Abstract
The Drosophila somatic sex-determination regulatory pathway has been well studied, but little is known about the target genes that it ultimately controls. In a differential screen for sex-specific transcripts expressed in fly heads, we identified a highly male-enriched transcript encoding Takeout, a protein related to a superfamily of factors that bind small lipophilic molecules. We show that sex-specific takeout transcripts derive from fat body tissue closely associated with the adult brain and are dependent on the sex determination genes doublesex (dsx) and fruitless (fru). The male-specific Doublesex and Fruitless proteins together activate Takeout expression, whereas the female-specific Doublesex protein represses takeout independently of Fru. When cells that normally express takeout are feminized by expression of the Transformer-F protein, male courtship behavior is dramatically reduced, suggesting that male identity in these cells is necessary for behavior. A loss-of-function mutation in the takeout gene reduces male courtship and synergizes with fruitless mutations, suggesting that takeout plays a redundant role with other fru-dependent factors involved in male mating behavior. Comparison of Takeout sequences to the Drosophila genome reveals a family of 20 related secreted factors. Expression analysis of a subset of these genes suggests that the takeout gene family encodes multiple factors with sex-specific functions.
Keywords: Sex determination, courtship behavior, fat body, gene family, doublesex, fruitless
Sexual differentiation affects the form and function of a wide variety of tissues in adult organisms. In Drosophila, somatic sexual identity is controlled by a well-studied pathway of regulatory genes in which globally acting factors (i.e., sis-a, sis-b, Sxl, tra, tra-2) determine the alternative sex-specific products that are synthesized from the fru and dsx genes. Sex-specific Dsx and Fru proteins then enact sexual differentiation in distinct subsets of somatic tissues (Nagoshi et al. 1988; Ito et al. 1996; Ryner et al. 1996). The male-specific Fru protein is necessary for sexual differentiation within the CNS of the adult fly and appears to be a major factor controlling the sexual differentiation of behavior (Baker et al. 2001). XY fru mutants develop with the appearance of normal males, but do not perform normal male courtship behavior and fail to distinguish correctly between males and females (Hall 1994; Taylor et al. 1994; Ryner et al. 1996; Villella et al. 1997; Anand et al. 2001). Somatic sexual differentiation outside of the CNS is controlled primarily by the dsx gene. Two alternative sex-specific proteins encoded by this gene (Dsx-M and Dsx-F) specify male and female differentiation, respectively (Burtis and Baker 1989). In the absence of dsx function, sexual differentiation in both XX and XY flies is ambiguous, resulting in nearly identical intersexual adults that have both male and female characteristics (Baker and Ridge 1980). Dsx also appears to play a role in the differentiation of some tissues needed for sex-specific behavior, as dsx mutant flies have quantitatively reduced ability to perform various aspects of male courtship and lack the ability to produce the sine-song, humming sounds that are part of the courtship song (McRobert and Tompkins 1985; Taylor et al. 1994; Villella and Hall 1996).
Although the regulatory interactions within the sex-determination pathway are well understood, the interactions of these factors with the downstream target genes that the pathway ultimately controls are largely unexplored. The Fru and Dsx proteins both encode transcription factors that function at parallel terminal positions in the known regulatory hierarchy and, thus, are likely to directly control at least some targets. However, as yet, no genes regulated by Fru have been identified. Dsx has been shown to regulate the function of factors that control patterning in the genital disc (Keisman and Baker 2001; Keisman et al. 2001; Sanchez et al. 2001), the pigmentation of abdominal cells (Kopp et al. 2000), and the expression of a variety of factors associated with reproductive systems of adults (Chapman and Wolfner 1988), but its only clear direct targets are the female-specific yolk protein genes that are activated by Dsx-F and repressed by Dsx-M (Coschigano and Wensink 1993; Bownes 1994). On the basis of this example, it seems likely that the dsx gene can act as a bimodal molecular switch, oppositely affecting target gene expression in males and females. Identification of more targets of the sex-determination pathway would improve our understanding of the function of Dsx, Fru, and other downstream sex-determination regulators. Moreover, analysis of target gene function is likely to lead to insights into molecular processes that determine sex-specific traits. Here, we present evidence that the takeout gene is a tissue-specific target of regulation by both Fru and Dsx. Takeout is a member of a large family of secreted factors that bind small lipophiles and was identified previously in several molecular screens as a robust circadian-regulated gene (Sarov-Blat et al. 2000; Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Lin et al. 2002). Interestingly, Takeout is induced in adults by starvation and improves their tolerance to nutrient deprivation. (Sarov-Blat et al. 2000). Through analysis of takeout mutants and sexual mosaics, we find that it is also required for normal levels of male courtship behavior. These findings lead us to propose that takeout plays a role in integrating information about the organism's sex, nutritional status, and circadian cycle to affect adult male behavior.
Results
Takeout expression is sex-specific in brain-associated fat body and is regulated by Transformer-2
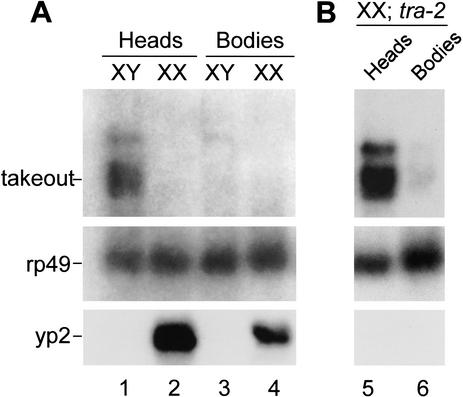
To identify genes under the control of the sex-determination regulatory pathway, we carried out a PCR-based subtractive hybridization screen for sex-specific RNAs expressed in adult fly heads. Head RNA of tra-2/tra-2+ phenotypically wild-type XX adult females was subtracted against the head RNA of sibling XX tra-2/tra-2 mutants, and vice versa. The latter flies are transformed into males both somatically and behaviorally (Watanabe 1975; Belote and Baker 1987). One cDNA clone that hybridized preferentially with sequences from phenotypic males was isolated and studied in more detail. Northern blot hybridizations confirmed that this sequence represents a highly male-specific 1.1-kb mRNA that was expressed primarily in adult heads (Fig. 1A). Expression of this mRNA was repressed by Tra-2 in females, as XX tra-2 mutants expressed levels similar to wild-type males (Fig. 1B). The sequence of the clone was later found to be identical to that of takeout, an independently identified gene responsive to circadian rhythms and starvation (Sarov-Blat et al. 2000). The takeout gene encodes a secreted protein related to circulating carrier proteins of lipophilic factors, such as the juvenile hormone-binding proteins of other insects. In our initial screen and in the experiments described below, we have used flies grown in parallel, nonstarved cultures that were not entrained to a light-dark cycle. Analysis of RNA prepared from these cultures at different times during the day failed to reveal any significant variation in takeout levels, presumably due to their asynchrony (data not shown).
Figure 1.
The takeout gene is expressed specifically in male heads and is derepressed in tra-2 mutant chromosomal females. (Top) Takeout expression in head and body RNA from nonstarved Canton-S male (XY) and female (XX) flies (A) and in tra-2PM6/ra-2PM7 mutant females (B) was examined by Northern analysis. The major band corresponding to takeout mRNA is indicated. The higher molecular weight band corresponds in size to takeout pre-mRNA. (Middle) Ribosomal protein 49 (rp49) hybridization to the same blot as a control for amount of RNA loaded. (Bottom) Hybridization with a probe for transcripts from the yolk protein 2 gene, which is expressed in female fat body.
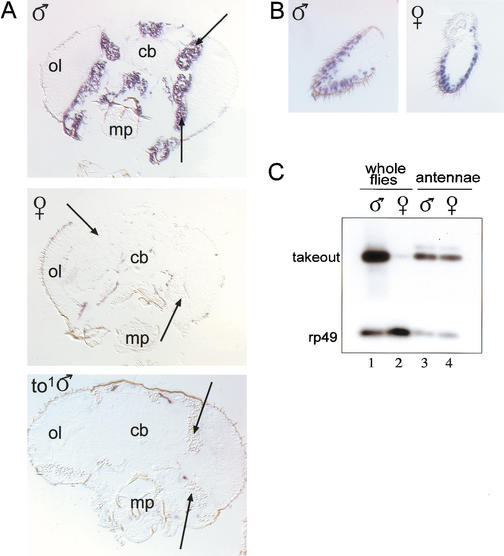
Takeout expression has been reported in the adult brain as well as in the cardia and other segments of the digestive system, in which it is induced by starvation (Sarov-Blat et al. 2000). To identify tissues giving rise to male-specific takeout transcripts, we performed RNA in situ hybridizations on parallel serial sections of adult male and female heads. Surprisingly, we did not detect takeout RNA within the adult brain using either of two probes from different regions of the takeout transcription unit, even when samples were overstained (see Materials and Methods). Instead, both probes detected high levels of RNA in the fat body that surrounds the brain as well as in a dispersed population of cells in the third antennal segment (Fig. 2A,B). Expression was not detected in males homozygous for the to1 mutation (Fig. 2A, bottom). Comparison of simultaneous hybridizations performed on sections from males and females revealed that fat-body expression was male specific, but that expression in the antennae was not. To confirm this, we dissected antennae from male and female heads and carried out low-cycle RT–PCR (Fig. 2C). This showed that although overall accumulation of takeout RNA in whole flies is highly male biased, no difference was apparent in male and female antennal RNA levels. We conclude that antenna-derived takeout messages account for only a small fraction of all takeout RNA, and that sex-specific expression of takeout in adult heads derives primarily from the fat body. Thus, the sex-specific regulation of takeout varies by tissue type.
Figure 2.
takeout RNA is expressed male specifically in brain-associated fat body, whereas expression in the antennae is not sex specific. (A,B) Frontal sections through male and female Canton-S heads were hybridized in situ with a takeout riboprobe originating from the 3′ untranslated region of the gene. Expression in the fat body (arrows) was observed in males, but not in females. Hybridization to the takeout probe was absent in takeout (to1) males. (B) Expression in the antennae was observed in both sexes. For reference, the positions of the central brain (cb), optical layers of the brain (ol), and mouthparts (mp) are indicated. (C) takeout RT–PCR on RNA from isolated antennae as well as whole male and female Canton-S flies is shown. Low-cycle PCR was performed with takeout and rp49 primers. The products were detected by Southern blotting and hybridization with an internal oligonucleotide probe.
Other members of the takeout gene family are expressed sex specifically
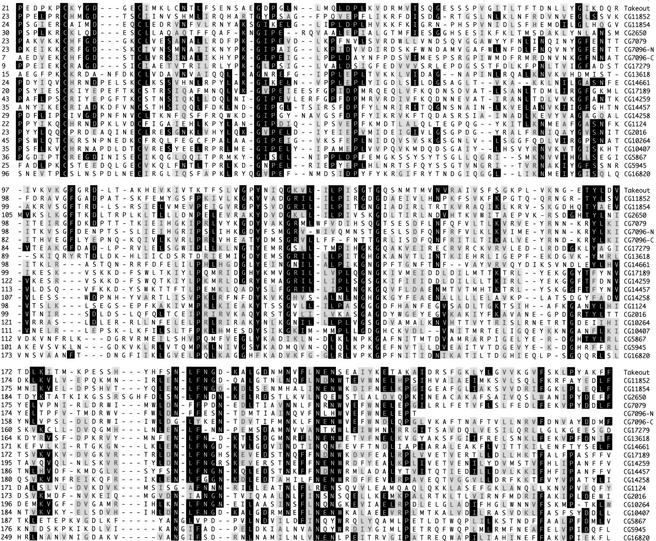
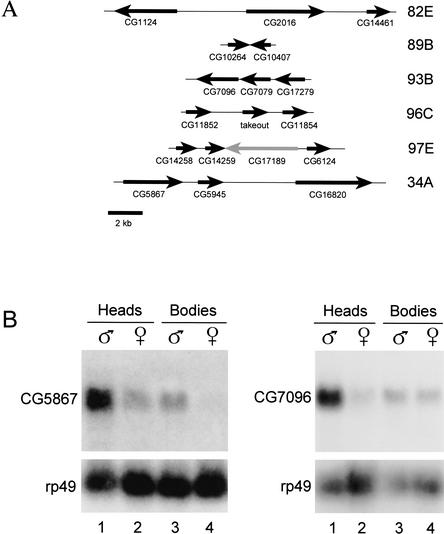
Takeout has been shown to have similarity to six other Drosophila proteins that are also under circadian control (So et al. 2000; Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Lin et al. 2002). When used to carry out BLAST-P searches of both the translated Drosophila genome sequence and the entire set of predicted Drosophila proteins in the Berkeley Drosophila Genome Project Database Takeout, Adams et al. (2000) identifies a family of 20 related proteins. An alignment of these proteins is shown in Figure 3. The sequences share interspersed regions of conservation that correspond to regions in Takeout noted previously (Sarov-Blat et al. 2000) to have similarity with circulating juvenile hormone-binding proteins (JHBPs) of other insects (Robertson et al. 1999; Vermunt et al. 2001). Although the takeout genes are dispersed to several locations in the genome, most are found in clusters of two or three closely linked genes (Fig. 4A). To determine whether other members of the takeout family are also expressed in a sex-specific manner, we surveyed the expression of five randomly selected family members (CG1124, CG2016, CG5867, CG7096, and CG11852) in adult males and females. Although transcripts from three of these genes accumulated equally in both sexes (data not shown), RNAs from CG5867 and CG7096 were found to be male enriched in adult heads (Fig. 4B,C). These results support the idea that members of the takeout gene family perform sex-specific functions.
Figure 3.
The Takeout family of proteins. An alignment is shown of the conserved protein-coding regions from the 20 different members of the takeout gene family encoded by Drosophila. Black shading indicates residues that are identical in at least eight members of the family, and gray shading indicates areas of similarity. CG7096-N and CG7096-C denote the N- and C-terminal sequences of the single ORF encoded by CG7096, in which the entire Takeout homology region is duplicated. In most cases, protein sequences are derived from translation of EST clones. In all other cases, sequences are based on the predicted genomic protein-coding regions as annotated by the Drosophila genome project (Adams et al. 2000). The sequences shown for CG14661, CG14457, and CG17279 are modified after splice junction reassignment on the basis of sequence alignment with other family members.
Figure 4.
Multiple members of the takeout family are expressed sex specifically. (A) Organization of takeout family gene clusters are diagrammed. Arrows denote the length of each coding region and transcriptional orientation. Exons and introns are not indicated. Names of genes are below the arrows. The gray arrow denotes a gene (CG17189) that interrupts a cluster but is not in the takeout family. The cytological position of each cluster in the Drosophila genome is given at right. Three genes at dispersed locations (CG2645, CG14457, and CG13618) are not shown. (B) RNA blot hybridization analysis of CG5867 and CG7096 RNA expression in heads and bodies from males and females is shown. CG5867 is male enriched in both tissues, whereas CG7096 is male enriched only in heads. Hybridization of the same blot with rp49 mRNA is shown at bottom.
Feminization of Takeout-expressing cells disrupts male courtship behavior
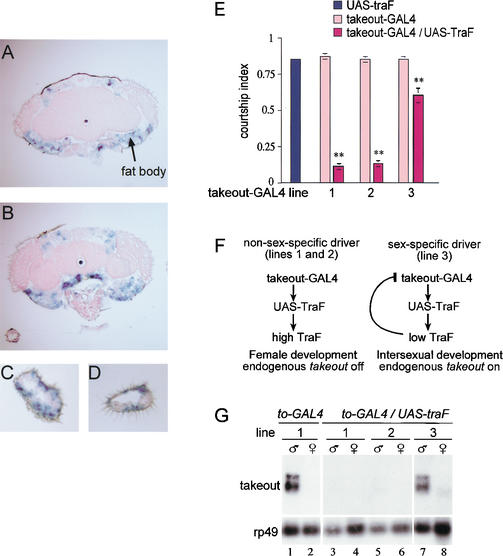
Because takeout identifies sexually differentiated cells within the adult head, we next asked whether male sexual identity within these cells is important for male-specific courtship behavior. Specific cell types can be sexually transformed from male to female by forcing them to express the female-specific transformer protein (TraF; Ferveur et al. 1995) by use of a tissue-specific promoter. Taking advantage of the yeast GAL4/UAS system, a 1.27-kb takeout promoter fragment was placed upstream of the Gal4 gene and used to drive expression of a UAS–TraF transgene. When expression of five independent takeout–GAL4 insertions was tested by crossing to a strain carrying a UAS–lacZ transgene, activity of the promoter was found consistently to be most prominently distributed in a pattern similar to that found in our in situ hybridization experiments. Within adult heads, activity was detected in fat body as well as in a subset of cells within the maxillary palps and antennae (Fig. 5A–D). In sections from whole adult fly bodies, a lower level of expression was also detected within the cardia and in fat cells dispersed throughout the abdomen and thorax (data not shown). Although expression of endogenous takeout RNA was not observed in maxillary palps, its detection in this experiment is likely due to higher sensitivity of the reporter method relative to RNA in situ hybridization. Notably, promoter activity was never observed in any part of the adult CNS, even when sections were purposely overstained. Whereas the spatial pattern of staining was consistent among the five strains we examined, the sex-specific expression was not. In only one of the lines examined was the expression observed in fat bodies clearly male specific. In other lines, significant levels of expression were observed in both males and females. Because each line represents an independent transgene insertion, we infer that the 1.27-kb takeout promoter fragment used is not sufficient to direct sex-specific expression at most insertion sites. This observation was fortunate as the non-sex-specific activity of these takeout–GAL4 insertions allowed us to circumvent artificial negative feedback regulation in our feminization strategy (see Fig. 5F) that would otherwise result from expression of TraF using a promoter under the negative control of the TraF protein.
Figure 5.
Feminization of Takeout-expressing cells disrupts male courtship behavior. GAL4 activity driven by the takeout promoter in sections from adult heads (A,B), antennae (C), and maxillary palps (D), was detected using a UAS–lacZ reporter gene. Frontal sections of takeout–GAL4/UAS–lacZ flies were stained with X-gal to detect β-galactosidase activity. Courtship indices of various males toward Canton-S virgin females are shown in E. Males carrying the UAS–traF (blue bar) or takeout–GAL4 (pink bars) transgenes individually have significantly higher courtship indices than the takeout–GAL4/UAS–traF males carrying both transgenes (red bars). The results from three different transgenic takeout–GAL4 lines are shown (lines 1,2,3). n = 10 for each group. ** indicates indices that were significantly different from those of parental strains (p < 0.001). (F) Diagram of the expected negative feedback loop set up in progeny from a cross of UAS–traF flies with the male-specific takeout–GAL4 line. (G) Northern analysis of endogenous takeout expression in takeout–Gal4/UAS–traF males and females as a measure for the feminization of takeout-expressing cells shows that takeout expression is drastically reduced in lines 1 and 2, but to a lesser degree in line 3. Expression of endogenous takeout RNA in the parental takeout–Gal4 adults from line 1 is shown for comparison (lanes 1,2).
Adult males expressing TraF under the direction of takeout–GAL4 transgenes were observed for their ability to court wild-type females in a mating chamber (Hall 1994; Greenspan 1995). Results are shown in Figure 5E, represented as the courtship index (CI), which is a measure of the time a male spends performing any of the steps of courtship during a fixed observation period. The takeout–GAL4/UAS–traF flies from all three driver lines tested gave markedly reduced courtship indices in relation to controls, reflecting the fact that these males spent much less time courting females. Feminization directed by the takeout promoter severely lowered the probability that a male would court or sustain courtship beyond the initial steps of orienting and following. Although, on occasion, all steps of courtship can be carried out by such males. These results indicate that the takeout gene is active in sexually differentiated cell types that play an important role in promoting male courtship behavior.
The requirement for male identity of takeout-expressing cells is further supported by comparison of the results obtained in the different driver lines tested. As an indicator of feminization, we looked at levels of endogenous takeout RNA (Fig. 5G). If takeout-expressing cell types are completely feminized, we expect that endogenous RNA expression will be greatly reduced. Feminization driven by nonsex-specific takeout–GAL4 drivers (Fig. 5G, lines 1,2) virtually eliminated takeout RNA expression. This argues that these lines are feminized in cell types that normally express takeout. In contrast, feminization by the male-specific takeout–GAL4 driver (Fig. 5G, line 3) was incomplete, and endogenous takeout RNA was only slightly reduced in amount. This is presumably due to the anticipated negative feedback regulation that reduces the level of TraF expressed in these flies (Fig. 5F). Feminization, as measured in this way, was well correlated with the ability of these flies to court females. Flies from lines 1 and 2 were more affected than those from line 3 (Fig. 5E, cf. red bars). We conclude that the degree of feminization of takeout-expressing cell types is related to the ability of males to perform courtship behavior.
A mutation in takeout interacts genetically with fruitless in male courtship
To determine whether takeout function is required for male mating behavior, we performed courtship assays on takeout mutant males. In a previous study (Sarov-Blat et al. 2000), it was shown that a rearranged mutant allele of takeout (to1) is fortuitously carried in a laboratory strain on the ry506 third chromosome. We obtained a ry506 mutant strain carrying an identical rearrangement, and found that it also fails to express takeout RNA (data not shown). Therefore, we refer to this allele as to1. PCR and Southern blotting experiments on to1 showed that the deletion found previously (Sarov-Blat et al. 2000) is associated with a chromosome rearrangement breakpoint and located entirely within a region between 39 nucleotides upstream and 494 nucleotides downstream of the takeout-translation stop codon (data not shown).
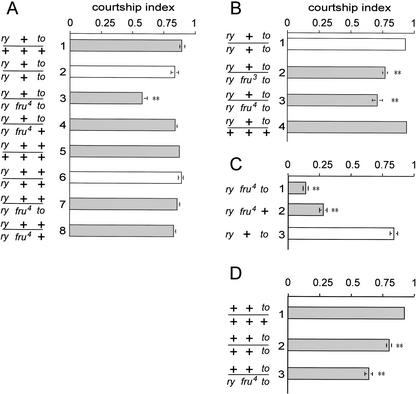
When tested for courtship behavior, ry506 to1 mutant males did not show a reduction in courtship relative to heterozygous siblings (Fig. 6A, cf. lanes 1 and 2). However, takeout mutant flies that were also heterozygous for fru (ry506 fru4 to1/ry506 fru+ to1and ry506 fru3 to1/ry506 fru+ to1) showed a significant reduction in courtship relative to a variety of control genotypes tested (Fig. 6A, lane 3, B, lanes 2,3). No effects of ry506 alone on courtship were observed (Fig. 6A). To determine whether the observed effect of takeout might be explained by any general sluggishness of the affected genotypes, we carried out short-term activity assays to measure the movement of males in the same chambers used for courtship assays, but without a female present (see Materials and Methods for details). In these assays, the activities of the courtship-defective genotypes ry to1/ry fru3 to1 (89 ± 4, n = 10) and ry to1/ry fru4 to1 (90 ± 4, n = 10) were found to be equal to or slightly greater than that of either ry to1 double homozygous (84 ± 5, n = 10) or heterozygous (76 ± 5, n = 10) control flies that were tested in parallel.
Figure 6.
Mutations in takeout and fruitless interact to affect male courtship. Four sets of experiments are shown. (A) Courtship indices (± S.E.M.) of test males toward wild-type virgin females. The genotypes of males tested are indicated beside each bar. The different genotypes were generated by crossing various strains with ry506 to1 (lanes 1–4) or ry506 to+ (lanes 5–8). (Lane 1) takeout heterozygous males; (lane 2) homozygous mutant takeout males are not reduced in courtship; (lane 3) males homozygous for takeout and heterozygous for fru4 show a statistically significant reduction in courtship (marked by **, p < 0.001); (lane 4) males heterozygous for both takeout and fru4 show normal courtship. To control for a potential effect on courtship of the ry506 mutation on the takeout chromosome, parallel assays were performed on a ry506 strain that carries a wild-type takeout allele (to+ry; lanes 5–8). No effects of ry506 alone on courtship were observed. (B) Both the fru3 and fru4 allele interact with takeout to affect male courtship. ry phenotypes are indicated by white bars, ry+ phenotypes by dark bars. The fru4 and fru3 mutations are caused by independent P[ry+] insertion. n = 10 for each genotype. (C) The courtship index of to1 fru4 double homozygous mutant males toward females is lower than that of fru4 single mutant males alone (n = 19, p < 0.001). The courtship index of takeout single mutants is shown for comparison. (D) Males from a takeout1 strain that was outcrossed to the wild-type Canton-S strain [to1 (CS)] show a reduction in courtship (n = 9, p < 0.005).
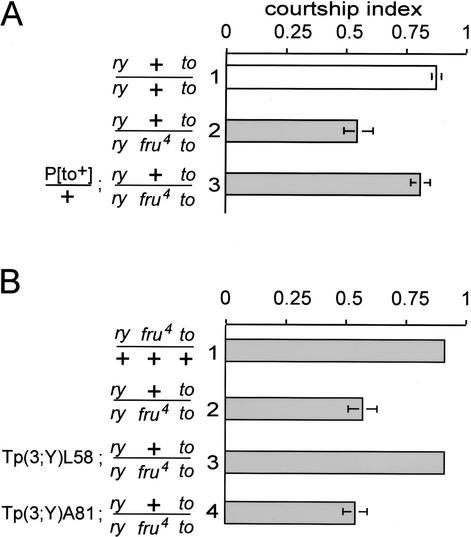
Although courtship of the above mutant flies is quantitatively reduced, it is not absent. The mutant males are capable of all steps of courtship, but perform them less frequently, and seem to lack motivation to court. Unlike homozygous fru mutants, takeout mutants did not display male-chaining behavior (data not shown), suggesting that these males are capable of distinguishing between males and females as potential mates. To verify that the mutation in takeout, rather than any other feature of the chromosome, is responsible for the reduced courtship, we introduced a P element carrying a wild-type genomic fragment encompassing the takeout transcription unit into this strain. When this transgene was crossed into the ry506 to1/ry506 fru4 to1 genotype, normal levels of male courtship were restored (Fig. 7A). Due to the size and complexity of the fru gene, similar transgene rescue experiments with it were impractical; however, we found that introduction of a duplication of a chromosomal segment carrying the fru+ allele also restored normal courtship behavior to the ry506 to1/ry506 fru4 to1 genotype (Fig. 7B). Taken together, these results confirm that a simultaneous reduction of fruitless and takeout function interferes with male courtship.
Figure 7.
Rescue of the to1/to1, fru/+ courtship defect by wild-type takeout or fruitless. (A) A genomic takeout+ transgene rescues the mutant phenotype (n = 8, p < 0.05). (B) Likewise, a duplication containing the wild-type fruitless gene (breakpoints at cytological locations 88D, 93D), rescues the courtship defect (lane 3; n = 6, p < 0.001), whereas a control duplication of similar origin (breakpoints at cytological locations 75D, 80) does not (lane 4; n = 6, p < 0.001).
The original fru3 and fru4 mutant alleles were generated in a ry506 genetic background. Prior to the above studies, we noticed that the third chromosomes in our fru3 and fru4 strains carry not only these fru and ry mutations, but also the to1 mutant allele (data not shown). This led us to test whether previously observed courtship phenotypes associated with these fru alleles (Ryner et al. 1996; Villela et al. 1997) might have been enhanced by the presence of the takeout mutation. We compared the courtship of the double mutant flies (to1, fru4) with a recombinant fru4 line (to+, fru4) carrying the allele of takeout from the wild-type strain Canton-S (Fig. 6C). We again found that the presence of the takeout mutation caused a statistically significant reduction in courtship index. However, this effect was small in relation to that of the fru4 mutation alone.
The to1 strains used in all of the above analysis were maintained in homozygous condition for many generations before the discovery of the mutant allele. This led us to wonder whether selection for fertility had resulted in the accumulation of modifiers that suppress an effect of takeout on courtship behavior. We therefore outcrossed the to1 chromosome to wild-type (Canton-S) individuals for four generations, allowing free recombination between the Canton-S and takeout chromosomes (see Materials and Methods for details). When to1 homozygous mutants with the Canton-S background were recovered and tested for their ability to perform courtship behavior, a statistically significant reduction in courtship index was observed (Fig. 6D). Thus, takeout function is required for normal levels of male courtship in this background.
Expression of takeout RNA is regulated by both Doublesex and Fruitless
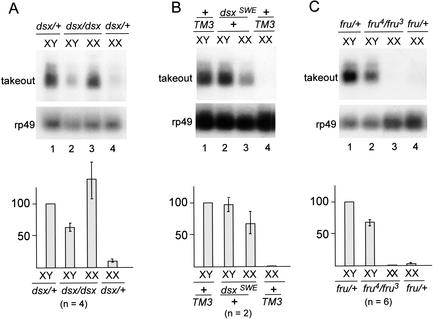
We next examined how Dsx and Fru affect takeout expression. As dsx is known to affect sexual differentiation in males and females, it might either activate takeout in males, repress it in females, or both. Figure 8A shows takeout expression in dsx homozygous mutant animals compared with their heterozygous siblings. In repeated blot hybridization experiments, XY dsx individuals were found to have takeout RNA levels reduced by 37% relative to XY dsx/+ flies, indicating that the male-specific Dsx-M product functions to activate takeout expression. In chromosomally XX individuals, dsx mutations had an opposite effect. In comparison with XX dsx/+ siblings, XX dsx/dsx animals have levels of takeout mRNA increased by 13-fold, indicating that Dsx-F normally functions to repress takeout expression. Thus, the differential expression in males and females is achieved (at least in part) by dsx-dependent repression in females and activation in males. Curiously, XX dsx/dsx intersexes have more takeout RNA than do XY dsx/dsx intersexes (Fig. 8A), suggesting that sex-specific factors other than dsx also affect overall takeout expression.
Figure 8.
Expression of takeout is affected by both doublesex and fruitless. RNA from whole flies was analyzed. (A) Northern analysis of takeout expression in dsx1 mutant flies shows that takeout expression in dsx1 mutant males is reduced (lane 2), and is derepressed in dsx1 females (lane 3) when compared with the expression in dsx1/+ siblings (lanes 1,4). (B) Forced expression of male-specific forms of dsx induce takeout in XX individuals. Females expressing the male form of dsx from the dominant mutation dsxSWE show activation of takeout (lane 3) compared with control females (lane 4). These males are dsxSWE/dsx+ and produce both Dsx-F and Dsx-M. There is no effect of dsxSWE/dsx+ on takeout expression in males (cf. lanes 1 and 2). (C) takeout expression is reduced in fru4/fru3 males (lane 2), but unaltered in fru4/fru3 females (lane 3) compared with their heterozygous siblings (lanes 1,4). (Bottom) Quantitation on the basis of several independent experiments (number indicated by n-value below each group). RNA levels were normalized to XY control males and rp49 controls.takeout expression in these males was assigned a value of 100 for each blot.
We also examined the effect of the dominant dsxSWE allele on takeout expression. Due to a deletion in the female-specific exon that results in constitutive male-specific splicing of the dsx pre-mRNA, this allele produces only Dsx-M (Nagoshi and Baker 1990). XY flies carrying this allele are phenotypically normal males, and did not have reduced takeout expression. However, in XX; dsxSWE/+ animals, the presence of Dsx-M antagonizes Dsx-F function, resulting in intersexual flies that are similar in phenotype to those produced by dsx null mutations. We observed that takeout was derepressed to intermediate levels in such intersexes (Fig. 8B), further supporting the idea that takeout is controlled by dsx.
Analysis of fru mutants demonstrated that it affects takeout expression only in males. As shown in Figure 8C, repeated Northern analysis of RNA from XY fru adults showed that the levels of takeout RNA present in these individuals was consistently reduced by about 32% relative to fru/+ males. Expression of takeout was not increased by loss of fru function in XX females. This is consistent with the recent finding that functional sex-specific Fru protein is not present in females (Lee et al. 2000; Usui-Aoki et al. 2000). Taken together, the above results indicate that both Fru and Dsx function to specify male-specific expression of takeout RNA.
Discussion
Differences in the morphology, physiology, and behavior of males and females undoubtedly reflects the sex-specific activities of numerous genes. Identification of these genes and analysis of their function is likely to lead to insights into the molecular events that underlie a variety of sex-specific processes affecting mating and reproduction. In Drosophila, the control of sexual differentiation is best understood in the soma, in which it is regulated by the combined action of the transcription factors Dsx and Fru, which are among the most terminal known components in the hierarchy. Functional analysis of these two factors has led to the recent suggestion that they have distinct and complementary roles, with Fru specifying sexual identity of tissues in the CNS that are responsible for courtship behavior, and Dsx specifying sex in other somatic tissues (Baker et al. 2001). However, given the observation that dsx mutants also have minor effects on courtship behavior (Villella and Hall 1996), we believe that a clear delineation of the roles played by Dsx and Fru will require more information about the specific genes and cell types whose sexual identity these factors specify.
Here, we have presented evidence that the takeout gene is a target of regulation by the somatic sex-determination pathway. Although takeout expression in some tissues is nonsex-specific, the vast majority of takeout RNA derives from fat body within the adult head and is specific to males. Surprisingly, our analysis of RNA from mutant flies indicates that sex-specific takeout expression depends on the function of both Dsx and Fru. Although this would seem to contradict the expected restriction of Fru function to the CNS, it is worth noting that the effect of Fru on takeout expression could be mediated indirectly by diffusible factors. In situ hybridization studies localized fru RNA to a variety of specific neurons (Ryner et al. 1996; Lee et al. 2000), but not the fat body cells in which male-specific takeout RNA is most prominently expressed. Interestingly, the only other instance of sexual differentiation outside of the CNS, where sex-specific Fru function is known to be required, is in the formation of the Muscle-of-Lawrence, a male-specific abdominal muscle in which sexual fate is determined through inductive signals that originate from the innervating motor neuron (Lawrence and Johnston 1986; Gailey et al. 1991; Usui-Aoki et al. 2000). In a previous study (So et al. 2000), circadian-regulated takeout RNA expression was found in several areas of the adult brain that were not detected in either our in situ hybridization studies or by reporters driven with the takeout–GAL4 transgene, despite repeated attempts. Moreover, when this GAL4 construct was used to drive TraF-mediated feminization of males, virtually all endogenous takeout expression was eliminated, arguing that takeout is not normally expressed outside of the regions in which the driver is active.
Both the dsx and fru genes encode alternatively spliced transcripts that encode distinct forms of the Dsx and Fru proteins in males and females. Thus, both genes could potentially play a role in either activating takeout in males or repressing it in females. We found that full activation of takeout is not achieved in either dsx null or fru hypomorphic mutant XY individuals and, instead, takeout RNA is present at levels intermediate between those found in males and females. In chromosomal females, we found that only Dsx is required for repression of takeout. The fact that fru mutants do not affect takeout expression is consistent with experiments suggesting that the female-specific form of fru mRNA is not translated into a functional protein (Lee et al. 2000; Usui-Aoki et al. 2000). Moreover, all sex-specific Fru functions so far identified have been found in males (Gailey and Hall 1989; Villella et al. 1997). Therefore, although a sex-specific Fru mRNA is produced in females that potentially encodes a protein, there is currently no evidence that it functions to regulate sexual differentiation.
The fact that dsx is capable of both activating and repressing takeout expression reflects the dsx gene's unusual ability to perform opposite functions in males and females by producing distinct proteins in the two sexes through alternative pre-mRNA splicing (Burtis and Baker 1989; Coschigano and Wensink 1993). The male-specific (Dsx-M) and the female-specific (Dsx-F) proteins share a common DM domain, which is required for DNA binding. The two proteins differ at their C termini, a region promoting dimerization in both forms (An et al. 1996; Erdman et al. 1996). The opposite activities of Dsx-M and Dsx-F at the molecular level have been most clearly shown in the case of the female-specific yp2 gene, which like takeout, is expressed primarily in the adult fat body (Burtis et al. 1991; Coschigano and Wensink 1993; An and Wensink 1995). Both Dsx-F and Dsx-M bind the yp2 promoter and affect its transcription in opposite ways. Bound Dsx-F activates yp2 transcription, whereas bound Dsx-M represses its transcription. Thus, the effects of Dsx proteins on the yp2 gene are reversed from those we observe on takeout. Three potential Dsx-binding sites (Erdman et al. 1996) are located within 1 kb upstream of the takeout translation initiation codon, but further studies will be required to determine whether Dsx proteins associate directly with the takeout promoter. Taken together, the results presented here and the findings from studies on yolk proteins suggest that Dsx-F and Dsx-M can each either activate or repress the activity of downstream genes. Presumably, the effect Dsx has on any particular gene is also determined by other regulators interacting with the gene's promoter. Consistent with this idea, Dsx-binding sites in the yp2 promoter overlap those of other transcription factors necessary for tissue-specific activation/repression of yp2 expression (An and Wensink 1995).
A possible role for Takeout and the fat body in mating behavior
The male-specific expression of its RNA in tissues closely associated with the adult brain suggested to us that secreted Takeout protein might affect male-specific behaviors that occur during courtship and mating. After outcrossing it from its original genetic background, we found that a takeout mutation reduced the ability of males to court and mate with wild-type females. Moreover, a significant synergistic effect on male courtship behavior was observed when to1 was combined with either of two strong hypomorphic mutations in the fru gene to produce flies simultaneously reduced in both takeout and fruitless function. In fru heterozygotes, which have normal male courtship, reduction in takeout function caused a significant reduction in courtship index. Likewise, the effects of fru homozygous mutants on courtship were enhanced by reduction in takeout function. The fact that takeout only affects courtship in certain genetic backgrounds and the observed interaction of takeout and fru mutations, suggests the possibility that the takeout protein acts redundantly on courtship with genes under the control of fru. Redundantly functioning genes might include those from the takeout family itself, some of which we have found here to also be male-specifically expressed in adult heads. Consistent with this idea, the feminization of takeout-expressing cells in XY individuals using a takeout promoter driven Tra-F cDNA results in a reduction of courtship behavior of males toward females. This effect on behavior clearly exceeds that observed in takeout mutants, suggesting that other sex-specific factors involved in behavior are affected in the feminized tissues.
Studies on Drosophila sexual mosaics have identified a region in the posterior brain as the primary tissue in which male differentiation is required for courtship behavior (Hall 1979). However, these studies did not exclude the possibility that the male identity of other tissues are also necessary, and subsequent studies support this idea (Greenspan and Ferveur 2000). The effects on behavior that we observe when Tra-F is driven by a takeout promoter fragment suggest that tissues outside of the adult brain affect behavior. The takeout–GAL4 transgene that we used to drive Tra-F was unable to produce detectable activity in the CNS. As these males were strongly reduced in courtship behavior, it seems likely that male differentiation is necessary in a takeout-expressing tissue outside of the CNS. It is worth noting here, however, that we cannot exclude the possibility that an undetectable level of TraF expression under control of the GAL4 driver in the CNS or in other cell types contributes to the observed effects.
Cells within the maxillary palp and third antennal segments in which the takeout promoter is active, could potentially mediate the perception of female pheromones. However, the requirement for these tissues in the courtship of females by males is unclear. The ablation of a large proportion of chemosensory sensillae in the antennae does not result in impaired courtship (Stocker and Gendre 1989), and other studies indicate that chemosensory organs on the proximal legs mediate the pheromonal response that stimulates courtship (Venard et al. 1989). Feminization of the antennal lobes, brain structures to which the antennal neurons project, leads to nondiscriminatory courtship of males toward both males and females, but does not lower courtship toward females (Ferveur et al. 1995; O'Dell et al. 1995). Current evidence for the involvement of maxillary palps in pheromone perception indicates that they mediate inhibitory rather than stimulatory effects on courtship (Stocker and Gendre 1989).
The only Takeout-expressing tissue in which there is clear evidence for sexual differentiation is the fat body surrounding the male brain, suggesting that it is responsible for the observed effect on courtship. How might fat body affect courtship behavior? These cells are an important source of products secreted into the hemolymph, which circulate throughout the adult body (Bownes and Hames 1977; Haunerland 1996; Meister et al. 1997). The juvenile hormone-binding proteins from other insects, to which Takeout is related, are synthesized in the fat body and secreted into the hemolymph, where they are thought to carry juvenile hormone or other small lipophilic ligands to target cells (Nowock et al. 1975; Glinka et al. 1995; Wojtasek and Prestwich 1995). It is not known whether juvenile hormone plays any role in Drosophila male courtship behavior, but it has been shown that juvenile hormone stimulates the synthesis of accessory gland proteins (Herndon et al. 1997). A role for juvenile hormone has also been found in the reproductive behavior of the Caribbean fruit fly Anastrepha suspense (Teal et al. 2000). In this species, mating was found to be accompanied by an increase in the juvenile hormone levels of males and was correlated with competitive mating advantage of mated over unmated males. Topical exposure to juvenile hormone or related compounds caused virgin males from this species to release pheromones and to mate precociously, suggesting that juvenile hormone affects the propensity of males to initiate courtship behavior. If sex-specific proteins from the takeout family associate with juvenile hormone or other similar ligands, they could potentially exert sex-specific effects on the fly's physiology and behavior. There is already significant evidence that diffusible factors can affect reproductive abilities in Drosophila. For instance, dissatisfaction, a gene encoding a nuclear receptor has been shown to affect male and female reproductive behavior (Finley et al. 1998). Also, several proteins present in male seminal fluid have been shown to enter the female hemolymph and induce changes in oviposition as well as their receptivity to subsequent courting males (Monsma et al. 1990; Kubli 1992; Wolfner 1997; Lung and Wolfner 1999; Ottiger et al. 2000).
Starvation, mating, and circadian rhythms
In a previous study, takeout was shown to become induced in starving flies and to prolong their survival (Sarov-Blat et al. 2000). This raises the intriguing issue of how the starvation response might be related to mating behavior. One possibility is that takeout is involved in a mechanism governing how males expend their energy when faced with nutrient deprivation. It is easy to imagine that pathways exist for managing the choice between foraging and courtship behavior that are critical for the male's survival and reproductive success. The observation that, in addition to sex and nutrition, takeout also responds to circadian rhythms (Sarov-Blat et al. 2000; So et al. 2000; Claridge-Chang et al. 2001; McDonald and Rosbash 2001; Lin et al. 2002) suggests that it integrates a variety of signals that affect the adult male's behavior.
Materials and methods
RNA subtraction
Poly(A+) RNA from isolated heads was prepared as described (Chandler et al. 2001). To create tra-2 null flies, we used two overlapping deficiencies, tra-2PM6 and tra-2PM7 (McGuffin et al. 1998). RNA was isolated from XX; tra-2PM6/tra-2PM7, which are transformed into males, and from their tra-2PM6/CyO or tra-2PM7/CyO heterozygous female siblings. The Clontech PCR-select cDNA subtraction kit (catalog no. K1804-1) was used for subtraction. PCR fragments were subcloned, and inserts with preferential expression in males were identified by Northern analysis as described (Mattox et al. 1996). The inserts from clones of interest were used to probe an adult head cDNA library (Palazzolo et al. 1990; Hamilton et al. 1991), a gift from P. Hardin (University of Houston, TX).
Fly strains
Flies were kept on standard cornmeal/sugar-based food at 25°C under noncontrolled light conditions. The fru4/TM3,Sb,ry and fru3/TM3,Sb,ry strains were a gift from B. Baker (Stanford University, Stanford, CA). The takeout mutant used was identified in Bloomington stock number 2541 (snw; ry506). The UAS–traF strain (stock no. 4590) and the to+, ry506 control strain (stock no. 225), as well as the Tp(3;Y)L58, y+/TM6,Ubx; C(1)RM, y1/C(1;Y)1, y1 (Baker 1980; stock no. 2914) and Tp(3;Y)A81, y+/TM6,Ubx; C(1)RM, y1/C(1;Y)1, y1 (stock no. 2787) strains were obtained from the Bloomington stock center. The genotypes of other strains used were as follows: w1118/BSY; pp dsx1/TM3, Sb and w1118/BSY; dsxSWE/Df(dsx)/TM3, Sb.
Takeout1 outcrossed flies were generated by backcrossing of the original snw; ry506 to1 strain to Canton-S for four generations. Individual chromosomes were isolated over a TM3 balancer of similar background and the presence of to1 was verified by PCR using a single nucleotide polymorphism in the coding region.
Generation of recombinant chromosomes
The to mutant alleles present in the fru3, fru4, and dsx1 chromosomes were replaced with the wild-type allele from Canton-S by meiotic recombination. Balanced males from lines carrying potential recombinant chromosomes were tested by PCR for the presence of the P[PZ] transposon insertion associated with both of the fru mutations (Castrillon et al. 1993). The takeout mutant and toCS alleles were identified in amplified products using a single nucleotide polymorphism in the coding region. The presence of the toCS allele was verified by observing expression of takeout RNA on Northern blots.
RNA Blot hybridizations and RT–PCR
DNAse-treated RNA from Canton-S flies was reverse transcribed using the Superscript II kit (GIBCO) with oligo(dT)12–18 as primer. Amplifications of takeout and rp49-derived cDNAs were performed simultaneously in 50 mM potassium chloride, 10 mM Tris-HCl (pH 8.3 at 25°C), 0.1% Triton X-100, 1.5 mM magnesium chloride, 0.6 μM primer (each), 0.1 mM dNTPs, and 5 units of Taq polymerase (Roche). Amplifications were carried out by first incubating for 2 min at 94°C, then 17 cycles of 30 sec at 94°C, 45 sec at 55°C, 90 sec at 72°C, followed by 10 min at 72°C. Half of the reaction was analyzed by Southern hybridization using end-labeled oligonucleotides. RNA equivalent to 15 isolated antennae and 0.6 μg RNA from whole flies was used per PCR reaction.
Hybridizations were done at 42°C in 1 M sodium chloride, 0.4% polyvinyl pyrollidone, 0.4% Ficoll, 0.4% BSA, 0.1 M PIPES (pH 7.0), 0.2% SDS, and 100 μg of salmon sperm DNA/mL. Blots were washed twice in 6× standard saline citrate (SSC) at 25°C for 10 min, then once in 6× SSC at 50°C for 20 min. The PCR primers used were as follows: ACCTTCTCACTCGTTG GAC and AATGTTTTTATGGTATTTTACAGA for takeout; AGCATACAGGCCCAAGA and GTGTATTCCGACCAGGTT for rp49. The oligonucleotides used as 32P-labeled probes for hybridization were GTTGTCACCCAGCGCCTTGT for takeout and AGCATACAGGCCCAAGA for rp49. For Northern hybridizations of various takeout family members, probes were generated by amplification of genomic DNA using the primers described as follows: CG11852, (1)GAACGATGGAGCGGC AACA, (2)GTTCCTCAACGACAACTGGAC; CG2016, (1)CG AGTCGCGTGAAAGTTG, (2)ACGGAAGTTGGCTAAGATGG; CG1124, (1)GCCTTAATCCCTTAGATTGT, (2)CACAGGGT GAATACGACTCT; CG5867, (1)CGCATTTGCATCGGTCAG GT, (2)CTATTTCCGCTGCCAAACATAAGC; CG7096, (1)CC TTGCAACGATCCGATGAT, (2)CAAGTAGAGCTTTCAAC AAAACC.
Northern blots were analyzed by phosphorimaging and quantitation using Kodak 1D 3.5 image quantitation software.
In situ hybridization to fly sections/X-gal staining
In situ hybridizations to frozen serial sections were performed using a modified version of the protocol of Han et al. (1992). In short, fresh flies were frozen in a fly collar. The 10-μm frozen sections were cut on a Leitz cryostat, dried at room temperature for 30 min, fixed in 4% paraformaldehyde in PBS for 10 min, and then washed in PBS. The sections were deacetylated, washed in PBS, and prehybridized in a humid box for 2 h at 60°C. Hybridizations were performed overnight at 60°C. Prehybridization and hybdridization buffer was 5× SSC, 50% formamide, 5× Denhardt's, 250 μg/mL yeast tRNA, 500 μg/mL salm sperm DNA, 50 μg/mL heparin, 2.5 mM EDTA, 0.1% Tween 20, 0.25% CHABS, and 0.1% SDS. Slides were washed at 65°C in 2× SSC/50% formamide 2 × 30 min., 2× SSC 2 × 15 min., 0.2× SSC 3 × 20 min., eqilibrated in PBS + 0.1%Triton X-100 (PBX) 1× 10 min., and blocked for 1 h at room temperature with 10% goat serum in PBX. They were incubated in the same buffer with a 1:1000 dilution of anti-DIG antibody (Roche) at 4°C overnight, washed 3 × 30 min in PBX, and stained following the supplier's protocol (Genius kit, Roche), including 1 mM levamisole in the reaction buffer.
For X-gal staining, 10-μm frozen sections were cut on a Leitz cryostat, dried at room temperature for 5 min, fixed in 1% glutaraldehyde in PBS for 10 min, and washed in PBS. X-gal staining was done as described by Fischer and Maniatis (1988). Sections comparing males and females were prepared and stained together on the same slide.
Probes for takeout
Northern analysis of takeout expression was performed with a 250-bp fragment from the 3′ untranslated region of the takeout gene as a probe. Fragments were labeled with 32P by random priming (Roche) or by PCR following the protocol of Mertz and Rashtchian (1994). The primers used to obtain the fragment were CTTCAAAGTGGGCAGAGTC and AATGTTTTTATG GTATTTTACAGA. Probes from a full-length cDNA clone were also used with identical results. All dsx and fru mutant strains used for RNA preparations carried recombinant chromosomes in which the Canton-S allele of takeout was introduced.
For in situ hybridizations, the same 250-bp PCR fragment from the 3′ end of the gene was subcloned and used as a template to generate DIG-labeled riboprobes (Roche). Identical results were also obtained with a nonoverlapping 383 nucleotide probe from the N-terminal part of the takeout protein-coding region defined by the primers 5′-TAAGTATGGTGATGGC GAATGTAT-3′ and 5′-TACCTTGCCCTGGATGTTATAG-3′.
takeout–Gal4 transgenic flies
At 1.27 kb immediately upstream of the takeout translation, start codon were amplified by PCR (primers, 5′-ATAAGAAT GCGGCCGCTGCTAACACGTCTATAACT-3′ and 5′-ATAAG AATGCGGCCGCACTGGTTCTGCTTCTGCGG-3′) and cloned into the NotI site of pCaSpeR4-GATN. The construct was verified by sequencing. pCaSpeR4-GATN was made by subcloning the Gal4 coding region of pGATN (Brand and Perrimon 1993) as a KpnI/SpeI fragment into pCaSpeR4 (Pirrotta 1988). Transgenic lines were established in a w1118 background.
Courtship assays
Males were collected within 2–4 h of emergence under light CO2 anesthesia, and individually stored in food vials for 4–6 d. For observation of courtship behavior, individual males were aspirated without anesthesia into a chamber (diameter 0.8 cm) of a plastic mating wheel, together with a 2–4-h-old Canton-S virgin female. The courtship index was calculated as the fraction of time the male spent displaying any element of courtship behavior (orienting, following, wing extension, licking, attempted copulation, copulation) within a 10-min observation period (Taylor et al. 1994).
Short-term activity assay
Assays were performed as described by Anand et al. (2001). Males were collected within 2–4 h of emergence under light CO2 anesthesia, and were individually stored in food vials for 4–6 d. Individual males were aspirated without anesthesia into a chamber (diameter 0.8 cm) of a plastic mating wheel containing a filter paper with a single line dividing the chamber in half. After 2–3 min of acclimation time, the number of times the male crossed the center line within the 3-min observation time was counted.
Takeout and fruitless rescue experiments
The 6.4-kb genomic takeout rescue fragment, obtained from P1 phage DS02779 DNA by restriction digests with XhoI and EcoRV, was sucloned into the Carnegie20 transformation vector. Transgenic strains were established in snw; ry506to1 flies. Heterozygous transformants were crossed to obtain externally identical siblings both with and without the transgene. Following the courtship assay, genomic DNA was prepared individually from each tested male (Gloor and Engels 1992) and the presence of the takeout+ transgene assessed by PCR.
Males carrying a duplication of the fruitless gene were produced by crossing Tp(3;Y)L58, y+/to1/TM6, Ubx; C(1)RM, y1 females to y1; to1, fru4/TM3, Sb males. Control flies were generated by crossing of Tp(3;Y)L58, y+/to1/TM6, Ubx; C(1)RM, y1 females to Canton-S males and by crossing of to1, fru4/TM3, Sb females to to1 males. Control males carrying a duplication unrelated to the fruitless locus (Tp(3;Y)A81) but with similar origin, were produced in the same way.
Statistical analysis
Analyses were performed with JMP 2.0 statistical software (SAS Institute, Inc.). Following initial analysis of variance (ANOVA), comparisons between groups were by unplanned multiple comparisons using Tukey-Kramer analysis at α = 0.05.
Acknowledgments
We thank Mitzi Kuroda, Barb Taylor, Richard Behringer, and Jean-François Ferveur for helpful comments on this manuscript; Bruce Baker, Paul Hardin, and Daisuke Yamamoto for flies and reagents; Ralph Greenspan for helpful suggestions with the courtship assays; and Randy Johnson for use of the cryostat. DNA sequencing was performed by the M.D. Anderson Cancer Center Sequencing Core, supported by a Cancer Center Support (Core) Grant, CA16672. This work was supported by grant no. S98-14 from the Whitehall Foundation to W.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wmattox@mdanderson.org; FAX (713) 794-4394
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1010302.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The Genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- An W, Wensink PC. Integrating sex- and tissue-specific regulation within a single Drosophila enhancer. Genes & Dev. 1995;9:256–266. doi: 10.1101/gad.9.2.256. [DOI] [PubMed] [Google Scholar]
- An W, Cho S, Ishii H, Wensink PC. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol. 1996;16:3106–3111. doi: 10.1128/mcb.16.6.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Villella A, Ryner LC, Carlo T, Goodwin SF, Song HJ, Gailey DA, Morales A, Hall JC, Baker BS, et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158:1569–1595. doi: 10.1093/genetics/158.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. Report of B. Baker. Drosophila Information Service. 1980;55:197. [Google Scholar]
- Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Taylor BJ, Hall JC. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell. 2001;105:13–24. doi: 10.1016/s0092-8674(01)00293-8. [DOI] [PubMed] [Google Scholar]
- Belote JM, Baker BS. Sexual behavior: Its genetic control during development and adulthood in Drosophila melanogaster. Proc Natl Acad Sci. 1987;84:8026–8030. doi: 10.1073/pnas.84.22.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M. The regulation of the yolk protein genes, a family of sex differentiation genes in Drosophila melanogaster. BioEssays. 1994;16:745–752. doi: 10.1002/bies.950161009. [DOI] [PubMed] [Google Scholar]
- Bownes M, Hames BD. Accumulation and degradation of three major yolk proteins in Drosophila melanogaster. J Exp Zool. 1977;200:149–156. doi: 10.1002/jez.1402000118. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, Wasserman SA. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: Characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler DS, McGuffin ME, Mattox W. Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing. Nucleic Acids Res. 2001;29:3012–3019. doi: 10.1093/nar/29.14.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KB, Wolfner MF. Determination of male-specific gene expression in Drosophila accessory glands. Dev Biol. 1988;126:195–202. doi: 10.1016/0012-1606(88)90253-9. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes & Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur JF, Stortkuhl KF, Stocker RF, Greenspan RJ. Genetic feminization of brain structures and changed sexual orientation in male Drosophila. Science. 1995;267:902–905. doi: 10.1126/science.7846534. [DOI] [PubMed] [Google Scholar]
- Finley KD, Edeen PT, Foss M, Gross E, Ghbeish N, Palmer RH, Taylor BJ, McKeown M. Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron. 1998;21:1363–1374. doi: 10.1016/s0896-6273(00)80655-8. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Maniatis T. Drosophila Adh: A promoter element expands the tissue specificity of an enhancer. Cell. 1988;53:451–461. doi: 10.1016/0092-8674(88)90165-1. [DOI] [PubMed] [Google Scholar]
- Gailey DA, Hall JC. Behavior and cytogenetics of fruitless in Drosophila melanogaster: Different courtship defects caused by separate, closely linked lesions. Genetics. 1989;121:773–785. doi: 10.1093/genetics/121.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailey DA, Taylor BJ, Hall JC. Elements of the fruitless locus regulate development of the muscle of Lawrence, a male-specific structure in the abdomen of Drosophila melanogaster adults. Development. 1991;113:879–890. doi: 10.1242/dev.113.3.879. [DOI] [PubMed] [Google Scholar]
- Glinka AV, Braun RP, Edwards JP, Wyatt GR. The use of a juvenile hormone binding protein for the quantitative assay of juvenile hormone. Insect Biochem Mol Biol. 1995;25:775–781. doi: 10.1016/0965-1748(94)00098-3. [DOI] [PubMed] [Google Scholar]
- Gloor G, Engels W. Single-fly DNA preps for PCR. Drosophila Information Service. 1992;71:148–149. [Google Scholar]
- Greenspan RJ. Understanding the genetic construction of behavior. Sci Am. 1995;272:72–78. doi: 10.1038/scientificamerican0495-72. [DOI] [PubMed] [Google Scholar]
- Greenspan RJ, Ferveur JF. Courtship in Drosophila. Annu Rev Genet. 2000;34:205–232. doi: 10.1146/annurev.genet.34.1.205. [DOI] [PubMed] [Google Scholar]
- Hall JC. Control of male reproductive behavior by the central nervous system of Drosophila: Dissection of a courtship pathway by genetic mosaics. Genetics. 1979;92:437–457. doi: 10.1093/genetics/92.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The mating of a fly. Science. 1994;264:1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Palazzolo MJ, Meyerowitz EM. Rapid isolation of long cDNA clones from existing libraries. Nucleic Acids Res. 1991;19:1951–1952. doi: 10.1093/nar/19.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han PL, Levin LR, Reed RR, Davis RL. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. [DOI] [PubMed] [Google Scholar]
- Haunerland NH. Insect storage proteins: Gene families and receptors. Insect Biochem Mol Biol. 1996;26:755–765. doi: 10.1016/s0965-1748(96)00035-5. [DOI] [PubMed] [Google Scholar]
- Herndon LA, Chapman T, Kalb JM, Lewin S, Partridge L, Wolfner MF. Mating and hormonal triggers regulate accessory gland gene expression in male Drosophila. J Insect Physiol. 1997;43:1117–1123. doi: 10.1016/s0022-1910(97)00062-0. [DOI] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keisman EL, Baker BS. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development. 2001;128:1643–1656. doi: 10.1242/dev.128.9.1643. [DOI] [PubMed] [Google Scholar]
- Keisman EL, Christiansen AE, Baker BS. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophila genital imaginal disc. Dev Cell. 2001;1:215–225. doi: 10.1016/s1534-5807(01)00027-2. [DOI] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Kubli E. The sex-peptide. BioEssays. 1992;14:779–784. doi: 10.1002/bies.950141111. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P. The muscle pattern of a segment of Drosophila may be determined by neurons and not by contributing myoblasts. Cell. 1986;45:505–513. doi: 10.1016/0092-8674(86)90282-5. [DOI] [PubMed] [Google Scholar]
- Lee G, Foss M, Goodwin SF, Carlo T, Taylor BJ, Hall JC. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O, Wolfner MF. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem Mol Biol. 1999;29:1043–1052. doi: 10.1016/s0965-1748(99)00078-8. [DOI] [PubMed] [Google Scholar]
- Mattox W, McGuffin ME, Baker BS. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics. 1996;143:303–314. doi: 10.1093/genetics/143.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- McGuffin ME, Chandler D, Somaiya D, Dauwalder B, Mattox W. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics. 1998;149:1477–1486. doi: 10.1093/genetics/149.3.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobert SP, Tompkins L. The effect of transformer, doublesex and intersex mutations on the sexual behavior of Drosophila melanogaster. Genetics. 1985;111:89–96. doi: 10.1093/genetics/111.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M, Lemaitre B, Hoffmann JA. Antimicrobial peptide defense in Drosophila. BioEssays. 1997;19:1019–1026. doi: 10.1002/bies.950191112. [DOI] [PubMed] [Google Scholar]
- Mertz LM, Rashtchian A. Nucleotide imbalance and polymerase chain reaction: Effects on DNA amplification and synthesis of high specific activity radiolabeled DNA probes. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- Monsma SA, Harada HA, Wolfner MF. Synthesis of two Drosophila male accessory gland proteins and their fate after transfer to the female during mating. Dev Biol. 1990;142:465–475. doi: 10.1016/0012-1606(90)90368-s. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, Baker BS. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes & Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Nowock J, Goodman W, Bollenbacher WE, Gilbert LI. Synthesis of juvenile hormone binding proteins by the fat body of Manduca sexta. Gen Comp Endocrinol. 1975;27:230–239. doi: 10.1016/0016-6480(75)90238-5. [DOI] [PubMed] [Google Scholar]
- O'Dell KM, Armstrong JD, Yang MY, Kaiser K. Functional dissection of the Drosophila mushroom bodies by selective feminization of genetically defined subcompartments. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Ottiger M, Soller M, Stocker RF, Kubli E. Binding sites of Drosophila melanogaster sex peptide pheromones. J Neurobiol. 2000;44:57–71. [PubMed] [Google Scholar]
- Palazzolo MJ, Hamilton BA, Ding DL, Martin CH, Mead DA, Mierendorf RC, Raghavan KV, Meyerowitz EM, Lipshitz HD. Phage λ cDNA cloning vectors for subtractive hybridization, fusion-protein synthesis and Cre-loxP automatic plasmid subcloning. Gene. 1990;88:25–36. doi: 10.1016/0378-1119(90)90056-w. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez RL, Denhardt DT, editors. Vectors: A survey of molecular cloning vectors and their uses. Boston, MA: Butterworth; 1988. pp. 437–456. [Google Scholar]
- Robertson HM, Martos R, Sears CR, Todres EZ, Walden KK, Nardi JB. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol Biol. 1999;8:501–518. doi: 10.1046/j.1365-2583.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Sanchez L, Gorfinkiel N, Guerrero I. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development. 2001;128:1033–1043. doi: 10.1242/dev.128.7.1033. [DOI] [PubMed] [Google Scholar]
- Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- So WV, Sarov-Blat L, Kotarski CK, McDonald MJ, Allada R, Rosbash M. takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol Cell Biol. 2000;20:6935–6944. doi: 10.1128/mcb.20.18.6935-6944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Gendre N. Courtship behavior of Drosophila genetically or surgically deprived of basiconic sensilla. Behav Genet. 1989;19:371–385. doi: 10.1007/BF01066165. [DOI] [PubMed] [Google Scholar]
- Taylor BJ, Villella A, Ryner LC, Baker BS, Hall JC. Behavioral and neurobiological implications of sex-determining factors in Drosophila. Dev Genet. 1994;15:275–296. doi: 10.1002/dvg.1020150309. [DOI] [PubMed] [Google Scholar]
- Teal PE, Gomez-Simuta Y, Proveaux AT. Mating experience and juvenile hormone enhance sexual signaling and mating in male Caribbean fruit flies. Proc Natl Acad Sci. 2000;97:3708–3712. doi: 10.1073/pnas.060034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, et al. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol. 2000;2:500–506. doi: 10.1038/35019537. [DOI] [PubMed] [Google Scholar]
- Venard R, Antony C, Jallon JM. Drosophila chemoreceptors. In: Singh R, Strausfeld J, editors. Neurobiology of sensory systems. New York: Plenum; 1989. pp. 377–385. [Google Scholar]
- Vermunt AM, Kamimura M, Hirai M, Kiuchi M, Shiotsuki T. The juvenile hormone binding protein of silkworm haemolymph: Gene and functional analysis. Insect Mol Biol. 2001;10:147–154. doi: 10.1046/j.1365-2583.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- Villella A, Hall JC. Courtship anomalies caused by doublesex mutations in Drosophila melanogaster. Genetics. 1996;143:331–344. doi: 10.1093/genetics/143.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Gailey DA, Berwald B, Ohshima S, Barnes PT, Hall JC. Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics. 1997;147:1107–1130. doi: 10.1093/genetics/147.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe TK. A new sex transforming gene on the second chromosome of Drosophila melanogaster. Jpn J Genet. 1975;50:269–271. [Google Scholar]
- Wojtasek H, Prestwich GD. Key disulfide bonds in an insect hormone binding protein: cDNA cloning of a juvenile hormone binding protein of Heliothis virescens and ligand binding by native and mutant forms. Biochemistry. 1995;34:5234–5241. doi: 10.1021/bi00015a037. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. Tokens of love: Functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]