, this interaction is somehow stabilized by the splicing process.
Figure 4.
Identification of two cross-linked proteins as SRp20 and REF/Aly. (A) AdML-AG pre-mRNA containing a single 4-thioU and 32P label at position −20 was incubated under splicing conditions for 60 min prior to UV irradiation. Cross-linked samples were denatured by boiling in the presence of 0.1% SDS, digested with RNase A, and then immunoprecipitated with α-SRp20 antiserum. Similar results were obtained with AdML-AG containing 4-thioU at position −24 (data not shown). (B) Same as A except AdML-AG, AdML-GG, and control mRNA substrates containing a single 4-thioU and 32P label at position −24 were incubated with α-REF antiserum.
The p32 visible in all 4-thioU cross-linking reactions was identified as REF/Aly (Fig. 4B). Consistent with its previous identification as a component of the EJC (Le Hir et al. 2000b; Zhou et al. 2000), REF/Aly cross-linked most intensely to AdML-AG (Fig. 4B, lane 2). However, REF/Aly also cross-linked to both AdML-GG and intronless control AdML mRNA (Fig. 4B, lanes 4,6), albeit with diminished intensities. Furthermore, a p32 cross-linked band was clearly visible in the fractionation experiment containing intronless control PIP.B+10 mRNA (Fig. 3D). Thus, REF/Aly is capable of binding RNA in the absence of splicing (H complex), but its association with the −24 region is apparently strengthened upon exon ligation.
Mass spectrometric and Western analyses of spliceosomes and spliced mRNPs
The above identification of REF/Aly as a protein capable of associating with RNA in the absence of splicing is consistent with our recent finding that REF/Aly is a component of affinity-purified H complex (Jurica et al. 2002). In that paper, we reported purification of both H and C complexes from HeLa nuclear extract and complete mass spectrometric analysis of each. Purifications were carried out in 150 mM KCl and 5 mM EDTA to minimize the binding of nonspecific proteins as well as aggregation of purified complexes. Of the previously reported EJC components (see above), only REF/Aly was detected in the H complex, whereas REF/Aly, Y14, Magoh, and SRm160 were all clearly present in the C complex (Table 1; Jurica et al. 2002). Thus whereas REF/Aly can associate with pre-mRNA independent of splicing, other EJC components join the splicing machinery at some later point prior to exon ligation.
Table 1.
Mass spectrometric data for EJC proteins
| Protein
|
MW
|
AdML H complexa
|
AdML C complexa
|
AdML mRNP
|
PIP85.B mRNP
|
|---|---|---|---|---|---|
| REF/Aly | 26.9 | 1 | 1 | 2 | 2 |
| Y14 | 19.9 | 3 | 6 | 9 | |
| Magoh | 17.1 | 1 | 5 | 7 | |
| RNPS1 | 34.2 | 5 | 4 | ||
| SRm160 | 93.5 | 1 | |||
| SRp20 | 19.3 | 3 | 2 |
Numbers indicate unique tryptic peptides identifying that protein. No unique peptides were found for DEK or UAP56.
Data from Jurica et al (2002).
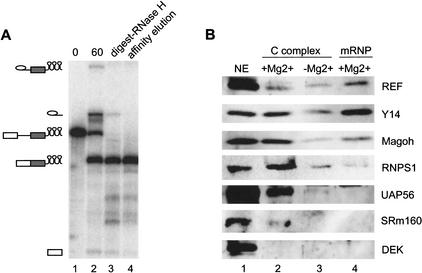
Here we report similar purification and mass spectrometric analysis of two spliced mRNPs: AdML and PIP85.B. For this purpose, MS2 sites were incorporated into the 3′ exon of each pre-mRNA substrate, allowing purification by MS2-MBP affinity chromatography (see Materials and Methods). Pilot studies revealed that mRNPs purified in the presence of EDTA contained very little protein (data not shown), indicating that the structural integrity of spliced mRNP is strongly Mg2+-dependent. Thus, both mRNP purifications were carried out in 2 mM Mg2+ without EDTA, but still in the presence of 150 mM KCl. AdML and PIP85.B mRNPs purified in this way were essentially free of pre-mRNA and splicing intermediates (Fig. 5A; data not shown), and Coomassie G staining showed that both mRNPs had defined protein compositions that were distinct from those of purified C and H complexes (data not shown).
Figure 5.
Affinity purification and Western analysis of spliced mRNP. (A) Denaturing gel (15% polyacrylamide) following labeled RNAs during splicing (0 and 60 min, lanes 1,2), RNase H digestion (lane 3), and affinity elution (lane 4). Positions of MS2-tagged pre-mRNA, intermediates, and spliced mRNA are indicated to the left. (B) Western analysis of purified mRNP and C complexes (+/−Mg2+). Lane 1 contains 1 μL of HeLa nuclear extract. Lanes 2–4 each contains ∼120 fmole of the indicated complex as estimated from the specific activity of splicing intermediates (C complexes) or mRNA (mRNP).
When analyzed by mass spectrometry, several peptides uniquely identifying REF/Aly, Y14, Magoh, and RNPS1 were detected in both spliced mRNPs (Table 1). Thus, these four proteins clearly associate with spliced mRNA, in agreement with previous reports. Additionally, SRp20 was detected in both spliced mRNPs. Its absence from the H and C complexes likely reflects a dependence on Mg2+ for binding, as no SR proteins were detected in either complex purified in the presence of EDTA (Jurica et al. 2002). Surprisingly, no peptides identifying SRm160, DEK, or UAP56 were observed in either spliced mRNP. Furthermore, neither DEK nor UAP56 was detected in our previous mass spectrometry analysis of the C complex (Jurica et al. 2002). There are several reasons that these proteins might have escaped detection here. First, some EJC components may dissociate under the conditions of our purification. Consistent with this was the observation that more proteins are associated with spliced mRNP in the presence of Mg2+ than in its absence (see above). Also, it should be noted that the buffer conditions used here differed somewhat from those used in our previous immunoprecipitation analyses (Le Hir et al. 2000b, 2001a). Second, the mass spectrometry technique we used entails analysis of SDS-PAGE gel slices, each containing numerous proteins; because of the complexity of the samples, it is possible that some proteins were simply missed, particularly in crowded regions of the gel. Third, some proteins are less amenable to mass spectrometry because they contain interfering protein modifications or few unique tryptic peptides.
To determine whether additional EJC components could be detected by an alternate method, and to confirm the mass spectrometry results, we performed Western blot analyses on purified AdML complexes. For better comparison between the C complex and mRNP, we included C complex purified in the presence of Mg2+, as well as in its absence. All complexes were loaded in equimolar amounts, as estimated by the specific activity of the in vitro-transcribed RNA contained within. REF/Aly, Y14, and Magoh were all evident in both C complex preparations, as well as in spliced mRNP (Fig. 5B). Thus, in agreement with the mass spectrometry results, these three EJC components all clearly join the splicing machinery prior to exon ligation and remain associated with spliced mRNA.
Although DEK was readily detectable in nuclear extract by Western blotting, it could not be detected in either the C complex or spliced mRNP. Taken together with the mass spectrometry results, we therefore conclude that DEK is not a stably bound EJC component. However, we and others previously reported that antisera against DEK could specifically coimmunoprecipitate spliced mRNA (Le Hir et al. 2000b, 2001a; McGarvey et al. 2000). Thus, either the association of DEK with splicing complexes is not stable enough to withstand the purification conditions used here, or the DEK antiserum recognizes an alternate epitope in the EJC. Consistent with the latter possibility, during our attempts to identify proteins that cross-link to the 5′ exon (see above), we observed that anti-DEK antisera precipitated an unidentified ∼20-kDa cross-linked protein (data not shown). Furthermore, recent work from the Maquat laboratory in which they analyzed the protein complement of endogenous nuclear mRNPs from COS cells indicated that although most of the other known EJC components were detectable in these complexes, DEK was not (Lejeune et al. 2002).
Western blotting confirmed that RNPS1 was present in spliced mRNP, but not in C complex purified in the absence of Mg2+. However, RNPS1 was readily detectable in C complex purified with Mg2+ (Fig. 5B). Thus, RNPS1 does associate with the spliceosome prior to exon ligation, but its binding is sensitive to buffer conditions. Similar sensitivities were observed for SRm160 and UAP56, both of which were readily apparent in C complex purified in the presence of Mg2+, but undetectable (SRm160) or barely visible (UAP56) in C complex purified in 5 mM EDTA. Furthermore, neither SRm160 nor UAP56 was discernable in spliced mRNP. We have subsequently learned that UAP56 association with spliced mRNP is quite sensitive to salt concentrations in excess of 60 mM (Z. Zhou, R. Reed, pers. comm.). Our observation that the association of SRm160 and UAP56 with C complex is less salt-sensitive than their associations with mRNP (Fig. 5B, cf. lanes 2 and 4) suggests that some structural alteration destabilizes their binding upon exon ligation. It is tempting to speculate that this is the same structural alteration that stabilizes REF/Aly association with the mRNP, as shown above.
Discussion
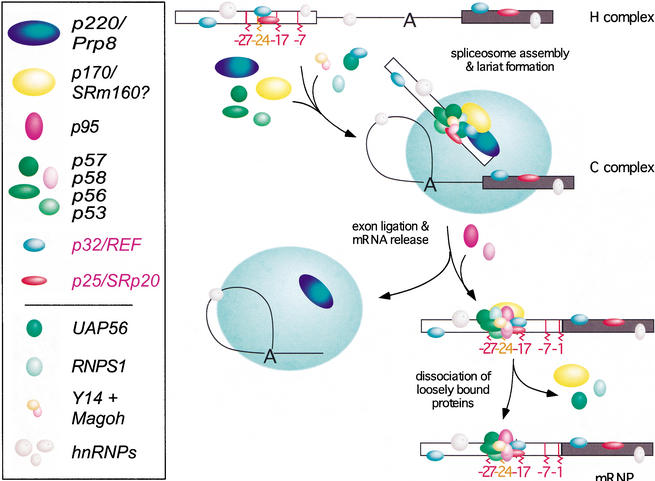
In this paper, we have begun to investigate the mechanism and timing of EJC assembly on spliced mRNA. Using a combination of RNase protection and photo-cross-linking assays, we showed that the 5′ exon is subject to numerous dynamic protein–RNA interactions over the course of splicing (Fig. 6). These interactions afford significant protection to the last 25–27 nt of the 5′ exon intermediate. Thus, between the two steps of splicing, interactions between the 5′ exon and the splicing machinery are much more extensive than originally thought. In the C complex we could detect at least nine proteins that cross-link across this region in a splicing-dependent manner. Coincident with exon ligation, interactions at the 3′ end of the 5′ exon disappear, and three new proteins (p170, p95, and p57) come into close proximity to position −24. Two other proteins that cross-link to this position in both unspliced and spliced RNAs were identified as REF/Aly and SRp20. Mass spectrometry and Western blotting confirmed that REF/Aly can interact with pre-mRNA prior to spliceosome assembly, whereas other known EJC components (Y14, Magoh, RNPS1, UAP56, and SRm160) are found in intermediate-containing spliceosomes. Upon exon ligation, association of RNPS1, UAP56, and SRm160 is destabilized, while REF/Aly, Y14 and Magoh remain stably bound to the spliced exons.
Figure 6.
A model for EJC assembly. EJC assembly is initiated as early as the H complex, when REF and SRp20 become associated with the 5′ exon. Additional EJC components join the complex prior to exon ligation. As the splicing reaction proceeds through exon ligation and mRNP release, at least three additional proteins become closely associated with the RNA, while other factors may remain with the spliceosome. Association of some EJC proteins becomes less stable after mRNP release. Proteins identified by cross-linking are named according to their apparent molecular weights, whereas p32/REF and p25/SRp20 were identified by denaturing immunoprecipitation. Proteins lacking a molecular-mass designation (bottom section) were shown to be present in the C complex or spliced mRNP by mass spectrometry and/or Western analysis.
Protection of the 5′ exon intermediate is extensive
Until recently, the splicing machinery was thought to make only minimal contacts with the 5′ exon. Other than a weakly conserved AG as the 3′-terminal dinucleotide, there are no apparent 5′ exon consensus sequences. The AG is complementary to C9 and U10 of U1 snRNA, and may thus contribute to early 5′ splice site recognition in the E/CC (early or commitment) complex (Seraphin and Kandels-Lewis 1993). Upon U4/U6.U5 tri-snRNP addition to form the B complex, a conserved loop in U5 snRNA (loop I) forms 2–3 promiscuous base pairs with the terminus of the 5′ exon, and strengthening this pairing can facilitate use of otherwise nonconsensus 5′ splice sites (Newman and Norman 1991; Wyatt et al. 1992). In Saccharomyces cerevisiae, the 5′ exon–U5 snRNA interaction is thought important for both retention of the 5′ exon intermediate and alignment of the 5′ and 3′ exons prior to exon ligation (Newman and Norman 1992). In mammalian extracts, however, U5 snRNA loop I can be deleted entirely with no ill effect on either step of splicing (O'Keefe et al. 1996; Segault et al. 1999). Thus, the mammalian spliceosome must use interactions in addition to any base pairings with U5 snRNA to maintain its grip on the 5′ exon intermediate and properly align the second-step reactants.
Although there are no conserved sequence elements in the 5′ exon beyond position −2, it has recently become apparent that the 5′ exon does become increasingly protected as spliceosome assembly proceeds. U1 snRNP addition to form E/CC complex results in protection of the last 5 nt, and subsequent U4/U6.U5 tri-snRNP addition extends the footprint out to 15 nt (Maroney et al. 2000). By the time of lariat formation, we find that the 5′ exon footprint is 25–27 nt long (Fig. 1C,D). Thus, it is now clear that much more of the 5′ exon is engaged in stable interactions with the splicing machinery than previously supposed. Yet, most of these 5′ exon interactions are clearly dispensable for splicing in vitro. In both yeast and human splicing extracts, no decrease in first-step splicing efficiency is observed when the 5′ exon is truncated to 2 nt, and even a 1-nt exon can support some lariat formation (Duchene et al. 1988; Hertel and Maniatis 1999). In our hands, an AdML substrate with a 6-nt 5′ exon underwent efficient lariat formation, but was significantly impaired for exon ligation (data not shown). A 14-nt 5′ exon did undergo exon ligation, but not as efficiently as longer sequences (data not shown). Similarly, efficient exon ligation in S. cerevisiae extracts requires the 5′ exon to be somewhere between 6 and 12 nt (Duchene et al. 1988). Thus, at least in vitro, truncation of the 5′ exon is of little or no consequence for the first step of splicing, but there is a minimum 5′ exon length requirement for the second step. Undoubtedly, some of the 5′ exon interactions required for the second step are needed to anchor the 5′ exon intermediate within the spliceosome, as Chanfreau et al. (1999) found that a yeast pre-mRNA with a 5-nt 5′ exon could support exon ligation to an alternate 5′ exon supplied in trans.
Enhanced 5′ exon association of REF/Aly and SRp20 upon exon ligation
Of the nine proteins that consistently cross-linked in a splicing-dependent or partially splicing-dependent manner, we were able to positively identify two as REF/Aly and SRp20. REF/Aly (p32) cross-linked to position −24 independent of splicing (Figs. 3D, left panel, 4B, lane 6). However, when the mRNA is spliced, REF/Aly cross-linked much more intensely (Figs. 3C,D, right panels, 4B, lane 2). These results agree with previous functional data showing that REF/Aly can interact weakly with and promote the nuclear export of unspliced RNAs, but is stably recruited to spliced RNAs as a component of the EJC (Le Hir et al. 2001a; Rodrigues et al. 2001). Interestingly, cross-linking of p32 was only barely detectable in reactions that were blocked for the second step and so accumulated C complex (Figs. 3C, left panel, 4B, lane 4) even though REF/Aly was clearly present in the C complex by mass spectrometry and Western blotting (Table 1; Fig. 5B). It seems possible that when REF/Aly is recruited to spliceosomes by UAP56 (Luo et al. 2001; Strasser and Hurt 2001), it is not initially positioned close to the 5′ exon. Upon exon ligation, and coincident with the same structural transition that destabilizes association of UAP56, REF/Aly may then become tightly associated with the EJC and so come into close proximity with 5′ exon position −24.
Identification of the p25 cross-link as SRp20 is consistent with a previous report from the Reed laboratory (Chiara et al. 1996) that SRp20 (and SRp30) could be cross-linked in the E/CC complex at positions −31 and −26 relative to the 5′ splice site of AdML pre-mRNA. We similarly reported that SRp20 cross-links to position −2 of PIP pre-mRNA independent of splicing (Le Hir et al. 2000a). In the 4-thioU reactions examined here, p25 clearly cross-linked both at early time points (Fig. 3A) and in the absence of splicing (control mRNA, Fig. 3D, left panel). Furthermore, p25 cross-links decreased in intensity upon addition of cold competitor. Taken together, these results confirm previous observations that SRp20 can bind to RNA independent of splicing (Cavaloc et al. 1999; Schaal and Maniatis 1999), but this binding is relatively weak. However, like REF/Aly, its association with the 5′ exon is apparently stabilized upon exon ligation, as evidenced by stronger p25 cross-linking to spliced RNAs (Fig. 3D, cf. left and right panels). Recently it was reported that SRp20, a known shuttling protein, can serve as an export adaptor for intronless mRNAs that contain several near-consensus SRp20-binding sites (Huang and Steitz 2001). In that case, SRp20 functions in collaboration with 9G8, another shuttling SR protein. Perhaps within the EJC, SRp20 similarly collaborates with REF/Aly to facilitate export of spliced RNAs.
As of this writing, the other cross-linked species observed in Figures 2 and 3 remain to be positively identified. The p220 that cross-linked to benzophenone at position −7 is most likely hPrp8p, as we and others have previously demonstrated that hPrp8p cross-links strongly near the end of the 5′ exon and at the 5′ splice site (Chiara et al. 1996 and references therein; Reyes et al. 1999; Le Hir et al. 2000a). Likewise, the p170 that cross-linked only after exon ligation may well be SRm160—using a different antiserum from that used here (see Materials and Methods), we have previously shown that SRm160 can become cross-linked to benzophenone incorporated adjacent to the exon–exon junction of spliced mRNA (Le Hir et al. 2000a). Lastly, it is possible that the 50–60-kD proteins that became cross-linked to benzophenone at positions −17 and −27 (Fig. 2) or p57 that cross-linked to 4-thioU at position −24 (Fig. 3) could include UAP56 or RNPS1.
Insights into EJC structure and assembly
Our previous characterizations of the protein complement of spliced mRNP and the EJC relied almost exclusively on coimmunoprecipitation analyses. To confirm and extend these findings, we have now analyzed affinity-purified H, C, and mRNP complexes by mass spectrometry and Western blotting. From these results we can begin to build an initial framework for both EJC assembly and its subsequent structural rearrangements (Fig. 6). The model includes both known EJC components and cross-linked proteins of unknown identity. Because some overlap likely remains between these two subsets (see above), future identification of cross-linked species may simplify this picture somewhat.
Even though REF/Aly and SRp20 can associate with the −20/−24 region independent of splicing, it is unclear whether this initial binding contributes to EJC assembly or simply reflects the general RNA-binding ability of these proteins. Recently it was reported that splicing-dependent recruitment of REF/Aly requires the putative RNA helicase UAP56 (Luo et al. 2001; Strasser and Hurt 2001), which has also been implicated in early spliceosome assembly (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). In yeast, it has been proposed that Sub2p (the UAP56 ortholog) serves to recruit Yra1p (the REF/Aly ortholog) to mRNAs but then must dissociate to allow interaction between Yra1p and Mex67p (the TAP ortholog; Strasser and Hurt 2001). UAP56 has been detected in spliced mRNPs purified under low salt conditions (60 mM NaCl; Luo et al. 2001). In contrast, when purifications were carried out in 150 mM KCl, we could only detect UAP56 in the C complex and not in spliced mRNP, whereas REF/Aly was present in both complexes (Fig. 5B; Table 1). Taken together, the biochemical data are consistent with the idea that UAP56 serves to recruit REF/Aly in a splicing-dependent manner and subsequently dissociates after mRNP release. Whether or not UAP56 also directly recruits other EJC components is unknown at present.
Like UAP56, SRm160 is not as stably associated with spliced mRNP in vitro as are other EJC components. Given that more proteins can be detected in association with the EJC in vivo than in vitro (see above), it is possible that the association of UAP56 and SRm160 with spliced mRNA is somewhat more stable in vivo. Nonetheless, once the EJC is assembled, neither UAP56 nor SRm160 is necessary for continued EJC association with spliced mRNA. Similarly, binding of RNPS1 appears to weaken upon exon ligation, so it is also unlikely to be at the EJC core. Candidates for core proteins include some of the unidentified cross-linked proteins in Figures 2 and 3, as well as REF/Aly, Y14, and Magoh, which all exhibit similar abundance in the C complex and mRNP (Fig. 5B). Yet, REF/Aly is known to dissociate from the EJC upon nuclear export of mRNA (Kim et al. 2001a), whereas Y14 and Magoh remain bound in the cytoplasm (Kataoka et al. 2000, 2001; Kim et al. 2001a; Le Hir et al. 2001a, 2001b). Y14 is bound so tightly that its removal requires transit of ribosomes across the EJC locus (Dostie and Dreyfuss 2002). Therefore, of the known EJC components, those most likely to constitute the core complex are Y14 and its binding partner Magoh. Curiously, however, even though Y14 contains an RNA recognition motif (RRM) and we could immunoprecipitate in vitro-translated Y14 under denaturing conditions (data not shown), we could not demonstrate that any of the cross-linked proteins observed here was Y14. Because the EJC binds independent of sequence, it is likely that the most crucial protein–RNA contacts are through the phosphodiester backbone. Future studies directed at understanding EJC structure should explore this possibility, as well as focus on identifying the complete complement of core EJC proteins, some of which are likely to be one or more of the unidentified cross-linked proteins described here.
Materials and methods
RNA substrates and antibodies
Uniformly labeled splicing substrates were prepared by standard transcription using T7 RNA polymerase and [α-32P]UTP. DNA templates for AdML (Gozani et al. 1994), PIP85.B (Query et al. 1994), and α-TM (Smith et al. 1989) were as previously described. DNA templates for corresponding control 5′ exons and mRNAs were constructed by PCR. RNAs containing single [32P] labels and/or photoactivatable cross-linking groups were constructed by splinted RNA–RNA ligations (Moore and Query 1998). Benzophenone-containing RNAs were prepared essentially as described (MacMillan et al. 1994; Moore and Query 1998) by ligating the 10-nt RNA oligonucleotide (GGUGUCGCdA*G) containing a single convertible deoxyadenosine (dA*) to a series of [32P]-phosphorylated 3′ RNA transcripts derived from AdML-AG and AdML-GG, each containing progressively longer 5′ exons. RNAs containing 4-thioU were constructed by initiating the 3′ RNA fragment with synthetic 4-thioUpG during in vitro transcription. This transcript was [32P]-phosphorylated and joined to a 5′ RNA capped with GpppG.
The following antibodies were kindly provided by the person indicated: α-REF, α-Y14, α-Magoh [E. Izaurralde (European Molecular Biology Laboratory, Heidelberg, Germany)]; α-RNPS1 [A. Mayeda (University of Miami, FL)]; α-UAP56 [R. Reed (Harvard Medical School, Boston, MA)]; α-DEK [G. Grosveld (St. Jude Children's Hospital, Memphis, TN)]; and α-SRp20 [K.Neugebauer (Max Planck Institute, Dresden, Germany)]. The α-SRm160 antiserum used in our previous study (Le Hir et al. 2000a) was no longer available; the α-SRm160 used here was purchased from Santa Cruz Biotechnology.
In vitro splicing, glycerol gradients, and RNase digestions
In vitro splicing reactions, analysis of splicing products, glycerol gradients, and RNase digestions (H, A, and T1) were performed as previously described (Le Hir et al. 2000a,b). For RNase H digestions, splicing reactions containing the AdML-GG and α-TM substrates were incubated for 90 min, whereas control 5′ exons were similarly incubated for 45 min prior to addition of cDNA oligonucleotides.
Cross-linking and immunoprecipitation
Benzophenone- and 4-thioU-derivatized substrates were incubated under standard splicing conditions for the times indicated. Cross-linking was carried out on ice by UV irradiation at 302 nm or 365 nm for benzophenone- and 4-thioU-derivatized RNAs, respectively. Reactions were then digested with 0.1 mg/mL RNase A (Sigma) at 37°C for 30 min. For competition experiments, splicing reactions were supplemented with cold competitor RNA to 0.5 mg/mL final concentration and incubated at 30°C for an additional 15 min prior to UV irradiation. Cross-linked proteins were separated on SDS-16% polyacrylamide (200:1 acrylamide:bis) gels and detected by autoradiography or with a Molecular Dynamics PhosphorImager.
Immunoprecipitations were carried out essentially as described (Le Hir et al. 2000a), except that reactions containing α-SRp20 were supplemented with a threefold excess of Triton X-100 (over SDS) following denaturation and prior to dilution for IP.
Purification and characterization of spliced mRNPs
Affinity purification of AdML +Mg2+ C complex was performed as previously described for −Mg2+ C complex (Jurica et al. 2002) except that the sizing column buffer contained 150 mM KCl, 2 mM MgCl2, 20 mM Tris (pH 7.9), and 0.5% NP-40; and the amylose column buffer contained 150 mM KCl, 2 mM MgCl2, and 20 mM Tris (pH 7.9). An MS2-MBP (maltose-binding protein) fusion protein [the construct was a gift from Josep Vilardell (Institute for Genomic Research, Barcelona, Spain)] served as an affinity tag. Substrates used to generate spliced mRNPs for purification were derivatives of AdML (HMS388, generously provided by Robin Reed) and PIP85.B (Query et al. 1994) that each contained three MS2 sites in the 3′ exon. mRNP purification followed essentially the same protocol as the +Mg2+ C complex except that after splicing, reactions were supplemented with five 12-nt DNA oligonucleotides (500 nM each) complementary to intron sequences to induce cleavage by endogenous RNase H of any remaining pre-mRNA substrate. The reaction was then fractionated on an S-300 sizing column (Amersham Pharmacia) equilibrated with 150 mM KCl, 2 mM MgCl2, 20 mM Tris (pH 7.9), and 8% glycerol. Amylose chromatography was performed with the same buffer, and complexes were eluted by addition of 10 mM maltose. For Western analysis, ∼120 fmole of each complex was separated on a 5%–15% gradient SDS-PAGE, transferred to a nitrocellulose membrane, and probed with the indicated antibody. Peptide sequence analysis by mass spectrometry was performed as described previously (Jurica et al. 2002).
Acknowledgments
We are grateful to those colleagues acknowledged in the text for providing constructs and antisera. We thank Fred LaRiviere, Ajit Nott, Charles Query, and Thomas Tange for critical reading of the manuscript. M.J.M. is an associate investigator with the Howard Hughes Medical Institute; M.S.J. is a Paul Sigler/Agouron Institute Fellow of the Helen Hay Whitney Foundation. This work was supported, in part, by National Institutes of Health Grant GM53007.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mmoore@brandeis.edu; FAX (781) 736-2337.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1030602.
References
- Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes & Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Gouyette C, Schwer B, Jacquier A. Trans-complementation of the second step of pre-mRNA splicing by exogenous 5′ exons. RNA. 1999;5:876–882. doi: 10.1017/s1355838299990544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Duchene M, Low A, Schweizer A, Domdey H. Molecular consequences of truncations of the first exon for in vitro splicing of yeast actin pre-mRNA. Nucleic Acids Res. 1988;16:7233–7239. doi: 10.1093/nar/16.15.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Maniatis T. Serine-arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc Natl Acad Sci. 1999;96:2651–2655. doi: 10.1073/pnas.96.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Jurica M, Licklider L, Gygi S, Grigorieff N, Moore M. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 2001a;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001b;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes & Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes & Dev. 2000a;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000b;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001a;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001b;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- MacMillan AM, Query CC, Allerson CR, Chen S, Verdine GL, Sharp PA. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J, Grosveld G, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC. Uses of site-specifically modified RNAs constructed by RNA ligation. In: Smith CWJ, editor. RNA–protein interactions: A practical approach. Oxford, UK: IRL Press; 1998. pp. 75–108. [Google Scholar]
- Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: The 2‘-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- ————— U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Query CC, Moore MJ, Sharp PA. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes & Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: Identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segault V, Will CL, Polycarpou-Schwarz M, Mattaj IW, Branlant C, Luhrmann R. Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol Cell Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Kandels-Lewis S. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993;73:803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- Smith CWJ, Porro EB, Patton JG, Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- Zhang M, Green MR. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes & Dev. 2001;15:30–35. doi: 10.1101/gad.851701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]